Pro-Oxidant Activity of an ALS-Linked SOD1 Mutant in Zn-Deficient Form
Abstract
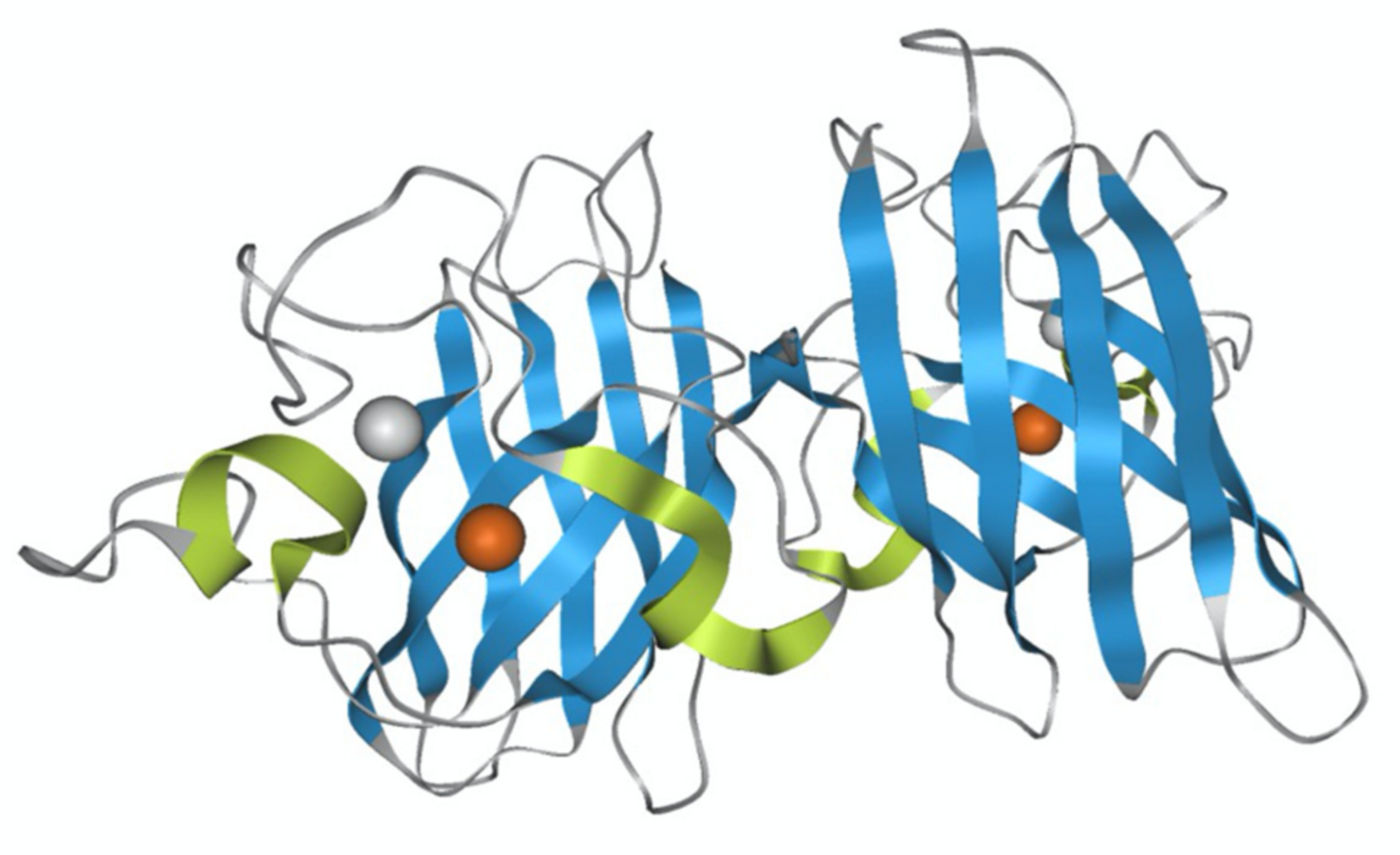
:1. Introduction
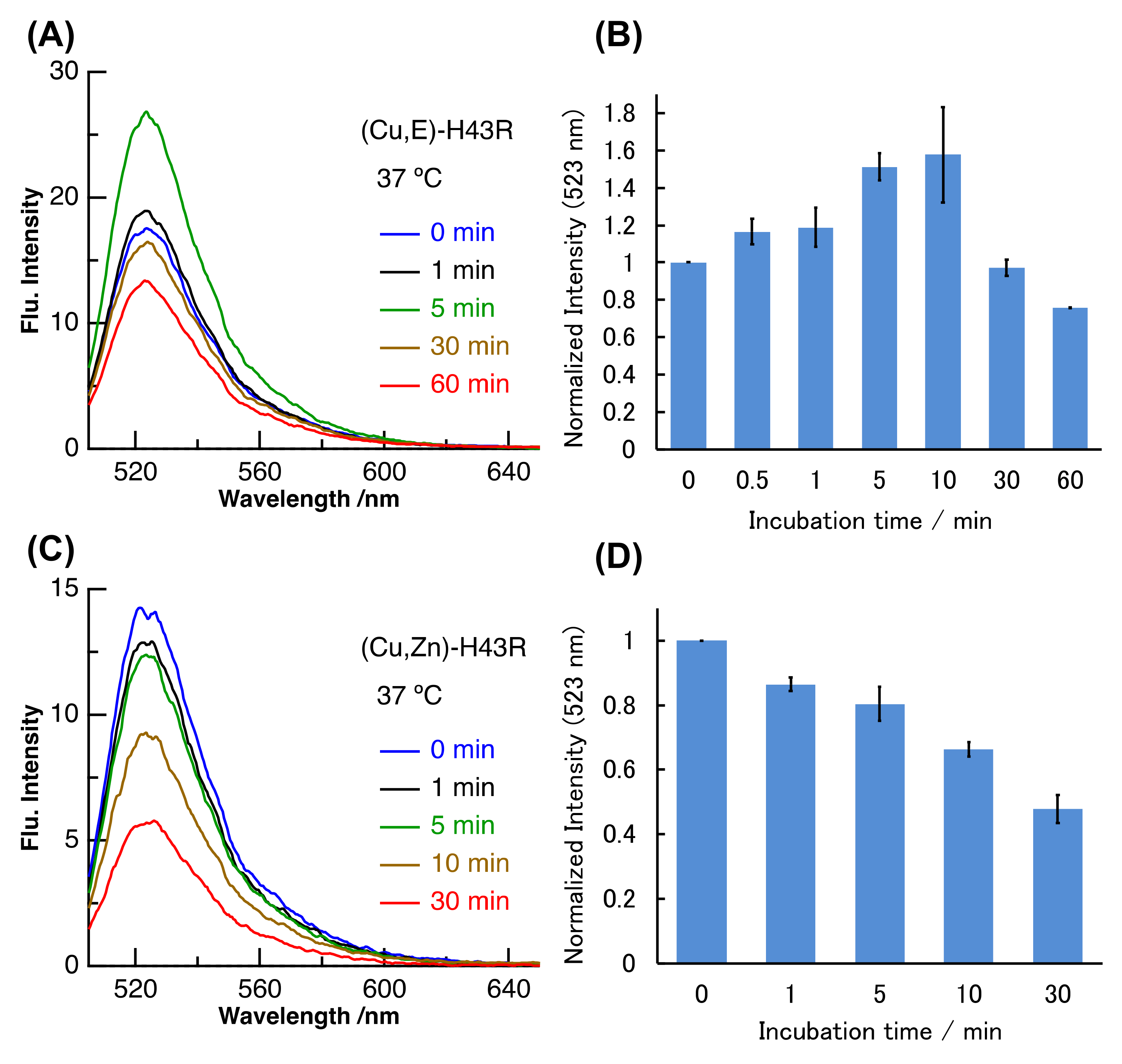
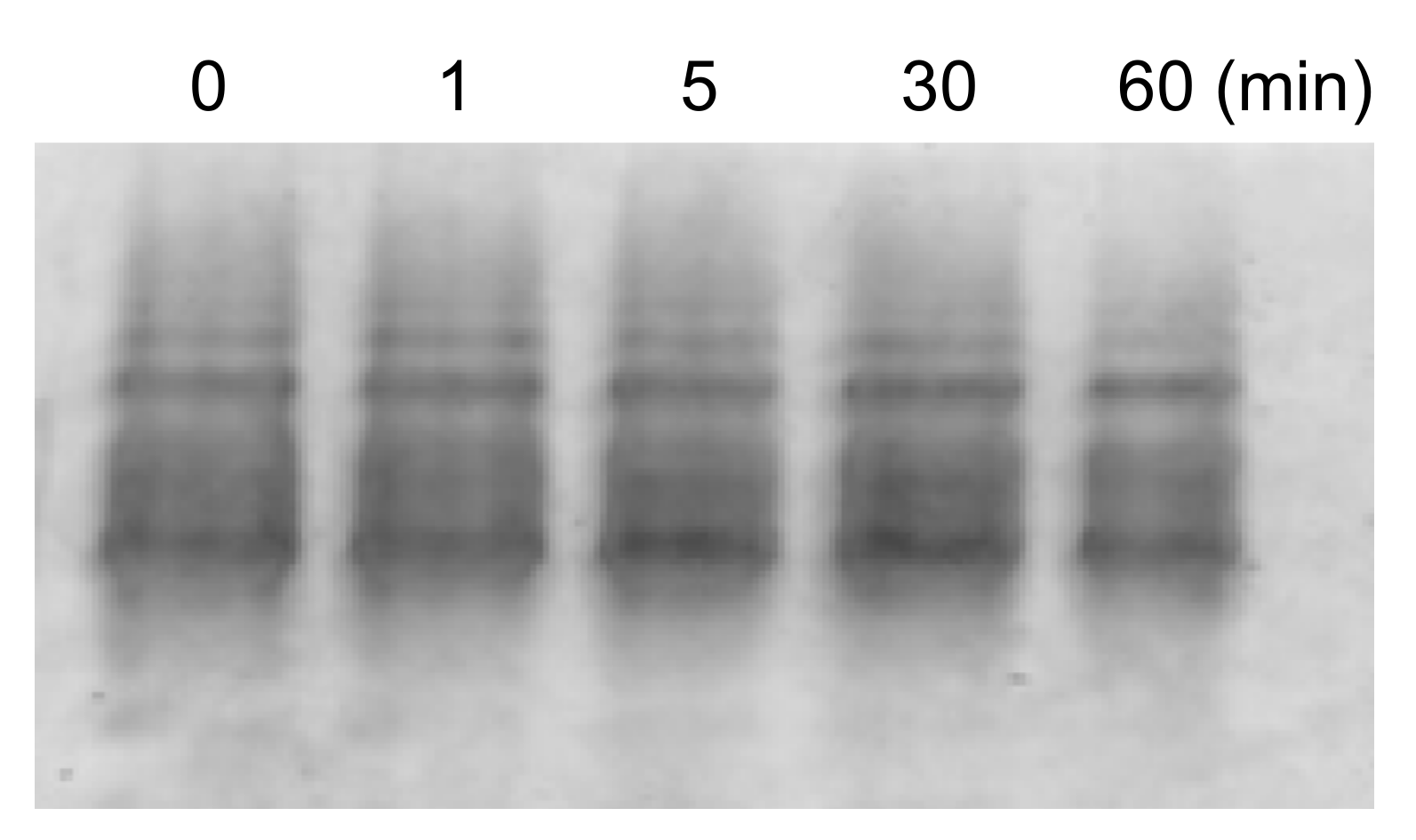
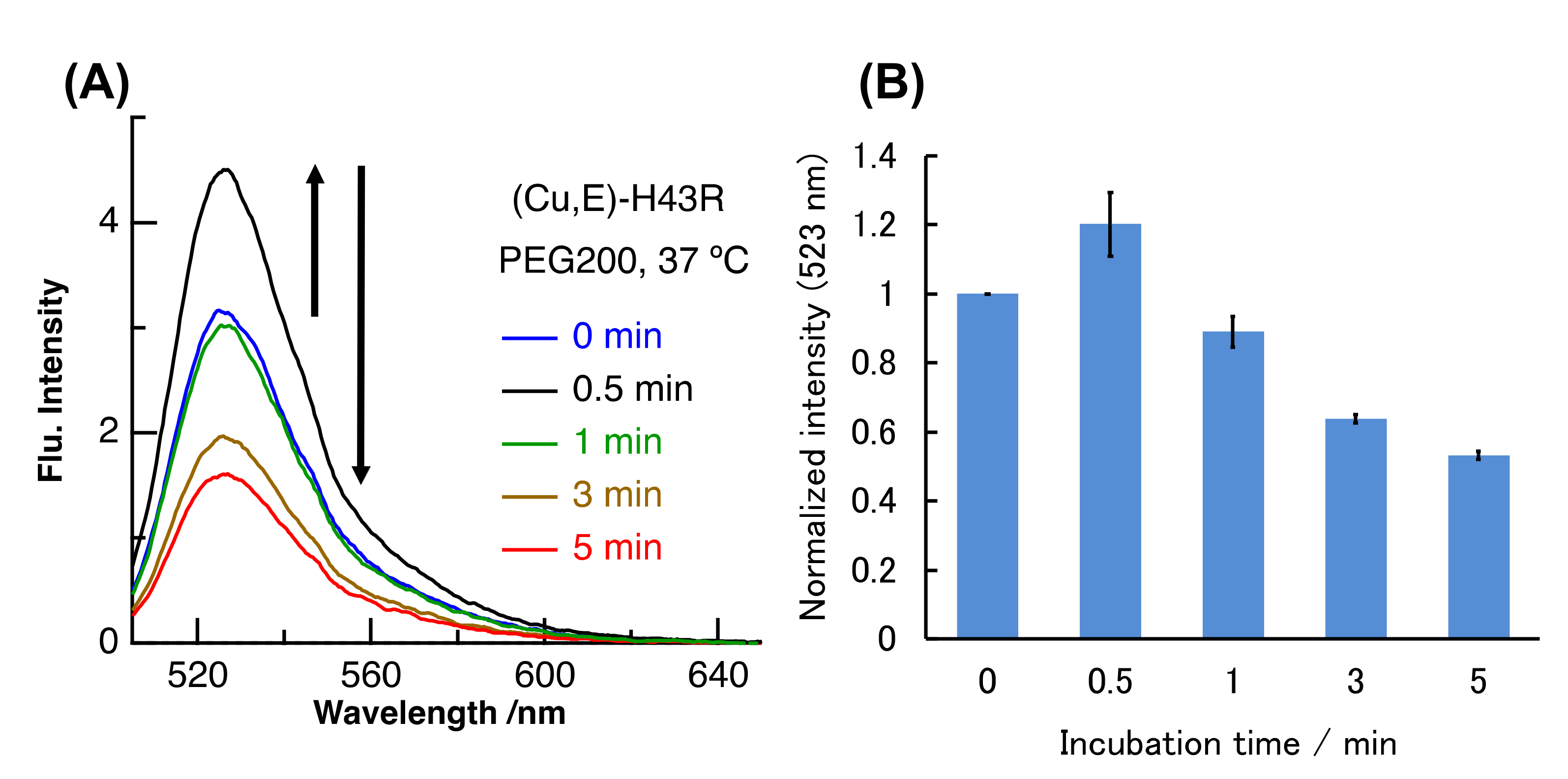
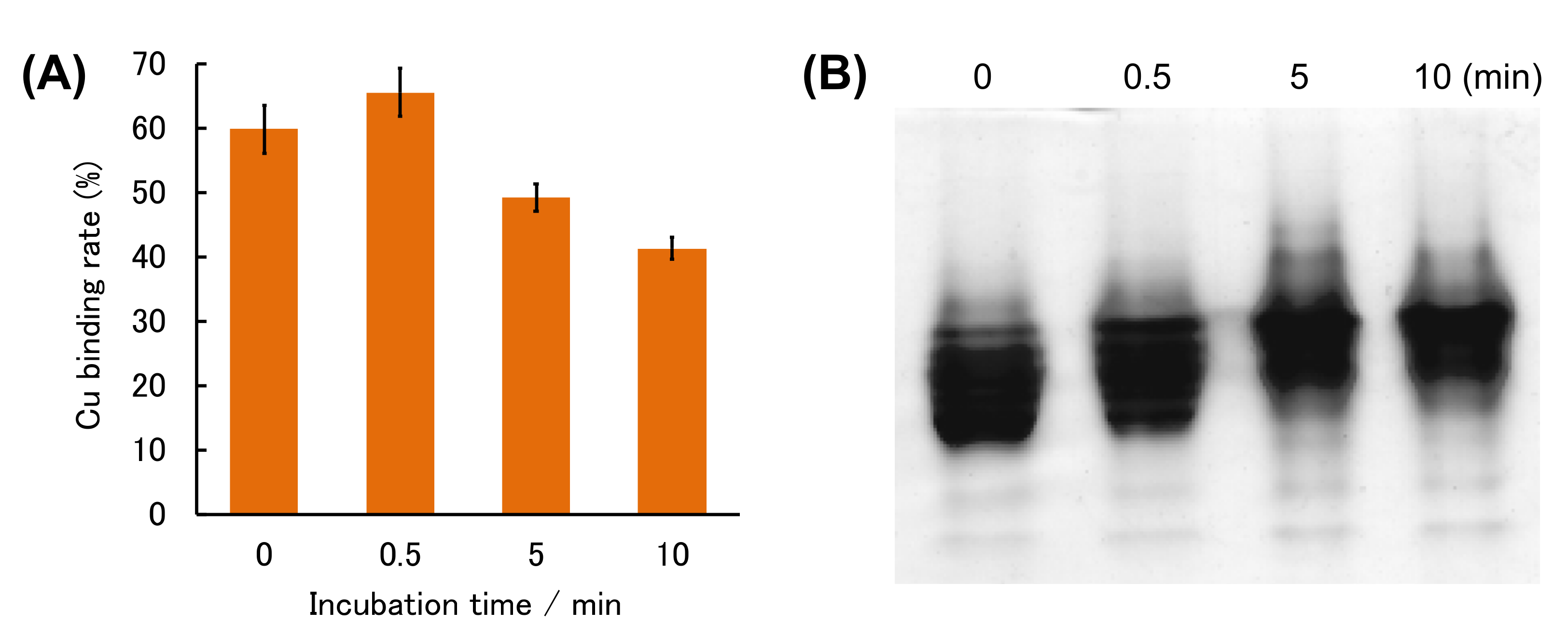
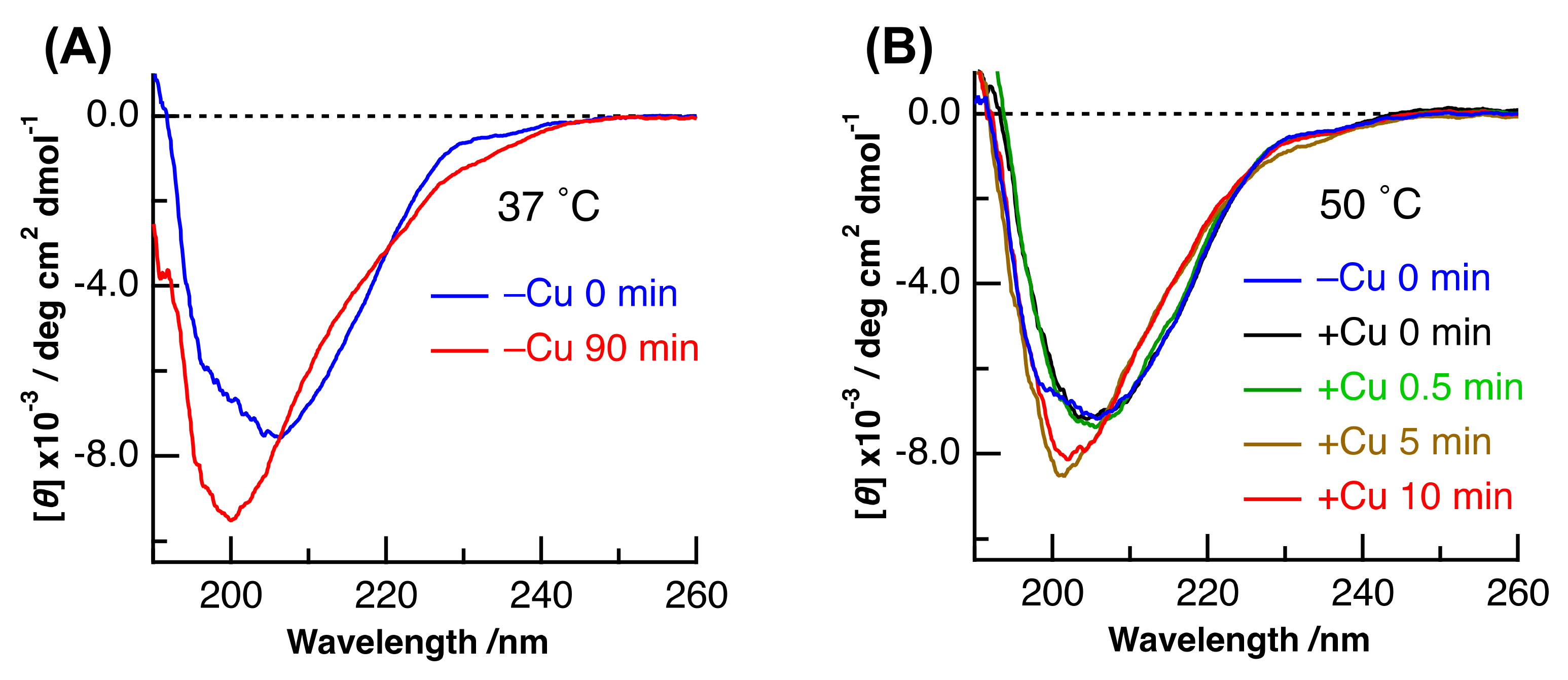
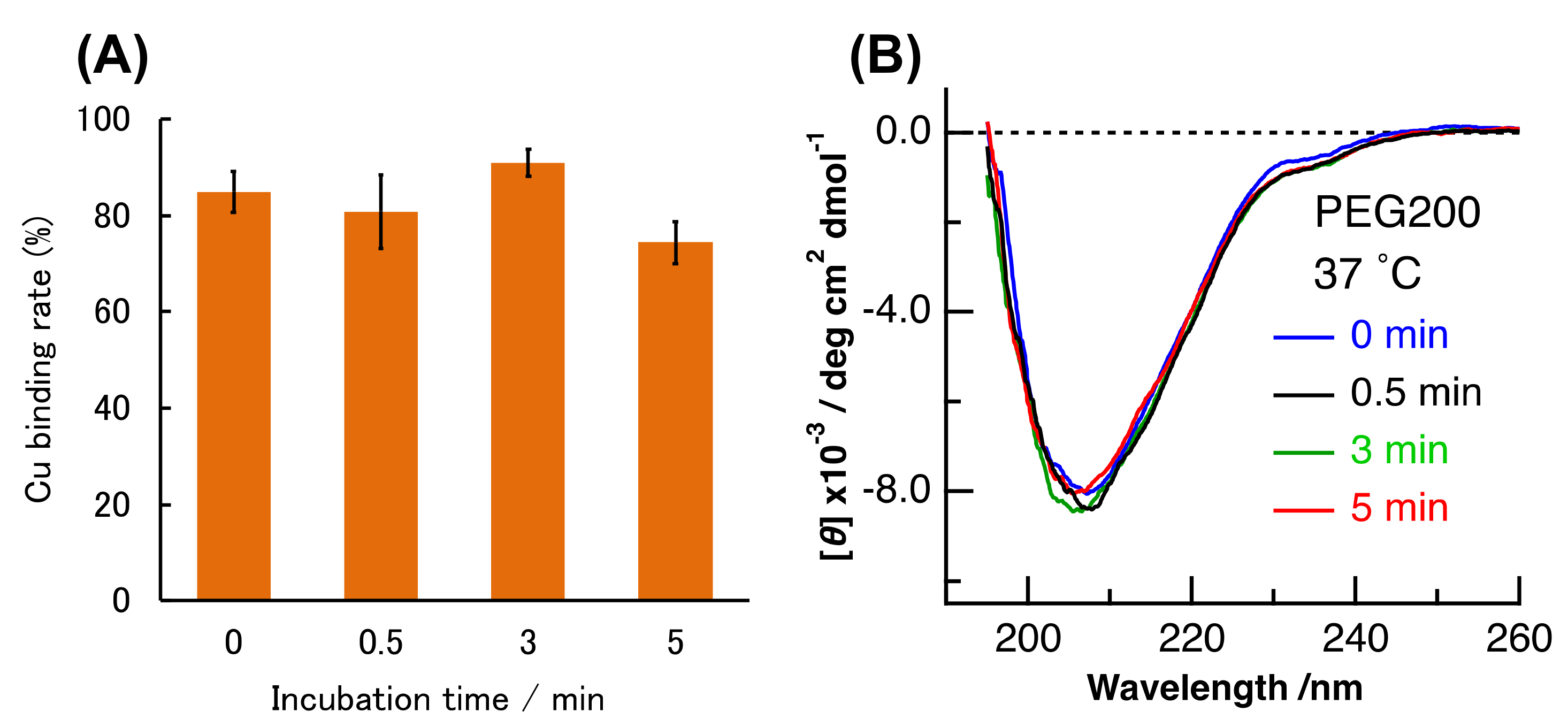
2. Results and Discussion
3. Materials and Methods
3.1. Sample Preparation
3.2. Analyses of Sample Solution
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Cleveland, D.W.; Rothstein, J.D. From Charcot to Lou Gehrig: Deciphering Selective Motor Neuron Death in ALS. Nat. Rev. Neurosci. 2001, 2, 806–819. [Google Scholar] [CrossRef]
- Taylor, J.P.; Brown, R.H.; Cleveland, D.W. Decoding ALS: From genes to mechanism. Nature 2016, 539, 197–206. [Google Scholar] [CrossRef] [Green Version]
- Jaiswal, M.K. Riluzole and edaravone: A tale of two amyotrophic lateral sclerosis drugs. Med. Res. Rev. 2019, 39, 733–748. [Google Scholar] [CrossRef]
- Mackenzie, I.R.A.; Rademakers, R.; Neumann, M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010, 9, 995–1007. [Google Scholar] [CrossRef]
- Elden, A.C.; Kim, H.J.; Hart, M.P.; Chen-Plotkin, A.S.; Johnson, B.S.; Fang, X.; Armakola, M.; Geser, F.; Greene, R.; Lu, M.M.; et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 2010, 466, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Al-Chalabi, A.; Jones, A.; Troakes, C.; King, A.; Al-Sarraj, S.; van den Berg, L.H. The genetics and neuropathology of amyotrophic lateral sclerosis. Acta Neuropathol. 2012, 124, 339–352. [Google Scholar] [CrossRef] [PubMed]
- Aulas, A.; Vande-Velde, C. Alterations in stress granule dynamics driven by TDP-43 and FUS: A link to pathological inclusions in ALS? Front. Cell. Neurosci. 2015, 9, 423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boeynaems, S.; Bogaert, E.; Kovacs, D.; Konijnenberg, A.; Timmerman, E.; Volkov, A.; Guharoy, M.; de Decker, M.; Jaspers, T.; Ryan, V.H.; et al. Phase Separation of C9orf72 Dipeptide Repeats Perturbs Stress Granule Dynamics. Mol. Cell 2017, 65, 1044–1055. [Google Scholar] [CrossRef]
- Becker, L.A.; Huang, B.; Bieri, G.; Ma, R.; Knowles, D.A.; Jafar-Nejad, P.; Messing, J.; Kim, H.J.; Soriano, A.; Auburger, G.; et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 2017, 544, 367–371. [Google Scholar] [CrossRef] [Green Version]
- Maurel, C.; Dangoumau, A.; Marouillat, S.; Brulard, C.; Chami, A.; Hergesheimer, R.; Corcia, P.; Blasco, H.; Andres, C.R.; Vourc’h, P. Causative Genes in Amyotrophic Lateral Sclerosis and Protein Degradation Pathways: A Link to Neurodegeneration. Mol. Neurobiol. 2018, 55, 6480–6499. [Google Scholar] [CrossRef]
- Marrone, L.; Drexler, H.C.A.; Wang, J.; Tripathi, P.; Distler, T.; Heisterkamp, P.; Anderson, E.N.; Kour, S.; Moraiti, A.; Maharana, S.; et al. FUS pathology in ALS is linked to alterations in multiple ALS-associated proteins and rescued by drugs stimulating autophagy. Acta Neuropathol. 2019, 138, 67–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghadge, G.D.; Lee, J.P.; Bindokas, V.P.; Jordan, J.; Ma, L.; Miller, R.J.; Roos, R.P. Mutant superoxide dismutase-1-linked familial amyotrophic lateral sclerosis: Molecular mechanisms of neuronal death and protection. J. Neurosci. 1997, 17, 8756–8766. [Google Scholar] [CrossRef] [PubMed]
- Brasil, A.A.; Magalhães, R.S.S.; de Carvalho, M.D.C.; Paiva, I.; Gerhardt, E.; Pereira, M.D.; Outeiro, T.F.; Eleutherio, E.C.A. Implications of fALS mutations on SOD1 function and oligomerization in cell models. Mol. Neurobiol. 2018, 55, 5269–5281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Johnson, J.L.; Agar, N.Y.R.; Agar, J.N. Protein aggregation and protein instability govern familial amyotrophic lateral sclerosis patient survival. Plos Biol. 2008, 6, 170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- di Donato, M.; Craig, L.; Huff, M.E.; Thayer, M.M.; Cardoso, R.M.F.; Kassmann, C.J.; Lo, T.P.; Bruns, C.K.; Powers, E.T.; Kelly, J.W.; et al. ALS mutants of human superoxide dismutase form fibrous aggregates via framework destabilization. J. Mol. Biol. 2003, 332, 601–615. [Google Scholar] [CrossRef]
- Chattopadhyay, M.; Nwadibia, E.; Strong, C.D.; Gralla, E.B.; Valentine, J.S.; Whitelegge, J.P. The disulfide bond; but not zinc or dimerization; controls initiation and seeded growth in amyotrophic lateral sclerosis-linked Cu/Zn superoxide dismutase (SOD1) fibrillation. J. Biol. Chem. 2015, 290, 30624–30636. [Google Scholar] [CrossRef] [Green Version]
- Leal, S.S.; Cristóvão, J.S.; Biesemeier, A.; Cardoso, I.; Gomes, C.M. Aberrant zinc binding to immature conformers of metal-free copper–zinc superoxide dismutase triggers amorphous aggregation. Metallomics 2015, 7, 333–346. [Google Scholar] [CrossRef]
- Banerjee, V.; Shani, T.; Katzman, B.; Vyazmensky, M.; Papo, N.; Israelson, A.; Engel, S. Superoxide Dismutase 1 (SOD1)-Derived Peptide Inhibits Amyloid Aggregation of Familial Amyotrophic Lateral Sclerosis SOD1 Mutants. ACS Chem. Neurosci. 2016, 7, 1595–1606. [Google Scholar] [CrossRef]
- Tokuda, E.; Furukawa, Y. Abnormal protein oligomers for neurodegeneration. Oncotarget 2017, 8, 39943–39944. [Google Scholar] [CrossRef]
- Mattiazzi, M.; D’Aurelio, M.; Gajewski, C.D.; Martushova, K.; Kiaei, M.; Flint Beal, M.; Manfredi, G. Mutated human SOD1 causes dysfunction of oxidative phosphorylation in mitochondria of transgenic mice. J. Biol. Chem. 2002, 277, 29626–29633. [Google Scholar] [CrossRef] [Green Version]
- Estévez, A.C.; Crow, J.P.; Sampson, J.B.; Reiter, C.; Zhuang, Y.; Richardson, G.J.; Tarpey, M.M.; Barbeito, L.; Beckman, J.S. Induction of nitric oxide-dependent apoptosis in motor neurons by zinc-deficient superoxide dismutase. Science 1999, 286, 2498–2500. [Google Scholar]
- Liu, D.; Wen, J.; Liu, J.; Li, L. The Roles of Free Radicals in Amyotrophic Lateral Sclerosis: Reactive Oxygen Species and Elevated Oxidation of Protein, DNA, and Membrane Phospholipids. Faseb J. 1999, 13, 2318–2328. [Google Scholar] [CrossRef] [PubMed]
- Bakavayev, S.; Chetrit, N.; Zvagelsky, T.; Mansour, R.; Vyazmensky, M.; Barak, Z.; Israelson, A.; Engel, S. Cu/Zn-superoxide dismutase and wild-type like fALS SOD1 mutants produce cytotoxic quantities of H2O2 via cysteine-dependent redox short-circuit. Sci. Rep. 2019, 9, 10826. [Google Scholar] [CrossRef] [PubMed]
- Nishitoh, H.; Kadowaki, H.; Nagai, A.; Maruyama, T.; Yokota, T.; Fukutomi, H.; Noguchi, T.; Matsuzawa, A.; Takeda, K.; Ichijo, H. ALS-linked mutant SOD1 induces ER stress and ASK1-dependent motor neuron death by targeting Derlin-1. Genes Dev. 2008, 22, 1451–1464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rotunno, M.S.; Bosco, D.A. An emerging role for misfolded wild-type SOD1 in sporadic ALS pathogenesis. Front. Cell. Neurosci. 2013, 7, 253. [Google Scholar] [CrossRef] [Green Version]
- Medinas, D.B.; Rozas, P.; Martínez-Traub, F.; Woehlbier, U.; Brown, R.H.; Bosco, D.A.; Hetz, C. Endoplasmic reticulum stress leads to accumulation of wild-type SOD1 aggregates associated with sporadic amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA 2018, 115, 8209–8214. [Google Scholar] [CrossRef] [Green Version]
- Kitamura, F.; Fujimaki, N.; Okita, W.; Hiramatsu, H.; Takeuchi, H. Structural instability and Cu-dependent pro-oxidant activity acquired by the apo form of mutant SOD1 associated with amyotrophic lateral sclerosis. Biochemistry 2011, 50, 4242–4250. [Google Scholar]
- Fujimaki, N.; Kitamura, F.; Takeuchi, H. Pro-oxidant copper-binding mode of the apo form of ALS-linked SOD1 Mutant H43R denatured at physiological temperature. Biochemistry 2013, 52, 5184–5194. [Google Scholar]
- Fujimaki, N.; Miura, T.; Nakabayashi, T. The structural analysis of the pro-oxidant copper-binding site of denatured apo-H43R SOD1 and the elucidation of the origin of the acquisition of the pro-oxidant activity. Phys. Chem. Chem. Phys. 2016, 18, 4468–4475. [Google Scholar] [CrossRef]
- Fujimaki, N.; Nishiya, K.; Miura, T.; Nakabayashi, T. Acquisition of pro-oxidant activity of fALS-linked SOD1 mutants as revealed using circular dichroism and UV-resonance Raman spectroscopy. Chem. Phys. 2016, 479, 5–10. [Google Scholar] [CrossRef]
- Takahashi, A.; Nagao, C.; Murakami, K.; Kuroi, K.; Nakabayashi, T. Effects of molecular crowding environment on the acquisition of toxic properties of wild-type SOD1. Biochim. Biophys. Acta 2020, 1864, 129401. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, T.; Kuroi, K.; Wakabayashi, T.; Fujimaki, N.; Nakabayashi, T. Enhancement of Oxidative Reaction by the Intramolecular Electron Transfer between the Coordinated Redox-Active Metal Ions in SOD1. J. Phys. Chem. B 2020, 124, 2116–2123. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.A.; Valentine, J.S.; Eggers, D.K.; Roe, J.A.; Tiwari, A.; Brown, R.H., Jr.; Hayward, L.J. Familial amyotrophic lateral sclerosis-associated mutations decrease the thermal stability of distinctly metallated species of human copper/zinc superoxide dismutase. J. Biol. Chem. 2002, 277, 15932–15937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lelie, H.L.; Liba, A.; Bourassa, M.W.; Chattopadhyay, M.; Chan, P.K.; Gralla, E.B.; Miller, L.M.; Borchelt, D.R.; Valentine, J.S.; Whitelegge, J.P. Copper and zinc metallation status of copper-zinc superoxide dismutase from amyotrophic lateral sclerosis transgenic mice. J. Biol. Chem. 2011, 286, 2795–2806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rae, T.D.; Schmidt, P.J.; Pufahl, R.A.; Culotta, V.C.; O’Halloran, T.V. Undetectable intracellular free copper: The requirement of a copper chaperone for superoxide dismutase. Science 1999, 284, 805–808. [Google Scholar] [CrossRef] [Green Version]
- Homma, K.; Fujisawa, T.; Tsuburaya, N.; Yamaguchi, N.; Kadowaki, H.; Takeda, K.; Nishitoh, H.; Matsuzawa, A.; Naguro, I.; Ichijo, H. SOD1 as a molecular switch for initiating the homeostatic ER stress response under zinc deficiency. Mol. Cell 2013, 52, 75–86. [Google Scholar] [CrossRef] [Green Version]
- Kaneko, M.; Noguchi, T.; Ikegami, S.; Sakurai, T.; Kakita, A.; Toyoshima, Y.; Kambe, T.; Yamada, M.; Inden, M.; Hara, H.; et al. Zinc transporters ZnT3 and ZnT6 are downregulated in the spinal cords of patients with sporadic amyotrophic lateral sclerosis. J Neurosci. Res. 2015, 93, 370–379. [Google Scholar] [CrossRef] [Green Version]
- Strange, R.W.; Antonyuk, S.; Hough, M.A.; Doucette, P.A.; Rodriguez, J.A.; Hart, P.J.; Hayward, L.J.; Valentine, J.S.; Hasnain, S.S. The structure of holo and metal-deficient wild-type human Cu; Zn superoxide dismutase and its relevance to familial amyotrophic lateral sclerosis. J. Mol. Biol. 2003, 328, 877–891. [Google Scholar] [CrossRef]
- Furukawa, Y.; Anzai, I.; Akiyama, S.; Imai, M.; Cruz, F.J.C.; Saio, T.; Nagasawa, K.; Nomura, T.; Ishimori, K. Conformational disorder of the most immature Cu; Zn-superoxide dismutase leading to amyotrophic lateral sclerosis. J. Biol. Chem. 2016, 291, 4144–4155. [Google Scholar] [CrossRef] [Green Version]
- Nakano, S.; Miyoshi, D.; Sugimoto, N. Effects of Molecular Crowding on the Structures, Interactions, and Functions of Nucleic Acids. Chem. Rev. 2014, 114, 2733–2758. [Google Scholar] [CrossRef]
- Mittal, S.; Chowhan, R.K.; Singh, L.R. Macromolecular crowding: Macromolecules friend or foe. Biochim. Biophys. Acta Gen. Subj. 2015, 1850, 1822–1831. [Google Scholar] [CrossRef] [PubMed]
- Shahid, S.; Imtaiyaz-Hassan, M.; Islam, A.; Ahmad, F. Size-dependent studies of macromolecular crowding on the thermodynamic stability; structure and functional activity of proteins: In vitro and in silico approaches. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 178–197. [Google Scholar] [CrossRef] [PubMed]
- Crow, J.P.; Sampson, J.B.; Zhuang, Y.; Thompson, J.A.; Beckman, J.S. Decreased zinc affinity of amyotrophic lateral sclerosis-associated superoxide dismutase mutants leads to enhanced catalysis of tyrosine nitration by peroxynitrite. J. Neurochem. 1997, 69, 1936–1944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Sample Availability: H43R SOD1 mutant is available from the authors. |

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nagao, C.; Kuroi, K.; Wakabayashi, T.; Nakabayashi, T. Pro-Oxidant Activity of an ALS-Linked SOD1 Mutant in Zn-Deficient Form. Molecules 2020, 25, 3600. https://doi.org/10.3390/molecules25163600
Nagao C, Kuroi K, Wakabayashi T, Nakabayashi T. Pro-Oxidant Activity of an ALS-Linked SOD1 Mutant in Zn-Deficient Form. Molecules. 2020; 25(16):3600. https://doi.org/10.3390/molecules25163600
Chicago/Turabian StyleNagao, Chise, Kunisato Kuroi, Taiyu Wakabayashi, and Takakazu Nakabayashi. 2020. "Pro-Oxidant Activity of an ALS-Linked SOD1 Mutant in Zn-Deficient Form" Molecules 25, no. 16: 3600. https://doi.org/10.3390/molecules25163600
APA StyleNagao, C., Kuroi, K., Wakabayashi, T., & Nakabayashi, T. (2020). Pro-Oxidant Activity of an ALS-Linked SOD1 Mutant in Zn-Deficient Form. Molecules, 25(16), 3600. https://doi.org/10.3390/molecules25163600





