Modified Enzyme Substrates for the Detection of Bacteria: A Review
Abstract
:1. Introduction
2. Classification of Synthetic Enzyme Substrates
2.1. Classification by Type of Generated Signal
2.1.1. Chromogenic and Fluorogenic Substrates
Soluble Substrates
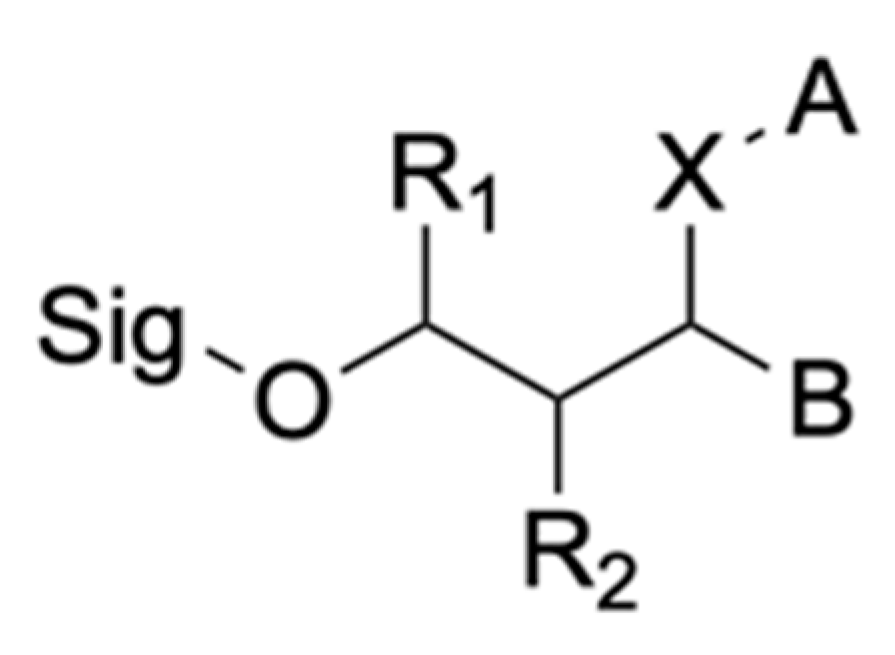
Insoluble Substrates
2.1.2. Luminogenic Substrates
Bioluminescent Substrates
Chemiluminescent Substrates
2.1.3. Electrogenic Substrates
2.2. Classification Based on Enzyme Triggering the Signal
2.2.1. Hydrolytic Enzymes
2.2.2. Non-Hydrolytic
2.2.3. Boolean Substrates
3. Emerging Trends for Enzyme-Based Bacterial Detection
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hameed, S.; Xie, L.; Ying, Y. Conventional and emerging detection techniques for pathogenic bacteria in food science: A review. Trends Food Sci. Technol. 2018, 81, 61–73. [Google Scholar] [CrossRef]
- Law, J.W.-F.; Ab Mutalib, N.-S.; Chan, K.-G.; Lee, L.-H. Rapid methods for the detection of foodborne bacterial pathogens: Principles, applications, advantages and limitations. Front. Microbiol. 2015, 5, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajapaksha, P.; Elbourne, A.; Gangadoo, S.; Brown, R.; Cozzolino, D.; Chapman, J. A review of methods for the detection of pathogenic microorganisms. Analyst 2019, 144, 396–411. [Google Scholar] [CrossRef] [PubMed]
- Váradi, L.; Luo, J.L.; Hibbs, D.E.; Perry, J.D.; Anderson, R.J.; Orenga, S.; Groundwater, P.W. Methods for the detection and identification of pathogenic bacteria: Past, present, and future. Chem. Soc. Rev. 2017, 46, 4818–4832. [Google Scholar] [CrossRef]
- Simner, P.J.; Antar, A.A.R.; Hao, S.; Gurtowski, J.; Tamma, P.D.; Rock, C.; Opene, B.N.A.; Tekle, T.; Carroll, K.C.; Schatz, M.C.; et al. Antibiotic pressure on the acquisition and loss of antibiotic resistance genes in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2018, 73, 1796–1803. [Google Scholar] [CrossRef]
- Menard, K.L.; Grossman, A.D. Selective pressures to maintain attachment site specificity of integrative and conjugative elements. PLoS Genet. 2013, 9. [Google Scholar] [CrossRef] [Green Version]
- Perry, J.D. A decade of development of chromogenic culture media for clinical microbiology in an era of molecular diagnostics. Clin. Microbiol. Rev. 2017, 30, 449–479. [Google Scholar] [CrossRef] [Green Version]
- Alouane, A.; Labruère, R.; Le Saux, T.; Schmidt, F.; Jullien, L. Self-immolative spacers: Kinetic aspects, structure–property relationships, and applications. Angew. Chem. Int. Ed. 2015, 54, 7492–7509. [Google Scholar] [CrossRef]
- Gnaim, S.; Shabat, D. Quinone-Methide Species, A gateway to functional molecular systems: From self-immolative dendrimers to long-wavelength fluorescent dyes. Acc. Chem. Res. 2014, 47, 2970–2984. [Google Scholar] [CrossRef]
- Manafi, M. Fluorogenic and chromogenic enzyme substrates in culture media and identification tests. Int. J. Food Microbiol. 1996, 31, 45–58. [Google Scholar] [CrossRef]
- Orenga, S.; James, A.L.; Manafi, M.; Perry, J.D.; Pincus, D.H. Enzymatic substrates in microbiology. J. Microbiol. Methods 2009, 79, 139–155. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.I. Bacterial Virulence Factors. In Bacterial Pathogens and Their Virulence Factors; Springer: Cham, Germany, 2018. [Google Scholar]
- Seidman, M.; Link, K. New Compounds. Substituted salicylaldehydes and derivatives. J. Am. Chem. Soc. 1950, 72, 4324. [Google Scholar] [CrossRef]
- Lederberg, J. The beta-d-galactosidase of Escherichia coli, strain K-12. J. Bacteriol. 1950, 60, 381–392. [Google Scholar] [CrossRef] [Green Version]
- Manafi, M.; Kneifel, W.; Bascomb, S. Fluorogenic and chromogenic substrates used in bacterial diagnostics. Microbiol. Rev. 1991, 55, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Manafi, M.; Kneifel, W. Rapid methods for differentiating Gram-positive from Gram-negative aerobic and facultative anaerobic bacteria. J. Appl. Bacteriol. 1990, 69, 822–827. [Google Scholar] [CrossRef] [PubMed]
- Dyer, D.L. Microbiological detection and identification system. U.S. Patent 3,551,295A, 1970. [Google Scholar]
- Hesari, N.; Alum, A.; Elzein, M.; Abbaszadegan, M. A biosensor platform for rapid detection of E. coli in drinking water. Enzyme Microb. Technol. 2016, 83, 22–28. [Google Scholar] [CrossRef] [Green Version]
- Satoh, H.; Kikuchi, K.; Katayose, Y.; Tsuda, S.; Hirano, R.; Hirakata, Y.; Kitajima, M.; Ishii, S.; Oshiki, M.; Hatamoto, M.; et al. Simple and reliable enumeration of Escherichia coli concentrations in wastewater samples by measuring β-d-glucuronidase (GUS) activities via a microplate reader. Sci. Total Environ. 2020, 715, 136928. [Google Scholar] [CrossRef]
- Chilvers, K.F.; Perry, J.D.; James, A.L.; Reed, R.H. Synthesis and evaluation of novel fluorogenic substrates for the detection of bacterial β-galactosidase. J. Appl. Microbiol. 2001, 91, 1118–1130. [Google Scholar] [CrossRef] [Green Version]
- Váradi, L.; Hibbs, D.E.; Orenga, S.; Babolat, M.; Perry, J.D.; Groundwater, P.W. β-Alanyl aminopeptidase-activated fluorogenic probes for the rapid identification of Pseudomonas aeruginosa in clinical samples. RSC Adv. 2016, 6, 58884–58889. [Google Scholar] [CrossRef]
- Wildeboer, D.; Jeganathan, F.; Price, R.G.; Abuknesha, R.A. Characterization of bacterial proteases with a panel of fluorescent peptide substrates. Anal. Biochem. 2009, 384, 321–328. [Google Scholar] [CrossRef]
- Cellier-Rastit, M.; James, A.L.; Orenga, S.; Perry, J.D.; Robinson, S.N.; Turnbull, G.; Stanforth, S.P. Fluorogenic l-alanylaminopeptidase substrates derived from 6-amino-2-hetarylquinolines and 7-amino-3-hetarylcoumarins and their potential applications in diagnostic microbiology. Bioorg. Med. Chem. Lett. 2019, 29, 1227–1231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eshghi, H.; Mirzaie, N.; Asoodeh, A. Synthesis of fluorescein aromatic esters in the presence of P2O5/SiO2 as catalyst and their hydrolysis studies in the presence of lipase. Dyes Pigm. 2011, 89, 120–126. [Google Scholar] [CrossRef]
- Guilini, C.; Baehr, C.; Schaeffer, E.; Gizzi, P.; Rufi, F.; Haiech, J.; Weiss, E.; Bonnet, D.; Galzi, J.-L. New fluorescein precursors for live bacteria detection. Anal. Chem. 2015, 87, 8858–8866. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Finney, N.S. Small-molecule fluorescent probes and their design. RSC Adv. 2018, 8, 29051–29061. [Google Scholar] [CrossRef] [Green Version]
- Yan, F.; Fan, K.; Bai, Z.; Zhang, R.; Zu, F.; Xu, J.; Li, X. Fluorescein applications as fluorescent probes for the detection of analytes. Trends Anal. Chem. 2017, 97, 15–35. [Google Scholar] [CrossRef]
- Datta, S.; Sherman, J.M.; Tovar, M.A.; Bravard, M.A.; Valencia, T.; Montoya, R.; Quino, W.; D’Arcy, N.; Ramos, E.S.; Gilman, R.H.; et al. Sputum microscopy with fluorescein diacetate predicts tuberculosis infectiousness. J. Infect. Dis. 2017, 216, 514–524. [Google Scholar] [CrossRef]
- Chrzanowski, T.H.; Crotty, R.D.; Hubbard, J.G.; Welch, R.P. Applicability of the fluorescein diacetate method of detecting active bacteria in freshwater. Microb. Ecol. 1984, 10, 179–185. [Google Scholar] [CrossRef]
- van Oosten, M.; Schäfer, T.; Gazendam, J.A.C.; Ohlsen, K.; Tsompanidou, E.; de Goffau, M.C.; Harmsen, H.J.M.; Crane, L.M.A.; Lim, E.; Francis, K.P.; et al. Real-time in vivo imaging of invasive- and biomaterial-associated bacterial infections using fluorescently labelled vancomycin. Nat. Commun. 2013, 4, 2584. [Google Scholar] [CrossRef]
- Breeuwer, P.; Drocourt, J.; Rombouts, F.M.; Abee, T. A novel method for continuous determination of the intracellular ph in bacteria with the internally conjugated fluorescent probe 5 (and 6-)-Carboxyfluorescein Succinimidyl ester. Appl. Environ. Microbiol. 1996, 62, 178–183. [Google Scholar] [CrossRef] [Green Version]
- Smith, E.L.; Bertozzi, C.R.; Beatty, K.E. An expanded set of fluorogenic sulfatase activity probes. ChemBioChem 2014, 15, 1101–1105. [Google Scholar] [CrossRef] [Green Version]
- Magro, G.; Bain, R.E.S.; Woodall, C.A.; Matthews, R.L.; Gundry, S.W.; Davis, A.P. Synthesis and application of resorufin β-d-glucuronide, a low-cost chromogenic substrate for detecting Escherichia coli in drinking water. Environ. Sci. Technol. 2014, 48, 9624–9631. [Google Scholar] [CrossRef] [PubMed]
- Burke, M.D.; Thompson, S.; Weaver, R.J.; Wolf, C.R.; Mayers, R.T. Cytochrome P450 specificities of alkoxyresorufin O-dealkylation in human and rat liver. Biochem. Pharmacol. 1994, 48, 923–936. [Google Scholar] [CrossRef]
- Hofmann, J.; Sernetz, M. Immobilized enzyme kinetics analyzed by flow-through microfluorimetry: Resorufin-β-d-galactopyranoside as a new fluorogenic substrate for β-galactosidase. Anal. Chim. Acta 1984, 163, 67–72. [Google Scholar] [CrossRef]
- Lavis, L.D.; Chao, T.-Y.; Raines, R.T. Synthesis and utility of fluorogenic acetoxymethyl ethers. Chem. Sci. 2011, 2, 521–530. [Google Scholar] [CrossRef] [Green Version]
- Genter, F.; Marks, S.J.; Clair-Caliot, G.; Mugume, D.S.; Johnston, R.B.; Bain, R.E.S.; Julian, T.R. Evaluation of the novel substrate RUG™ for the detection of Escherichia coli in water from temperate (Zurich, Switzerland) and tropical (Bushenyi, Uganda) field sites. Environ. Sci. Water Res. 2019, 5, 1082–1091. [Google Scholar] [CrossRef] [Green Version]
- James, A.L.; Perry, J.D.; Chilvers, K.; Robson, I.S.; Armstrong, L.; Orr, K.E. Alizarin-β-d-galactoside: A new substrate for the detection of bacterial β-galactosidase. Lett. Appl. Microbiol. 2000, 30, 336–340. [Google Scholar] [CrossRef] [Green Version]
- James, A.L.; Perry, J.D.; Ford, M.; Armstrong, L.; Gould, F.K. Evaluation of cyclohexenoesculetin-beta-D-galactoside and 8-hydroxyquinoline-beta-D-galactoside as substrates for the detection of beta-galactosidase. Appl. Environ. Microbiol. 1996, 62, 3868–3870. [Google Scholar] [CrossRef] [Green Version]
- James, A.L.; Perry, J.D.; Ford, M.; Armstrong, L.; Gould, F.K. Note: Cyclohexenoesculetin-β-D-glucoside: A new substrate for the detection of bacterial β-D-glucosidase. J. Appl. Microbiol. 1997, 82, 532–536. [Google Scholar] [CrossRef]
- Perry, J.D.; Morris, K.A.; James, A.L.; Oliver, M.; Gould, F.K. Evaluation of novel chromogenic substrates for the detection of bacterial β-glucosidase. J. Appl. Microbiol. 2007, 102, 410–415. [Google Scholar] [CrossRef]
- Swan, A. The use of a bile-aesculin medium and of Maxted’s technique of Lancefield grouping in the identification of enterococci (group D streptococci). J. Clin. Pathol. 1954, 7, 160–163. [Google Scholar] [CrossRef] [Green Version]
- Burton, M.; Perry, J.D.; Stanforth, S.P.; Turner, H.J. The synthesis of novel chromogenic enzyme substrates for detection of bacterial glycosidases and their applications in diagnostic microbiology. Bioorg. Med. Chem. 2018, 26, 4841–4849. [Google Scholar] [CrossRef] [PubMed]
- Kiernan, J.A. Indigogenic substrates for detection and localization of enzymes. Biotechn. Histochem. 2007, 82, 73–103. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.D.; Freydière, A.M. The application of chromogenic media in clinical microbiology. J. Appl. Microbiol. 2007, 103, 2046–2055. [Google Scholar] [CrossRef] [PubMed]
- Angelov, A.; Li, H.; Geissler, A.; Leis, B.; Liebl, W. Toxicity of indoxyl derivative accumulation in bacteria and its use as a new counterselection principle. Syst. Appl. Microbiol. 2013, 36, 585–592. [Google Scholar] [CrossRef]
- Holt, S.J. A new approach to the cytochemical localization of enzymes. Proc. R. Soc. Lond. B–Biol. Sci. 1954, 142, 160–169. [Google Scholar] [CrossRef]
- Voss, G.; Drechsler, M.; Eller, S.; Gradzielski, M.; Gunzelmann, D.; Mondal, S.; van Smaalen, S.; Voertler, C.S. Polymeric colorants: Statistical copolymers of indigo building blocks with defined structures. Helv. Chim. Acta 2009, 92, 2675–2697. [Google Scholar] [CrossRef]
- Spitz, U.; Wick, L.; Bayer, A.; Weymuth, C.; Schabert, G. Novel Indicator Platform. U.S. Patent EP2,427,431B1, 1 May 2013. [Google Scholar]
- Wick, L.; Spitz, U.; Weymuth, C.; Schabert, G.; Mayer, T.; Bayer, A. Novel Applications of Indoloindole and Indoloquinoline Dyes. U.S. Patent EP2,635,640A1, 4 November 2011. [Google Scholar]
- Wick, L.; Spitz, U. Novel Plating Media. U.S. Patent 2,010,029,159,1A1, 15 September 2010. [Google Scholar]
- Hananya, N.; Shabat, D. A Glowing Trajectory between bio-and chemiluminescence: From luciferin-based probes to triggerable dioxetanes. Angew. Chem. Int. Ed. 2017, 56, 16454–16463. [Google Scholar] [CrossRef]
- Kaskova, Z.M.; Tsarkova, A.S.; Yampolsky, I.V. 1001 lights: Luciferins, luciferases, their mechanisms of action and applications in chemical analysis, biology and medicine. Chem. Soc. Rev. 2016, 45, 6048–6077. [Google Scholar] [CrossRef]
- Fleiss, A.; Sarkisyan, K.S. A brief review of bioluminescent systems. Curr. Genet. 2019, 65, 877–882. [Google Scholar] [CrossRef] [Green Version]
- Fraga, H.; Esteves da Silva, J.C.G.; Fontes, R. Identification of luciferyl adenylate and luciferyl coenzyme a synthesized by firefly luciferase. ChemBioChem 2004, 5, 110–115. [Google Scholar] [CrossRef]
- Branchini, B.R.; Behney, C.E.; Southworth, T.L.; Fontaine, D.M.; Gulick, A.M.; Vinyard, D.J.; Brudvig, G.W. Experimental support for a single electron-transfer oxidation mechanism in firefly bioluminescence. J. Am. Chem. Soc. 2015, 137, 7592–7595. [Google Scholar] [CrossRef] [PubMed]
- Hauser, J.R.; Beard, H.A.; Bayana, M.E.; Jolley, K.E.; Warriner, S.L.; Bon, R.S. Economical and scalable synthesis of 6-amino-2-cyanobenzothiazole. Beilstein J. Org. Chem. 2016, 12, 2019–2025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCutcheon, D.C.; Porterfield, W.B.; Prescher, J.A. Rapid and scalable assembly of firefly luciferase substrates. Org. Biomol. Chem. 2015, 13, 2117–2121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Podsiadly, R.; Grzelakowska, A.; Modrzejewska, J.; Siarkiewicz, P.; Słowiński, D.; Szala, M.; Świerczyńska, M. Recent progress in the synthesis of firefly luciferin derivatives. Dyes Pigm. 2019, 170, 107627. [Google Scholar] [CrossRef]
- Li, J.; Chen, L.; Du, L.; Li, M. Cage the firefly luciferin!–A strategy for developing bioluminescent probes. Chem. Soc. Rev. 2013, 42, 662–676. [Google Scholar] [CrossRef]
- Masuda-Nishimura, I.; Fukuda, S.; Sano, A.; Kasai, K.; Tatsumi, H. Development of a rapid positive/absent test for coliforms using sensitive bioluminescence assay. Lett. Appl. Microbiol. 2000, 30, 130–135. [Google Scholar] [CrossRef] [Green Version]
- Heller, A.A.; Spence, D.M. A rapid method for post-antibiotic bacterial susceptibility testing. PLoS ONE 2019, 14, e0210534. [Google Scholar] [CrossRef]
- Dodeigne, C.; Thunus, L.; Lejeune, R. Chemiluminescence as diagnostic tool. A review. Talanta 2000, 51, 415–439. [Google Scholar] [CrossRef]
- Chen, G.; Jin, M.; Du, P.; Zhang, C.; Cui, X.; Zhang, Y.; Wang, J.; Jin, F.; She, Y.; Shao, H.; et al. A review of enhancers for chemiluminescence enzyme immunoassay. Food Agric. Immunol. 2017, 28, 315–327. [Google Scholar] [CrossRef] [Green Version]
- Hananya, N.; Shabat, D. Recent advances and challenges in luminescent imaging: Bright outlook for Chemiluminescence of Dioxetanes in water. ACS Cent. Sci. 2019, 5, 949–959. [Google Scholar] [CrossRef] [Green Version]
- Hananya, N.; Eldar Boock, A.; Bauer, C.R.; Satchi-Fainaro, R.; Shabat, D. Remarkable enhancement of chemiluminescent signal by dioxetane–fluorophore conjugates: Turn-On chemiluminescence probes with color modulation for sensing and imaging. J. Am. Chem. Soc. 2016, 138, 13438–13446. [Google Scholar] [CrossRef] [PubMed]
- Green, O.; Eilon, T.; Hananya, N.; Gutkin, S.; Bauer, C.R.; Shabat, D. Opening a gateway for chemiluminescence cell imaging: Distinctive methodology for design of bright chemiluminescent dioxetane probes. ACS Cent. Sci. 2017, 3, 349–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shabat, D.; Roth-Konforti, M.E.; Hananya, N.; Green, O.; Spitz, U.; Wick, L.; Ihssen, J.; Vorberg, R.; Cribiu, R.; Babjakova, Z.; et al. Dioxetane Compounds and their Use for the Detection of Microorganisms. U.S. Patent WO2,019,224,338A1, 28 November 2019. [Google Scholar]
- Roth-Konforti, M.; Green, O.; Hupfeld, M.; Fieseler, L.; Heinrich, N.; Ihssen, J.; Vorberg, R.; Wick, L.; Spitz, U.; Shabat, D. Ultrasensitive detection of Salmonella and Listeria monocytogenes by small-molecule chemiluminescence probes. Angew. Chem. Int. Ed. 2019, 58, 10361–10367. [Google Scholar] [CrossRef] [PubMed]
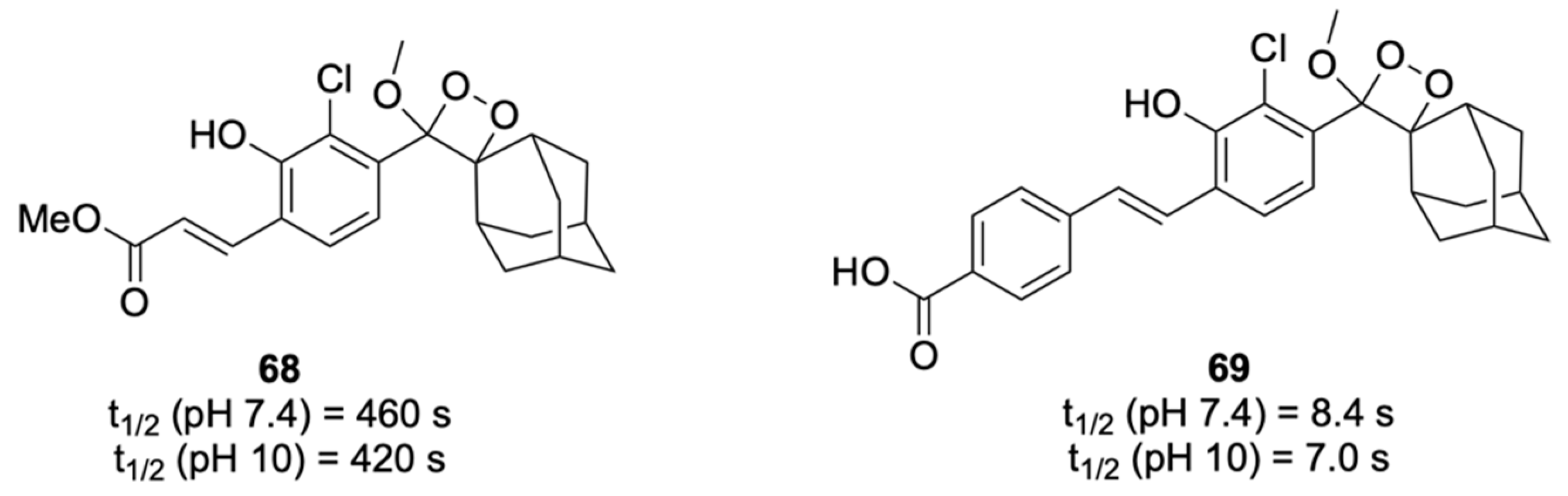
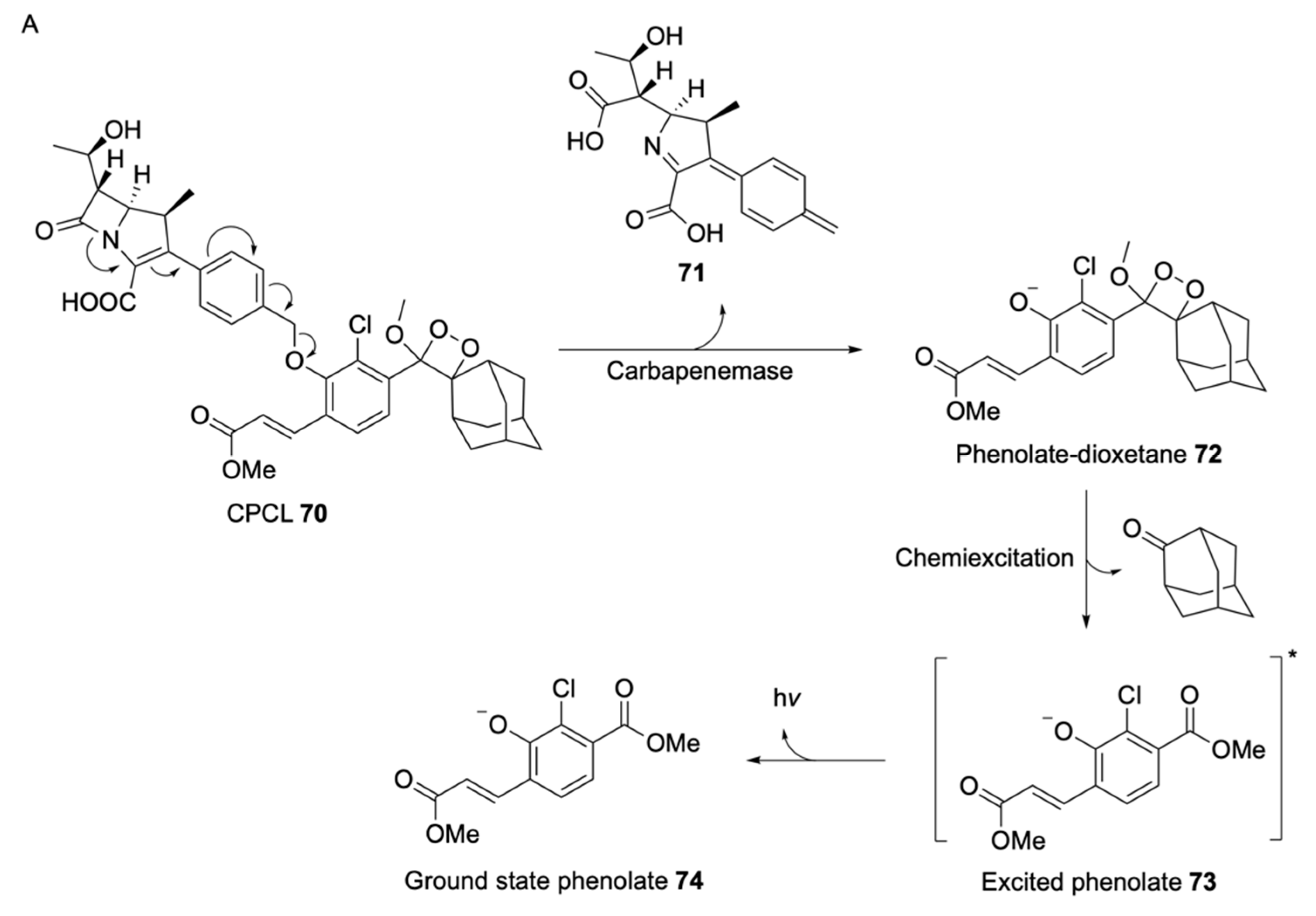
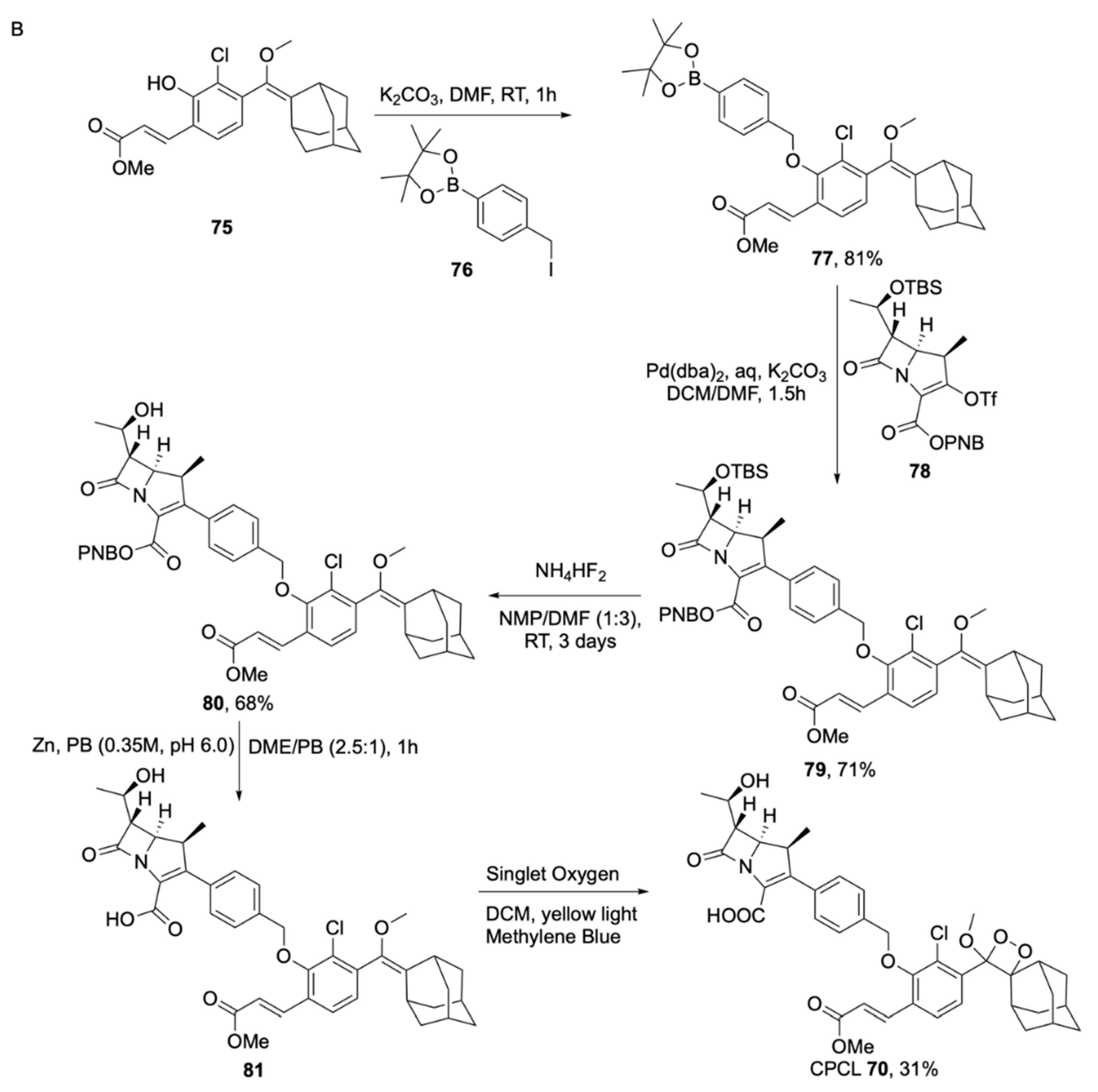
- Das, S.; Ihssen, J.; Wick, L.; Spitz, U.; Shabat, D. Chemiluminescent Carbapenem-Based Molecular Probe for Detection of Carbapenemase Activity in Live Bacteria. Chem. Eur. J. 2020, 26, 3647–3652. [Google Scholar] [CrossRef]
- Chen, J.; Andler, S.M.; Goddard, J.M.; Nugen, S.R.; Rotello, V.M. Integrating recognition elements with nanomaterials for bacteria sensing. Chem. Soc. Rev. 2017, 46, 1272–1283. [Google Scholar] [CrossRef] [PubMed]
- Kang, E.B.; Mazrad, Z.A.I.; Robby, A.I.; In, I.; Park, S.Y. Alkaline phosphatase-responsive fluorescent polymer probe coated surface for colorimetric bacteria detection. Eur. Polym. J. 2018, 105, 217–225. [Google Scholar] [CrossRef]
- Fanjul-Bolado, P.; González-García, M.B.; Costa-García, A. 3-Indoxyl phosphate as an electrochemical substrate for horseradish peroxidase. Electroanalysis 2004, 16, 988–993. [Google Scholar] [CrossRef]
- Fernández-Sánchez, C.; Costa-Garcı´a, A. 3-Indoxyl Phosphate: An alkaline phosphatase substrate for enzyme immunoassays with voltammetric detection. Electroanalysis 1998, 10, 249–255. [Google Scholar] [CrossRef]
- Rashid, J.I.A.; Yusof, N.A. The strategies of DNA immobilization and hybridization detection mechanism in the construction of electrochemical DNA sensor: A review. Sens. Bio-Sens. Res. 2017, 16, 19–31. [Google Scholar] [CrossRef]
- Chen, J.; Huang, Z.; Luo, Z.; Yu, Q.; Xu, Y.; Wang, X.; Li, Y.; Duan, Y. Multichannel-structured three-dimensional chip for highly sensitive pathogenic bacteria detection based on fast DNA-programmed signal polymerization. Anal. Chem. 2018, 90, 12019–12026. [Google Scholar] [CrossRef]
- Fanjul-Bolado, P.; Hernández-Santos, D.; González-García, M.B.; Costa-García, A. Alkaline phosphatase-catalyzed silver deposition for electrochemical detection. Anal. Chem. 2007, 79, 5272–5277. [Google Scholar] [CrossRef] [PubMed]
- Bugga, P.; Mrksich, M. Dynamic substrates for cell biology. Curr. Opin. Colloid Interface Sci. 2018, 38, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Yeo, W.-S.; Mrksich, M. Electroactive Self-assembled monolayers that permit orthogonal control over the adhesion of cells to patterned substrates. Langmuir 2006, 22, 10816–10820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neufeld, T.; Mittelman, A.S.; Buchner, V.; Rishpon, J. Electrochemical phagemid assay for the specific detection of bacteria using Escherichia coli TG-1 and the M13KO7 phagemid in a model system. Anal. Chem. 2005, 77, 652–657. [Google Scholar] [CrossRef] [PubMed]
- Neufeld, T.; Schwartz-Mittelmann, A.; Biran, D.; Ron, E.Z.; Rishpon, J. Combined phage typing and amperometric detection of released enzymatic activity for the specific identification and quantification of bacteria. Anal. Chem. 2003, 75, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.T.; Lunte, C.E.; Halsall, H.B.; Heineman, W.R. P-Aminophenyl phosphate: An improved substrate for electrochemical enzyme immnoassay. Anal. Chim. Acta 1988, 214, 187–195. [Google Scholar] [CrossRef]
- Amiri, M.; Bezaatpour, A.; Jafari, H.; Boukherroub, R.; Szunerits, S. Electrochemical methodologies for the detection of pathogens. ACS Sens. 2018, 3, 1069–1086. [Google Scholar] [CrossRef]
- Chen, J.; Jackson, A.A.; Rotello, V.M.; Nugen, S.R. Colorimetric detection of Escherichia coli based on the enzyme-induced metallization of gold nanorods. Small 2016, 12, 2469–2475. [Google Scholar] [CrossRef] [Green Version]
- Wick, L.; Mayer, T.; Spitz, U. Non-Hydrolytic Microbial Probes. U.S. Patent WO2,010,049,450A1, 6 May 2010. [Google Scholar]
- Conrad, M.J.; He, L. Fluorogenic Substrates. U.S. Patent 6,949,632B2, 23 September 2002. [Google Scholar]
- Ge, F.-Y.; Chen, L.-G.; Zhou, X.-L.; Pan, H.-Y.; Yan, F.-Y.; Bai, G.-Y.; Yan, X.-L. Synthesis and study on hydrolytic properties of fluorescein esters. Dyes Pigm. 2007, 72, 322–326. [Google Scholar] [CrossRef]
- McKenzie, D.A. Milk testing–a forward look. Int. J. Dairy Technol. 1962, 15, 207–212. [Google Scholar] [CrossRef]
- Chen, J.L.; Steele, T.W.J.; Stuckey, D.C. Metabolic reduction of resazurin; location within the cell for cytotoxicity assays. Biotechnol. Bioeng. 2018, 115, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Krainer, F.W.; Glieder, A. An updated view on horseradish peroxidases: Recombinant production and biotechnological applications. Appl. Microbiol. Biotechnol. 2015, 99, 1611–1625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
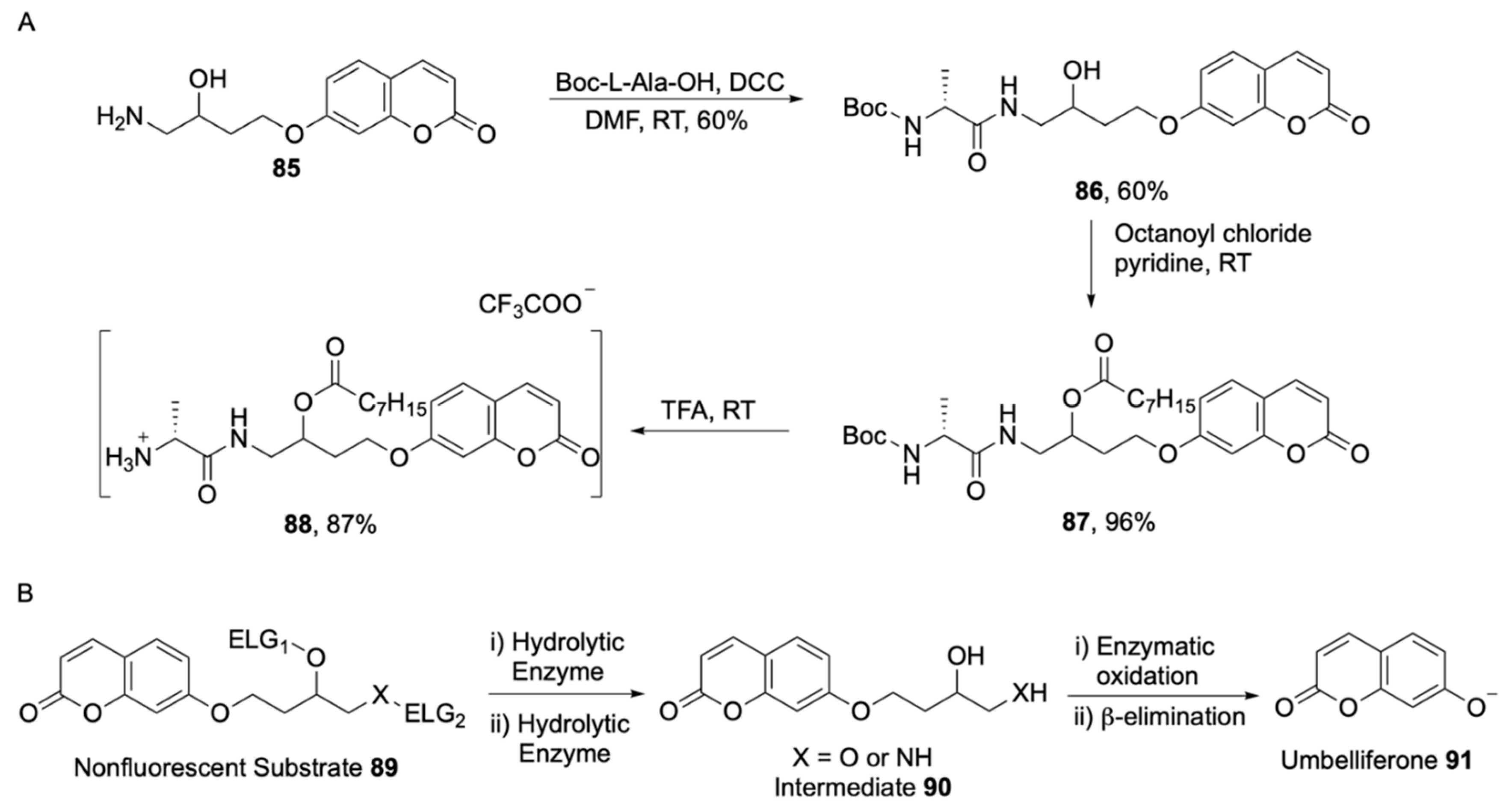
- James, A.L.; Perry, J.D.; Jay, C.; Monget, D.; Rasburn, J.W.; Gould, F.K. Fluorogenic substrates for the detection of microbial nitroreductases. Lett. Appl. Microbiol. 2001, 33, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Guo, L.; Shan, X.; Lin, X.; Gu, T.; Zhang, J.; Ge, J.; Li, W.; Ge, H.; Jiang, Q.; et al. An elegant nitroreductase responsive fluorescent probe for selective detection of pathogenic Listeria in vitro and in vivo. Talanta 2019, 198, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Krämer, C.E.M.; Wiechert, W.; Kohlheyer, D. Artificial fluorogenic substrates in microfluidic devices for bacterial diagnostics in biotechnology. J. Flow Chem. 2016, 6, 3–7. [Google Scholar] [CrossRef] [Green Version]
- Montes-Cebrián, Y.; Álvarez-Carulla, A.; Ruiz-Vega, G.; Colomer-Farrarons, J.; Puig-Vidal, M.; Baldrich, E.; Miribel-Català, P.L. Competitive USB-powered hand-held potentiostat for POC applications: An HRP detection case. Sensors (Basel) 2019, 19, 5388. [Google Scholar] [CrossRef] [Green Version]
- Furst, A.L.; Francis, M.B. Impedance-based detection of bacteria. Chem. Rev. 2019, 119, 700–726. [Google Scholar] [CrossRef]
- Abbasian, F.; Ghafar-Zadeh, E.; Magierowski, S. Microbiological sensing technologies: A review. Bioengineering (Basel) 2018, 5, 20. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.T.; Trinh, K.T.L.; Yoon, W.J.; Lee, N.Y.; Ju, H. Integration of a microfluidic polymerase chain reaction device and surface plasmon resonance fiber sensor into an inline all-in-one platform for pathogenic bacteria detection. Sens. Actua. B Chem. 2017, 242, 1–8. [Google Scholar] [CrossRef]
- Myziuk, L.; Romanowski, B.; Johnson, S.C. BVBlue Test for diagnosis of bacterial vaginosis. J. Clin. Microbiol. 2003, 41, 1925–1928. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Lin, X.; Hui, K.M.; Yang, S.; Wu, X.; Tan, Y.; Li, M.; Qin, A.-Q.; Wang, Q.; Zhao, Q.; et al. A biochemiluminescent sialidase assay for diagnosis of bacterial vaginosis. Sci. Rep. 2019, 9, 20024. [Google Scholar] [CrossRef] [PubMed]
- Romao, V.C.; Martins, S.A.M.; Germano, J.; Cardoso, F.A.; Cardoso, S.; Freitas, P.P. Lab-on-chip devices: Gaining ground losing size. ACS Nano 2017, 11, 10659–10664. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Malode, S.J.; Shetti, N.P.; Chandra, P. Modernization of Biosensing Strategies for the Development of Lab-on-chip Integrated Systems. In Bioelectrochemical Interface Engineering; John Wiley & Sons: Hoboken, NJ, USA, 2019; pp. 325–342. [Google Scholar] [CrossRef]





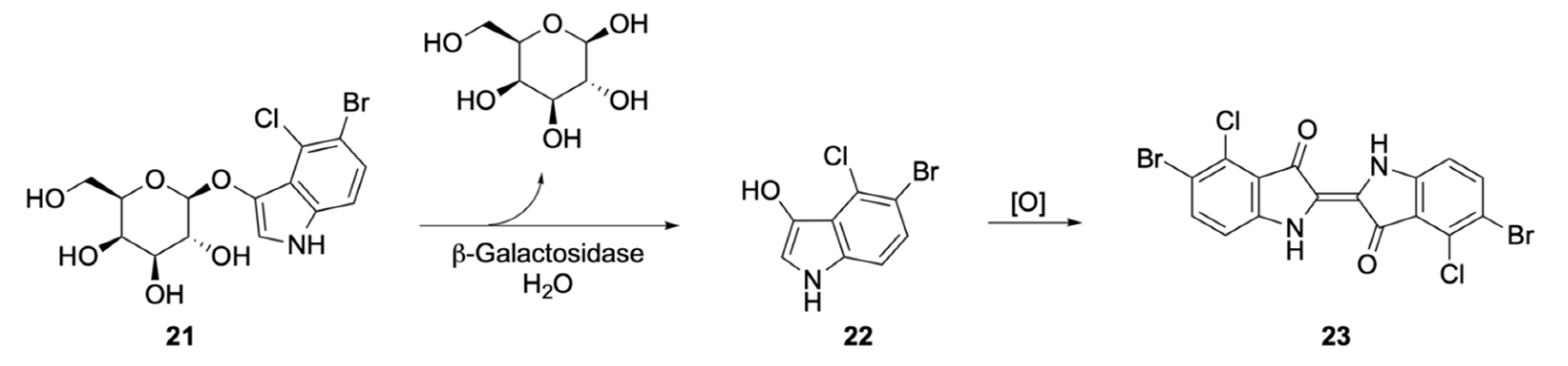



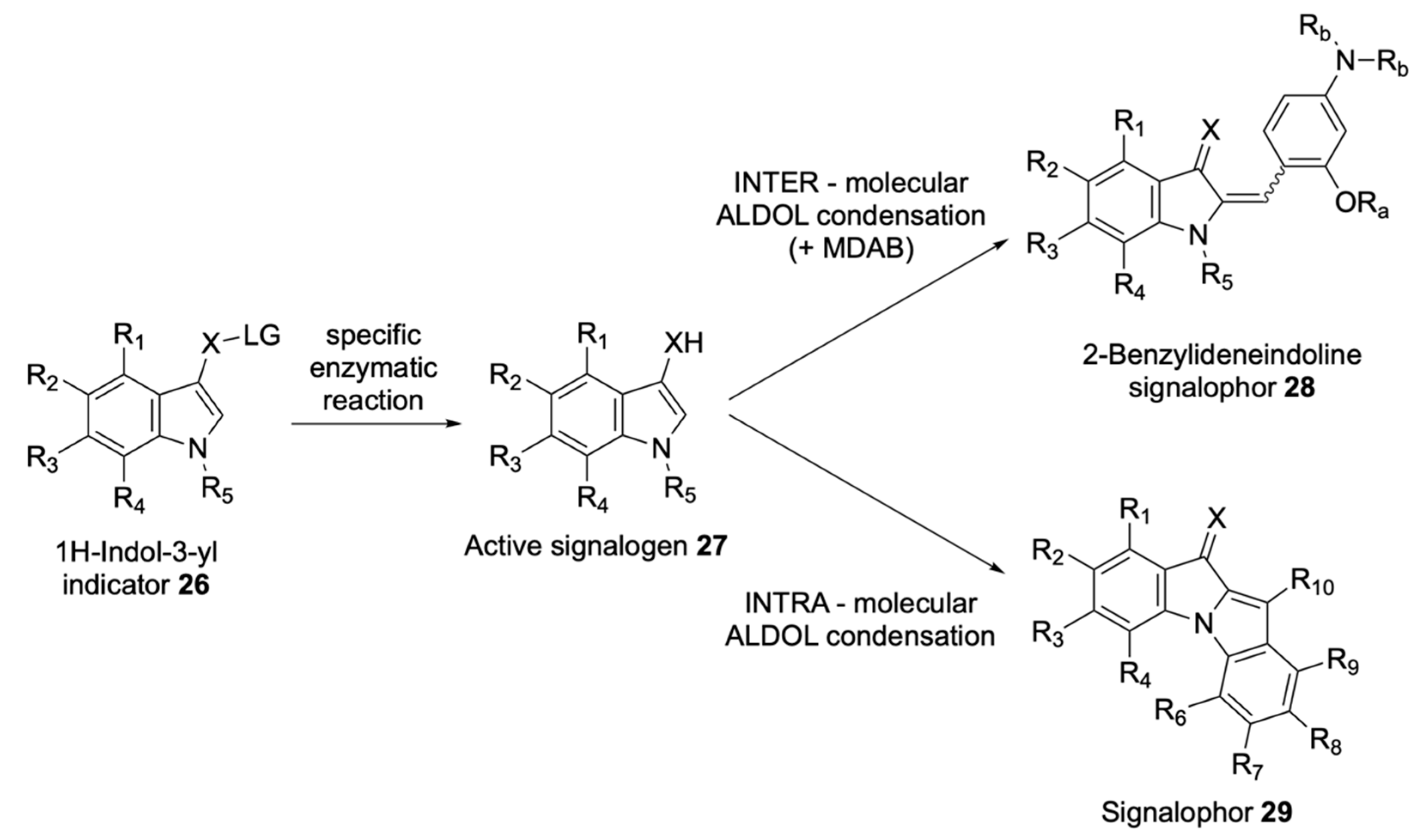



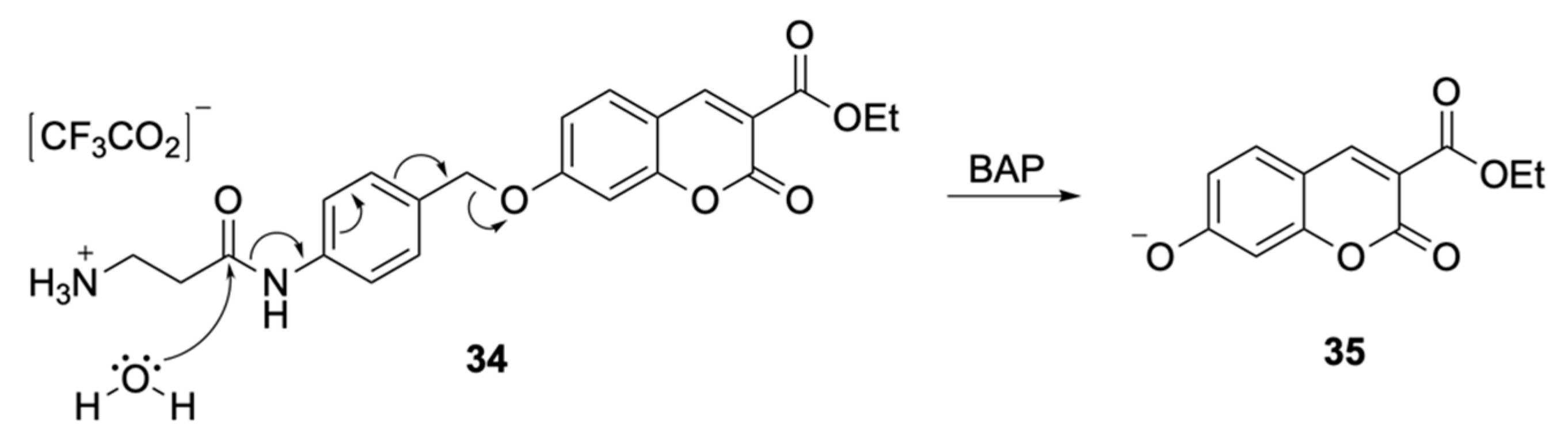

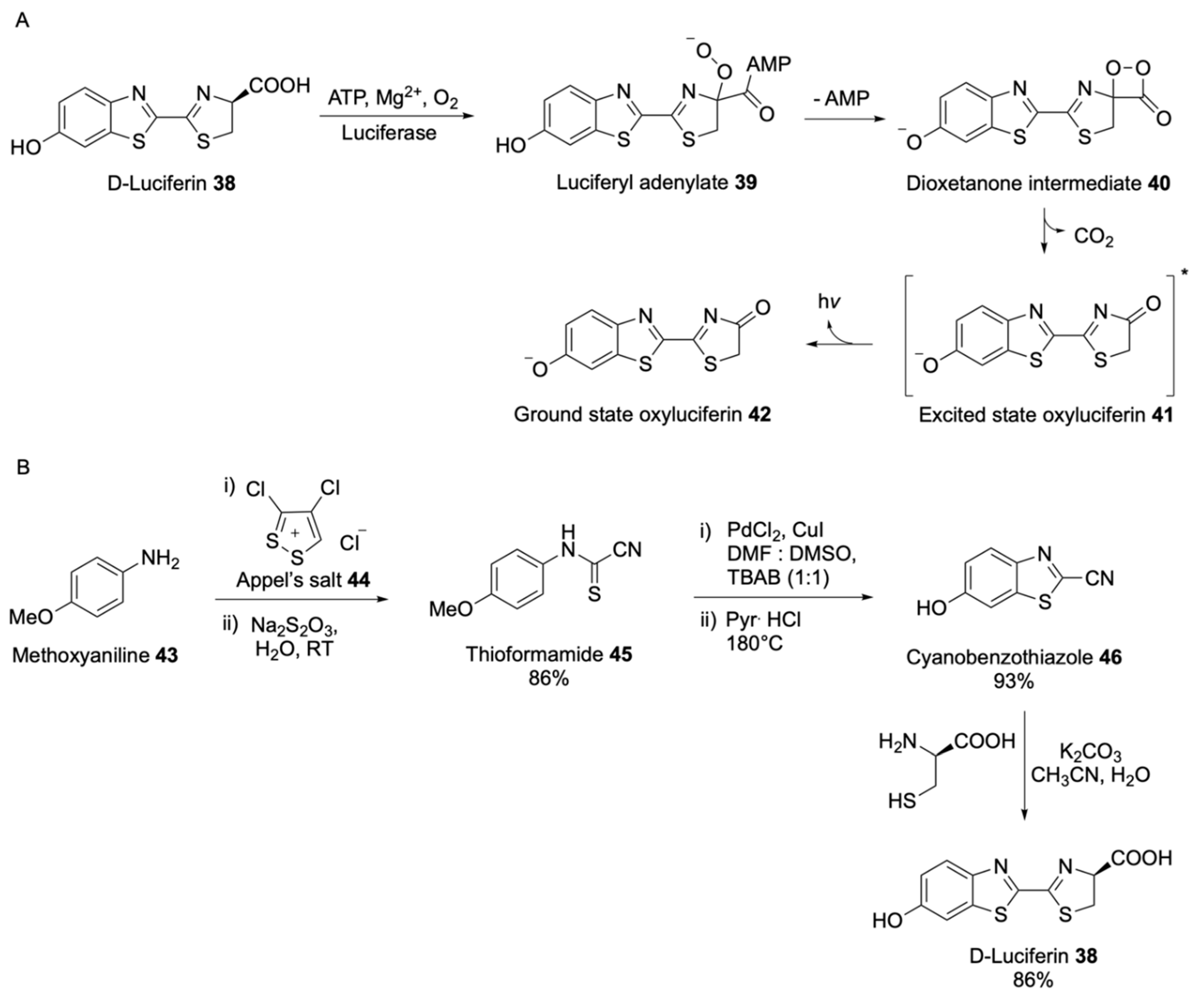





| Signalophor | Key Features | Drawbacks |
|---|---|---|
| Chromogenic and Fluorogenic Substrates | ||
| Nitrophenol | Soluble, chromogenic, yellow | Colour reduction at acidic pH, yellow colour overlapping with growth media |
| 4-Methylumbelliferone (4-MU) | Soluble, fluorogenic, low toxicity, easy hydrolysis | 4-MU esters have limited stability in water, blue emission overlaps with media fluorescence, relatively high pKa of 7.8 |
| 7-Amino-4-methylcoumarin (7-AMC) | Soluble, blue fluorescence, low toxicity, easy hydrolysis | Low fluorescence at low pH, signal declines over time |
| 7-Hydroxycoumarin-3-carboxylate (EHC) | Higher fluorescence at lower pH, lower toxicity compared to 4-MU | - |
| Resorufin | Soluble, sensitive, bright pink, relatively low pKa of 6.0 | Necessary presence of oxygen to avoid bleaching |
| Fluorescein | Soluble, sensitive, bright green fluorescence, biocompatible | Low photostability, hydrophilic |
| Dihydroxynaphthalenes | Insoluble, chromogenic, brown | Require presence of metal ions to form chelates |
| Indoxyl | Intense tuneable colour, insoluble, used in combination for multiple bacterial detection, versatile | Toxic to some bacterial species, limited to aerobic conditions |
| Aldols® | Chromogenic and fluorogenic properties, tuneable colour, insoluble (except one derivative), can be used in pairs for multiple bacterial detection, versatile, esters are stable, some soluble in plastic | - |
| ELF® | Insoluble, fluorogenic, used in anaerobic conditions, no need for auxiliary agents | - |
| Luminogenic Substrates | ||
| Pro-Luciferins-Luciferase | Bioluminogenic, highly sensitive, soluble | Low tissue and cell membrane permeability, low lipophilicity, inhibitory effects from intermediates |
| Luminol | Chemiluminescent, blue, soluble, broad application, low cost, ease of usage | Quenches with reducing agents, interference from metals such as Fe, Cu, Mn, and Co, dependent on signal enhancer, intensity reduction at lower pH, non-specific chemiluminescence as result of non-enzymatic hydrolysis |
| Dioxetanes | Chemiluminescent, stable up to several years at room temperature, highly sensitive | Low cell membrane permeability |
| Electrogenic Substrate | ||
| Indoxyl | Indigo based, colourless | - |
| p-Aminophenol | Effective with phosphatases | - |
| Redox Substrates | ||
| Formazan | Purple, formed by reduction of tetrazoles such as MTT. Used in viability assays | - |
| Resorufin | Bright pink, formed by reduction of resazurin | Possible further reduction forms colourless dihydroresorufin |
| Aldol® | Dye Solubility (Water) | Colour | Fluorescence |
|---|---|---|---|
| 458 | Soluble at pHβ6 | Yellow | Good in aqueous media (green) |
| 470 | Insoluble | Yellow | Medium (yellow) |
| 484 | Insoluble | Orange-red | - |
| 495 | Insoluble | Orange | Good in hydrophobic matrices (green to yellow) |
| 515/518 | Insoluble | Red | Good in hydrophobic matrices (red) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pala, L.; Sirec, T.; Spitz, U. Modified Enzyme Substrates for the Detection of Bacteria: A Review. Molecules 2020, 25, 3690. https://doi.org/10.3390/molecules25163690
Pala L, Sirec T, Spitz U. Modified Enzyme Substrates for the Detection of Bacteria: A Review. Molecules. 2020; 25(16):3690. https://doi.org/10.3390/molecules25163690
Chicago/Turabian StylePala, Laura, Teja Sirec, and Urs Spitz. 2020. "Modified Enzyme Substrates for the Detection of Bacteria: A Review" Molecules 25, no. 16: 3690. https://doi.org/10.3390/molecules25163690
APA StylePala, L., Sirec, T., & Spitz, U. (2020). Modified Enzyme Substrates for the Detection of Bacteria: A Review. Molecules, 25(16), 3690. https://doi.org/10.3390/molecules25163690






