Synthetic Analogues of Aminoadamantane as Influenza Viral Inhibitors—In Vitro, In Silico and QSAR Studies
Abstract
1. Introduction
2. Results and Discussion
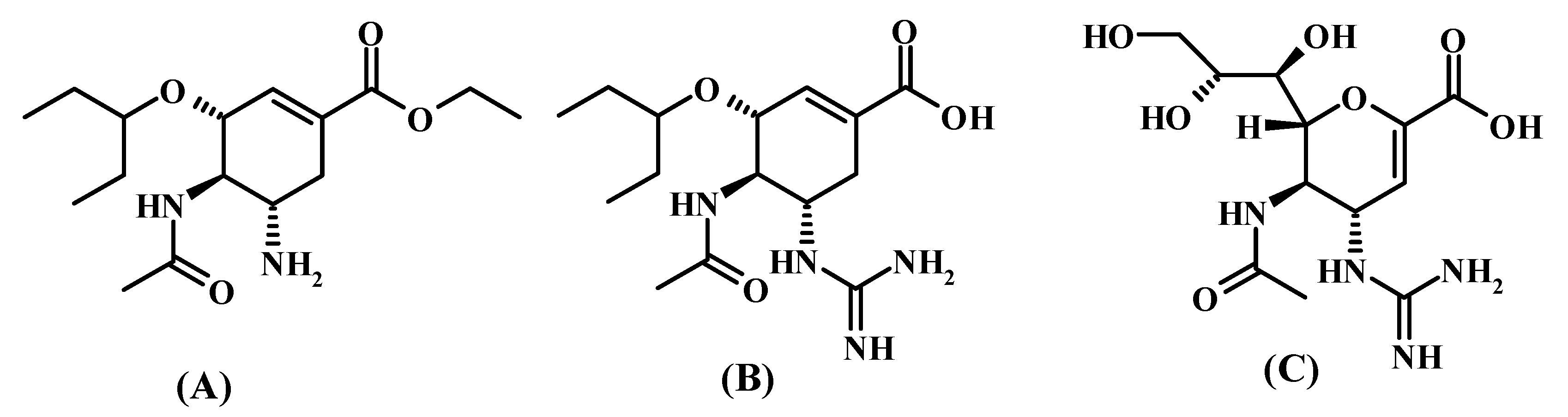
2.1. Synthesis of the Compounds
2.2. Biological Studies
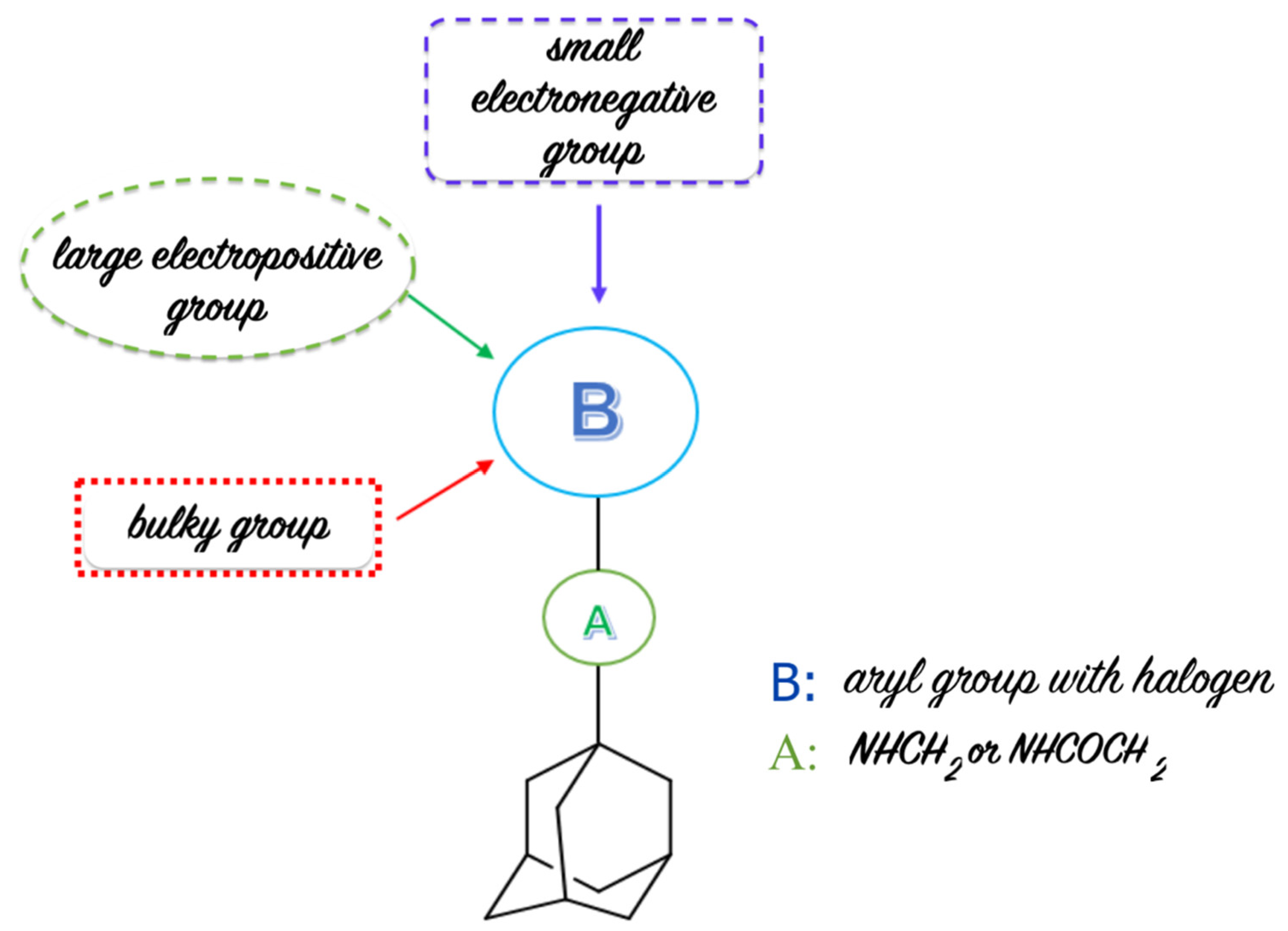
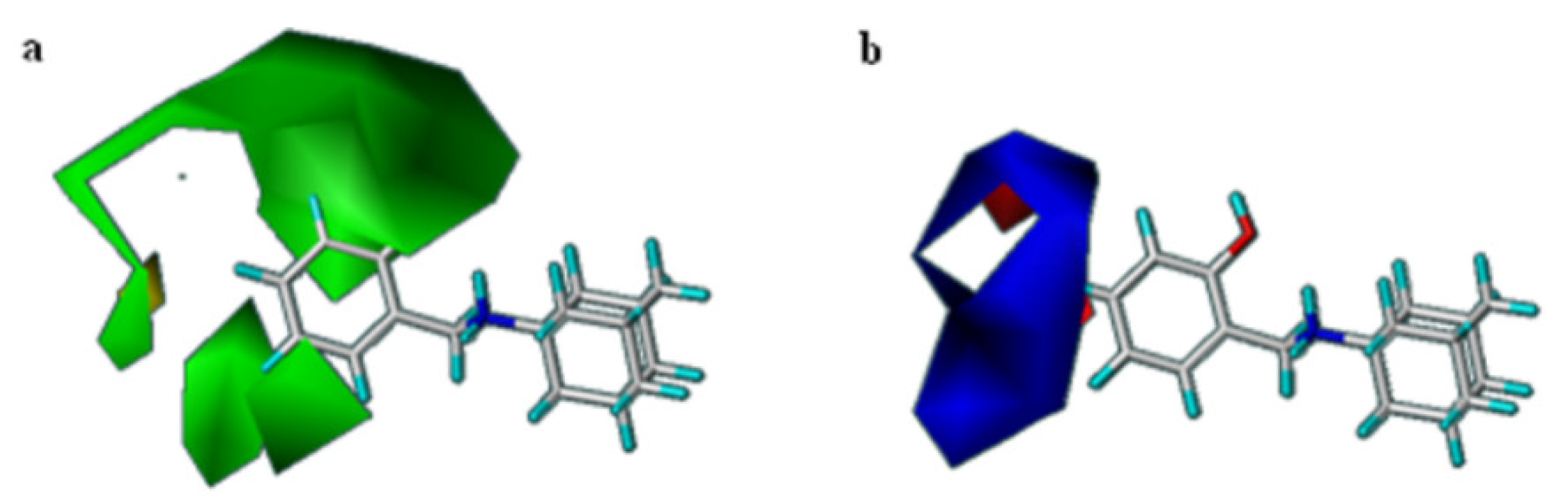
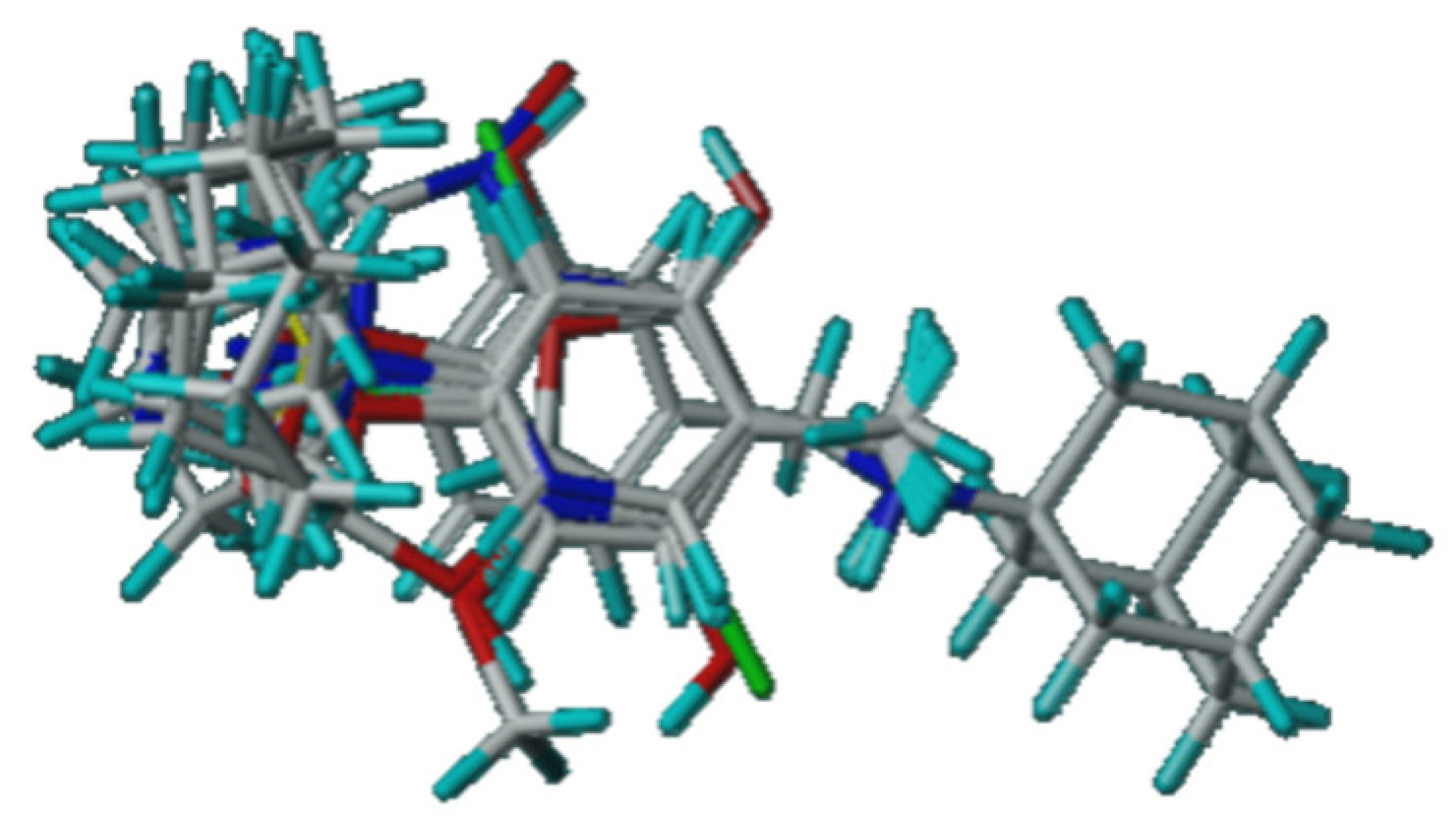
2.3. 3D-QSAR Studies
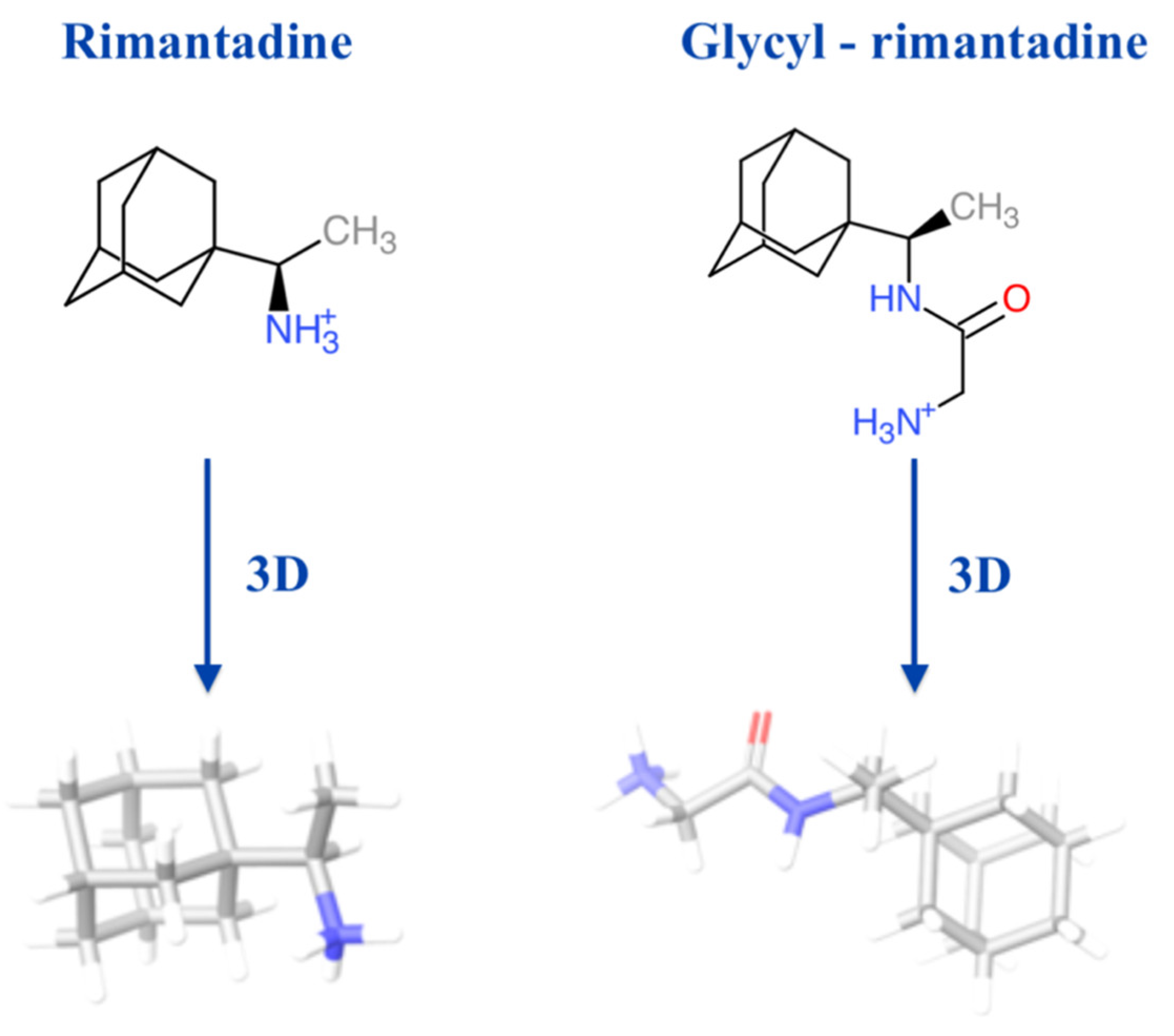
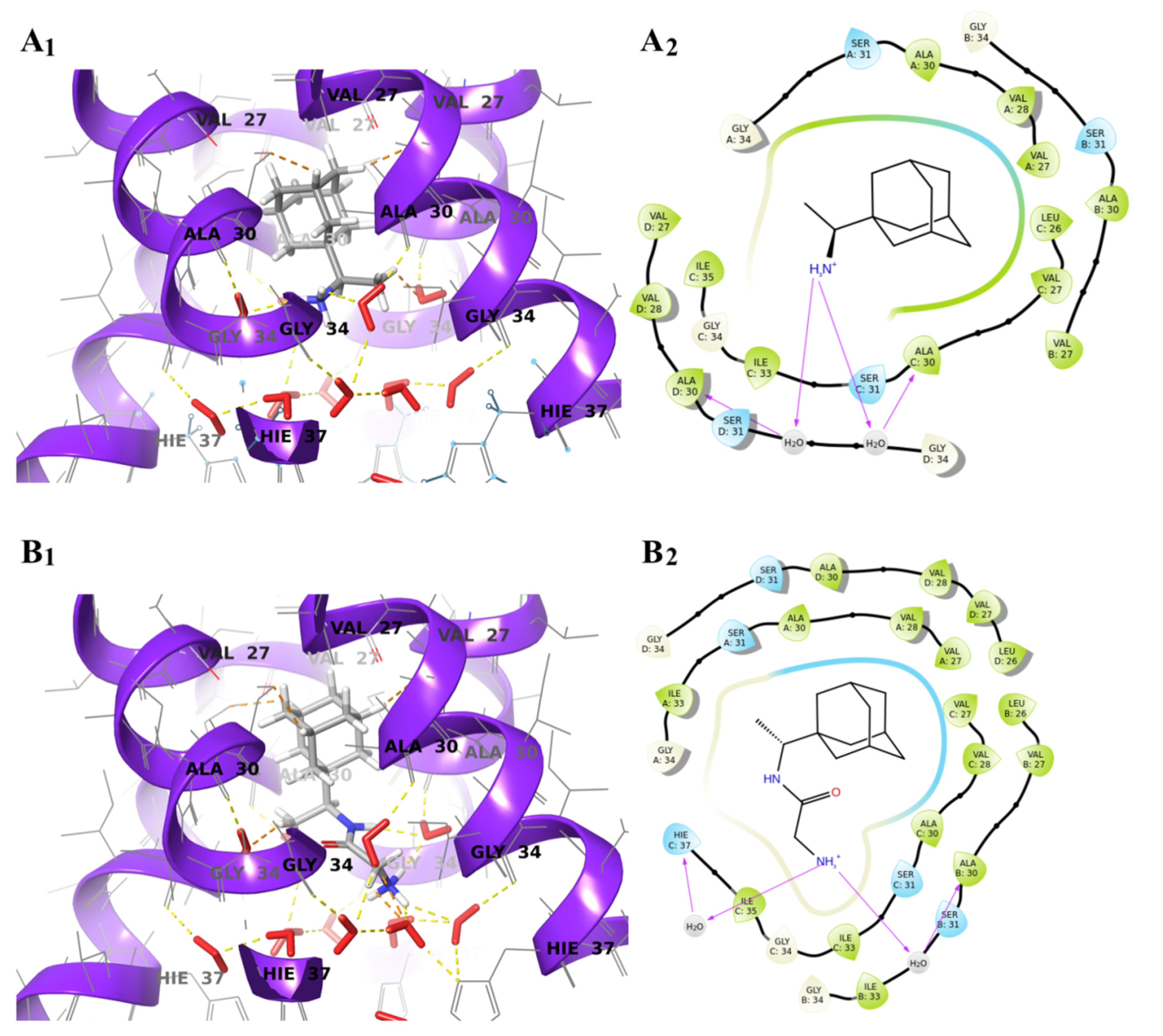
2.4. Molecular Docking Study of the Glycyl-Rimantadine
2.5. In Vitro Human Plasma Stability of Glycyl-Rimantadine (4b)
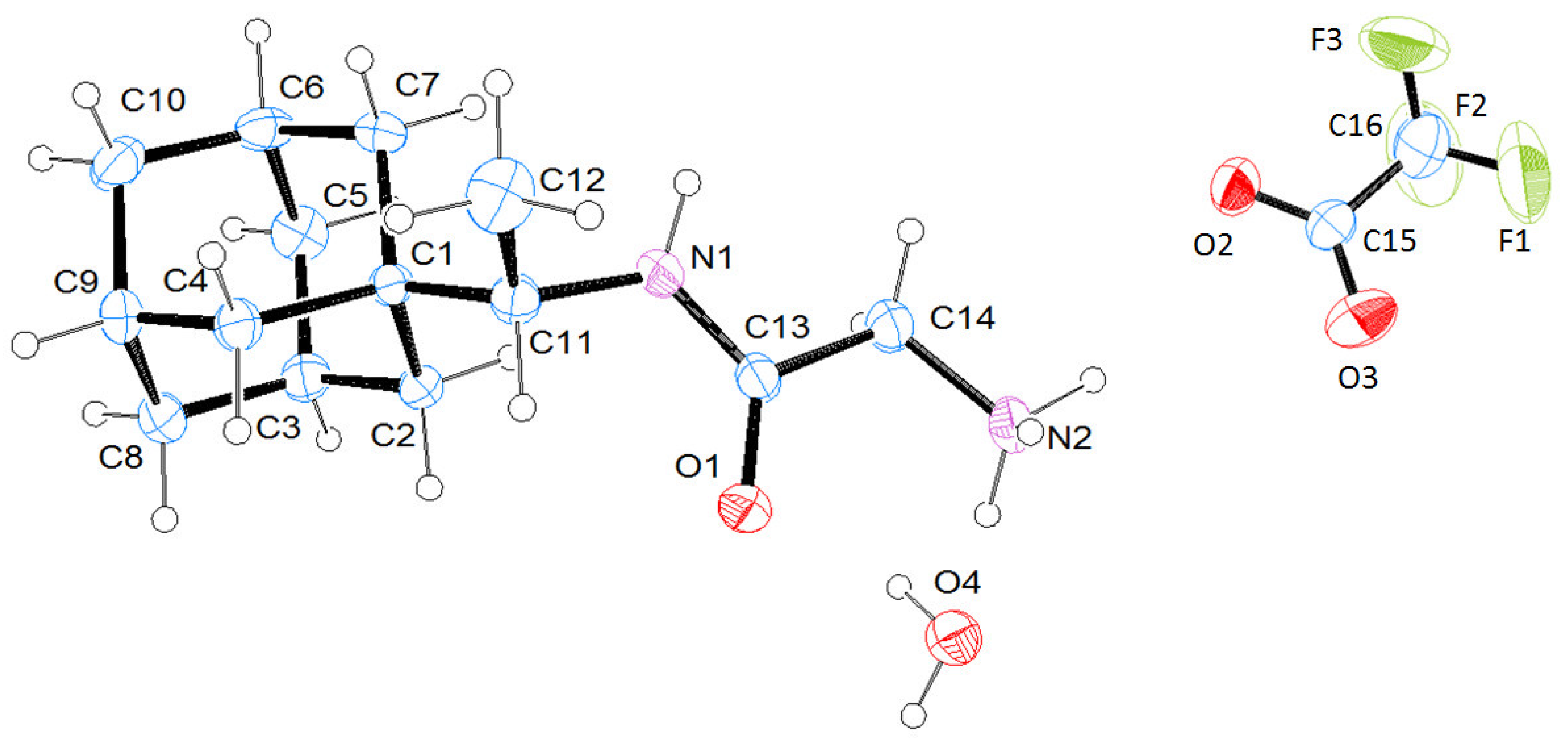
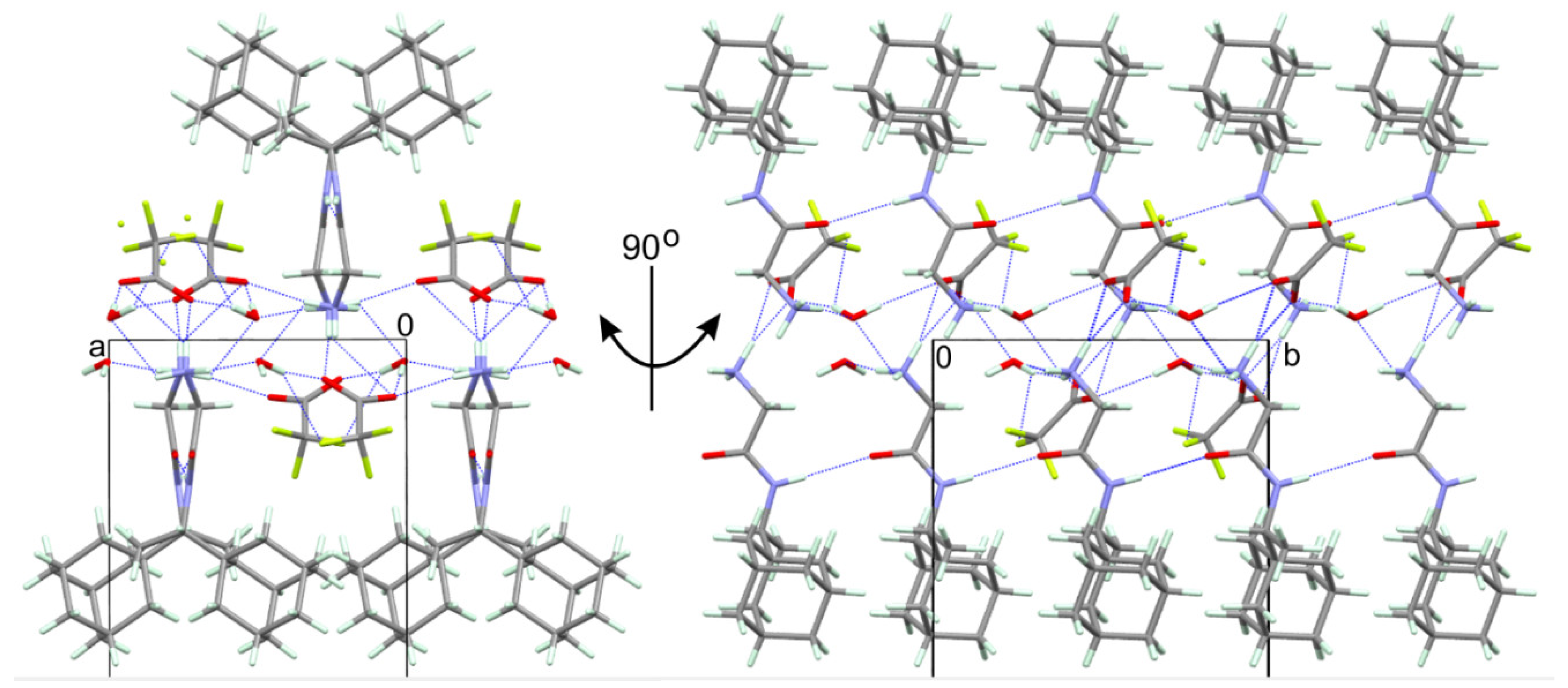
2.6. Elucidation of the Structure of Glycyl-Rimantadine (4b)
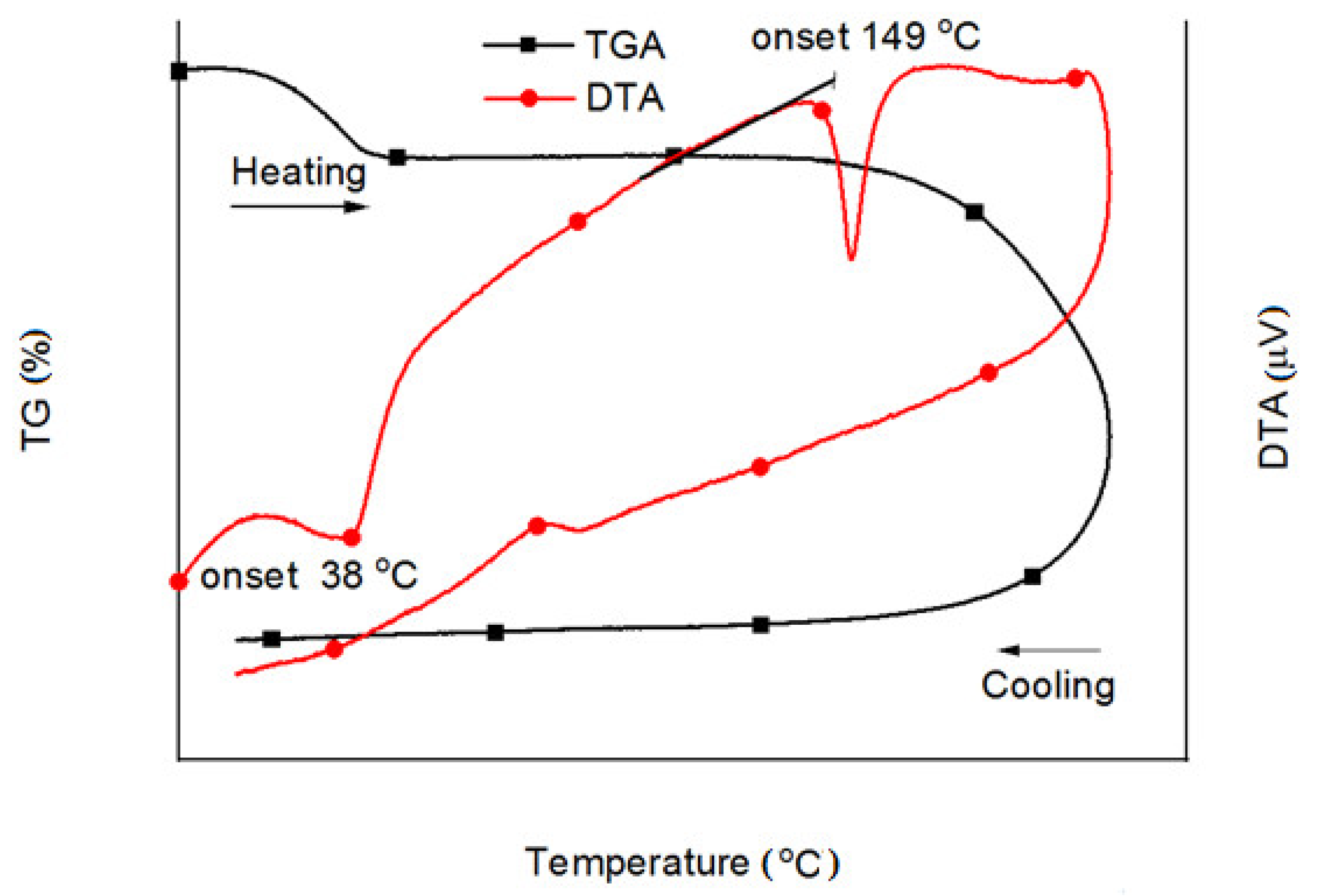
2.7. Thermal Analysis and Stability of Glycyl-Rimantadine (4b)
3. Materials and Methods
3.1. Chemicals and Reagents
3.1.1. General Procedure for Boc-Group Deprotection of the 4(a–j), 5a, 5e and 5g–h
3.1.2. Preparative Method of the 6a, 6i, 6j, 7a and 7h
3.2. Cytotoxicity
3.3. Antiviral Activity
3.4. 3D-QSAR Model Development
3.5. CoMFA Analysis
3.6. Partial Least Square (PLS) Analysis
3.7. 3D-QSAR CoMFA Model Validation
3.8. Molecular Docking of the Glycyl-Rimantadine (4b) in the Binding Site of Rimantadine Bound to the Influenza A M2 Transmembrane Domain
3.9. LC–MS/MS Plasma Stability Assay
3.9.1. LC–MS/MS Conditions
3.9.2. Stock, Working, Calibration Solutions and Extraction Procedure
3.9.3. In Vitro Plasma Stability
3.10. Thermal Analyses
3.11. Crystal Structure Analysis
3.11.1. Sample Crystallization
3.11.2. Data Collection and Crystal Structure Refinement
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Spilovska, K.; Zemek, F.; Korabecny, J.; Nepovimova, E.; Soukup, O.; Windisch, M.; Kuca, K. Adamantane-a lead structure for drugs in clinical practice. Curr. Med. Chem. 2016, 23, 3245–3266. [Google Scholar] [CrossRef] [PubMed]
- Shibnev, V.A.; Garaev, T.M.; Finogenova, M.P.; Shevchenko, E.S.; Burtseva, E.I. New adamantane derivatives can overcome resistance of influenza A(H1N1)pdm2009 and A(H3N2) viruses to remantadine. Bull. Exp. Biol. Med. 2012, 153, 233–235. [Google Scholar] [CrossRef]
- Deryabin, P.G.; Garaev, T.M.; Finogenova, M.P.; Botikov, A.G.; Shibnev, V.A. Amino acid derivatives of adamantane carbocycle are capable of inhibiting replication of highly virulent avian influenza A/H5N1 Virus. Bull. Exp. Biol. Med. 2014, 157, 62–65. [Google Scholar] [CrossRef] [PubMed]
- Shibnev, V.A.; Garaev, T.M.; Deryabin, P.G.; Finogenova, M.P.; Botikov, A.G.; Mishin, D.V. New carbocyclic amino acid derivatives inhibit infection caused by highly pathogenic influenza A virus strain (H5N1). Bull. Exp. Biol. Med. 2016, 161, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Hong, B.T.; Chen, C.L.; Fang, J.M.; Tsai, K.C.; Wang, S.Y.; Huang, W.I.; Cheng, Y.E.; Wong, C.H. Oseltamivir hydroxamate and acyl sulfonamide derivatives as influenza neuraminidase inhibitors. Bioorg. Med. Chem. 2014, 22, 6647–6654. [Google Scholar] [CrossRef]
- Jackson, R.J.; Cooper, K.L.; Tappenden, P.; Rees, A.; Simpson, E.L.; Read, R.C.; Nicholson, K.G. Oseltamivir, zanamivir and amantadine in the prevention of influenza: A systematic review. J. Infect. 2011, 62, 14–25. [Google Scholar] [CrossRef]
- FDA Approves Using Relenza to Prevent Flu, March 29, 2006. Available online: www.cidrap.umn.edu (accessed on 1 September 2020).
- Gaiday, A.V.; Levandovskiy, I.A.; Byler, K.G.; Shubina, T.E. Mechanism of influenza A M2 ion-channel inhibition: A docking and QSAR study. Comput. Sci. ICCS 2008, 5102, 360–368. [Google Scholar] [CrossRef]
- Knorr, R.; Trzeciak, A.; Annwarth, W.; Gillessen, D. New coupling reagents in peptide chemistry. Tetrahedron Lett. 1989, 30, 1927–1930. [Google Scholar] [CrossRef]
- Bernatowicz, M.; Wu, Y.; Matsueda, G. 1H-Pyrazole-1-carboxamidine hydrochloride an attractive reagent for guanylation of amines and its application to peptide synthesis. J. Org. Chem. 1992, 57, 2497–2502. [Google Scholar] [CrossRef]
- Schnell, J.R.; Chou, J.J. Structure and mechanism of the M2 proton channel of influenza A virus. Nature 2008, 451, 591–595. [Google Scholar] [CrossRef]
- Beauchamp, L.M.; Orr, G.F.; Demiranda, P.; Burnette, T.; Krenitsky, T.A. Amino-acid ester prodrugs of acyclovir. Antivir. Chem. Chemother. 1992, 3, 157–164. [Google Scholar] [CrossRef]
- Han, H.; de Vrueh, R.L.; Rhie, J.K.; Covitz, K.M.; Smith, P.L.; Lee, C.P.; Oh, D.M.; Sadee, W.; Amidon, G.L. 5′-Amino acid esters of antiviral nucleosides, acyclovir, and AZT are absorbed by the intestinal PEPT1 peptide transporter. Pharm. Res. 1998, 15, 1154–1159. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Kim, J.S.; Mitchell, S.; Kish, P.; Kijek, P.; Hilfinger, J. 5′-O-D-valyl ara A, a potential prodrug for improving oral bioavailability of the antiviral agent vidarabine. Nucleos. Nucleot. Nucl. Acids 2009, 28, 43–55. [Google Scholar] [CrossRef]
- Kim, C.U.; Lew, W.; Williams, M.A.; Wu, H.; Zhang, L.; Chen, X.; Escarpe, P.A.; Mendel, D.B.; Laver, W.G.; Stevens, R.C. Structure-activity relationship studies of novel carbocyclic influenza neuraminidase inhibitors. J. Med. Chem. 1998, 41, 2451–2460. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Tangallapally, R.; Kim, I.C.; Trovato, R.; Parker, D.; Patton, J.S.; Reeves, L.; Bradford, C.; Wettstein, D.; Baichwal, V.; et al. Discovery of an L-alanine ester prodrug of the Hsp90 inhibitor, MPC-3100. Bioorg. Med. Chem. Lett. 2015, 25, 5254–5257. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.I.; Heider, H.; Schroeder, C. Different modes of inhibition by adamantane amine derivatives and natural polyamines of the functionally reconstituted influenza virus M2 proton channel protein. J. Gen. Virol. 1997, 78, 767–774. [Google Scholar] [CrossRef]
- Leonov, H.; Astrahan, P.; Krugliak, M.; Arkin, I.T. How do aminoadamantanes block the influenza M2 channel, and how does resistance develop? J. Am. Chem. Soc. 2011, 133, 9903–9911. [Google Scholar] [CrossRef]
- Lamb, R.A.; Zebedee, S.L.; Richardson, C.D. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell 1985, 40, 627–633. [Google Scholar] [CrossRef]
- Nain, M.; Hinder, F.; Gong, J.H.; Schmidt, A.; Bender, A.; Sprenger, H.; Gemsa, D. Tumor necrosis factor-alpha production of influenza A virus-infected macrophages and potentiating effect of lipopolysaccharides. J. Immunol. 1990, 145, 1921–1928. [Google Scholar]
- Li, F.; Ma, C.; Hu, Y.; Wang, Y.; Wang, J. Discovery of potent antivirals against amantadine-resistant Influenza A viruses by targeting the M2-S31N proton channel. Acs Infect. Dis. 2016, 2, 726–733. [Google Scholar] [CrossRef]
- Wang, J.; Ma, C.; Wang, J.; Jo, H.; Canturk, B.; Fiorin, G.; Pinto, L.H.; Lamb, R.A.; Klein, M.L.; DeGrado, W.F. Discovery of novel dual inhibitors of the wild-type and the most prevalent drug-resistant mutant, S31N, of the M2 proton channel from influenza A virus. J. Med. Chem. 2013, 56, 2804–2812. [Google Scholar] [CrossRef] [PubMed]
- ChemBioDraw Ultra, version 14.0.; PerkinElmer. 2014. Available online: https://www.perkinelmer.com (accessed on 1 September 2020).
- Maestro, version 10.2.; Schroedinger, LLC: New York, NY, USA, 2015.
- LigPrep, version 3.4.; Schroedinger, LLC: New York, NY, USA, 2015.
- MacroModel, version 10.8.; Schroedinger, LLC: New York, NY, USA, 2015.
- Kaminski, G.A.; Friesner, R.A.; Tirado-Rives, J.; Jorgensen, W.L. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B 2001, 105, 6474–6487. [Google Scholar] [CrossRef]
- Vrontaki, E.; Melagraki, G.; Mavromoustakos, T.; Afantitis, A. Searching for anthranilic acid-based thumb pocket 2 HCV NS5B polymerase inhibitors through a combination of molecular docking, 3D-QSAR and virtual screening. J. Enzym. Inhib. Med. Chem. 2016, 31, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Tripos Associates. SYBYL QSAR and COMFA, version 8.0.; Tripos Associates: St. Louis, MO, USA, 2007. [Google Scholar]
- Gasteiger, J.; Marsili, M. Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 1980, 36, 3219–3228. [Google Scholar] [CrossRef]
- Purcell, P.; Singer, A. A brief review and table of semiempirical parameters used in the Hueckel molecular orbital method. J. Chem. Eng. Data 1967, 12, 235–246. [Google Scholar] [CrossRef]
- Clark, M.; Cramer, R.D.; Vanopdenbosch, N. Validation of the general-purpose Tripos 5.2 force-field. J. Comput. Chem. 1989, 10, 982–1012. [Google Scholar] [CrossRef]
- Cramer, R.D.; Patterson, D.E.; Bunce, J.D. Comparative molecular-field analysis (CoMFA).1. Effect of shape on binding of steroids to carrier proteins. J. Am. Chem. Soc. 1988, 110, 5959–5967. [Google Scholar] [CrossRef]
- Frank, I.; Feikema, J.; Constantine, N.; Kowalski, B. Prediction of product quality from spectral data using the partial least-squares method. J. Chem. Inf. Comput. Sci. 1984, 24, 20–24. [Google Scholar] [CrossRef]
- Rannar, S.; Lindgren, F.; Geladi, P.; Wold, S. A Pls kernel algorithm for data sets with many variables and fewer objects. 1. theory and algorithm. J. Chemometr. 1994, 8, 111–125. [Google Scholar] [CrossRef]
- Cramer, R.D.; Bunce, J.D.; Patterson, D.E.; Frank, I.E. Cross-validation, bootstrapping, and partial least-squares compared with multiple-regression in conventional QSAR studies. Quant. Struct-Act. Rel. 1988, 7, 18–25. [Google Scholar] [CrossRef]
- Hawkins, D.M.; Basak, S.C.; Mills, D. Assessing model fit by cross-validation. J. Chem. Inf. Comput. Sci. 2003, 43, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Tropsha, A. Best practices for QSAR model development, validation, and exploitation. Mol. Inform. 2010, 29, 476–488. [Google Scholar] [CrossRef] [PubMed]
- Melagraki, G.; Afantitis, A. Enalos KNIME nodes: Exploring corrosion inhibition of steel in acidic medium. Chemometr. Intell. Lab. 2013, 123, 9–14. [Google Scholar] [CrossRef]
- Golbraikh, A.; Tropsha, A. Beware of q2! J. Mol. Graph. Model. 2002, 20, 269–276. [Google Scholar] [CrossRef]
- LigPrep, version 2.5.; Schroedinger, LLC: New York, NY, USA, 2012.
- Protein Preparation Wizard, Epik version 2.3.; Schroedinger, LLC: New York, NY, USA, 2012.
- Impact, version 5.8.; Schroedinger, LLC: New York, NY, USA, 2012.
- Prime, version 3.1.; Schroedinger, LLC: New York, NY, USA, 2012.
- CrysAlis PRO version 171.36.20; Agilent Technologies, Ltd.: Yarnton, UK, 2011.
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. A 2008, 64, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Farrugia, L.J. WinGX and ORTEP for Windows: An update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Macrae, C.F.; Bruno, I.J.; Chisholm, J.A.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Rodriguez-Monge, L.; Taylor, R.; van de Streek, J.; Wood, P.A. Mercury CSD 2.0-new features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008, 41, 466–470. [Google Scholar] [CrossRef]
- Spek, A.L. Structure validation in chemical crystallography. Acta Crystallogr. D 2009, 65, 148–155. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available. |




| Compound | CC50 (μM) MDCK Cells | HNTC (μM) MDCK Cells | IC50 (μM) Against A/Hong Kong/68 |
|---|---|---|---|
| Ala-Rim (4a) | >100 | 14.65 | 1.53 |
| Gly-Rim (4b) | >100 | 13.89 | 0.11 |
| Ile-Rim (4c) | 58.35 | 5.33 | n/a |
| Leu-Rim (4d) | >100 | 11.66 | 0.75 |
| Phe-Rim (4e) | 21.93 | 4.75 | n/a |
| (4-F)-D-Phe-Rim (4f) | 19.91 | 6.23 | n/a |
| (4-F)-Phe-Rim (4g) | 15.46 | 4.37 | n/a |
| Val-Rim (4h) | 70.33 | 10.2 | n/a |
| β-Ala-Rim (4i) | >100 | 20.78 | 15.72 |
| Tyr-Rim (4j) | >100 | 9.97 | 0.52 |
| Ala-Am (5a) | >100 | >100 | 1.41 |
| Phe-Am (5e) | 89.4 | 15.1 | 0.75 |
| (4-F)-Phe-Am (5g) | 83.61 | 15.35 | 0.81 |
| Val-Am (5h) | >100 | >100 | 1.32 |
| Gua-Ala-Rim (6a) | >100 | >100 | 41.9 |
| Gua-β-Ala-Rim (6i) | >100 | >100 | n/a |
| Gua-Tyr-Rim (6j) | >100 | 36.69 | 8.19 |
| Gua-Ala-Am (7a) | >100 | >100 | 18.37 |
| Gua-Val-Am (7h) | >100 | >100 | 17.13 |
| Amantadine | >100 | >100 | 0.39 |
| Oseltamivir | >100 | >100 | 0.002 |
| CoMFA Model | |
|---|---|
| ONC | 4 |
| Q2 | 0.847 |
| SEP | 0.127 |
| R2 | 0.954 |
| SEE | 0.070 |
| F value | 123.383 |
| Field contribution | |
| Steric: Electrostatic | 0.694:0.306 |
| R2 test | 0.867 |
| Criterion | Assessment | Result |
|---|---|---|
| R2pred > 0.6 | PASS | R2pred = 0.862 |
| R2test > 0.5 | PASS | R2test = 0.867 |
| (R2 − R20)/R2 < 0.1 | PASS | (R2 − R20)/R2 = 0.015 |
| (R2 − R′20)/R2 < 0.1 | PASS | (R2 – R′20)/R2 = 0.0 |
| |(R2 − R20)|/R2< 0.3 | PASS | |(R2 − R20)|/R2 = 0.013 |
| 0.85 < k < 1.15 | PASS | k = 1.028 |
| 0.85 < k′ < 1.15 | PASS | k′ = 0932 |
| ID | Predicted Values CoMFA p(%) | IC50 (μM) | Active * | Prediction (Active) |
|---|---|---|---|---|
| 1 | 0.820 | 0.39 | Yes | Yes |
| 39(S) | 0.736 | 1.41 | Yes | Yes |
| 40(S) | 0.713 | 18.37 | Yes | Yes |
| 41(S) | 0.745 | 0.75 | Yes | Yes |
| 42(S) | 0.735 | 0.81 | Yes | Yes |
| 43(S) | 0.645 | 1.32 | Yes | Yes |
| 44(S) | 0.668 | 17.13 | Yes | Yes |
| 45(S) | 0.645 | 3.93 | Yes | Yes |
| 46(S) | −0.057 | 59.71 | No | No |
| D - | H… | A | d(D-H) (Å) | d(H…A) (Å) | d(D…A) (Å) | D-H…A (°) |
|---|---|---|---|---|---|---|
| N1 - | H1 | O11 | 0.89 (3) | 2.25 (3) | 3.135 (2) | 173 (2) |
| N2 - | H2A | O2 | 0.98 (3) | 1.78 (3) | 2.744 (2) | 169 (2) |
| N2 - | H2B | O32 | 0.89 (3) | 2.17 (3) | 2.922 (3) | 141 (2) |
| N2 - | H2B | O43 | 0.89 (3) | 2.64 (3) | 3.193 (3) | 121 (2) |
| C14 - | H14A | F254 | 0.95 (3) | 2.59 (3) | 3.285 (5) | 130.5 (2) |
| O4 - | H4C | O25 | 0.95 (4) | 1.88 (4) | 2.811 (3) | 166 (3) |
| O4 - | H4C | F265 | 0.95 (4) | 2.53 (3) | 3.152 (6) | 123 (2) |
| O4 - | H4D | O36 | 0.86 (4) | 2.00 (4) | 2.855 (3) | 175 (4) |
| N2 - | H2C | O4 | 1.00 (3) | 1.87 (4) | 2.847 (3) | 163 (3) |
| C11 - | H11 | O1 | 1.01 (2) | 2.35 (2) | 2.861 (3) | 110.0 (1) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chayrov, R.; Parisis, N.A.; Chatziathanasiadou, M.V.; Vrontaki, E.; Moschovou, K.; Melagraki, G.; Sbirkova-Dimitrova, H.; Shivachev, B.; Schmidtke, M.; Mitrev, Y.; et al. Synthetic Analogues of Aminoadamantane as Influenza Viral Inhibitors—In Vitro, In Silico and QSAR Studies. Molecules 2020, 25, 3989. https://doi.org/10.3390/molecules25173989
Chayrov R, Parisis NA, Chatziathanasiadou MV, Vrontaki E, Moschovou K, Melagraki G, Sbirkova-Dimitrova H, Shivachev B, Schmidtke M, Mitrev Y, et al. Synthetic Analogues of Aminoadamantane as Influenza Viral Inhibitors—In Vitro, In Silico and QSAR Studies. Molecules. 2020; 25(17):3989. https://doi.org/10.3390/molecules25173989
Chicago/Turabian StyleChayrov, Radoslav, Nikolaos A. Parisis, Maria V. Chatziathanasiadou, Eleni Vrontaki, Kalliopi Moschovou, Georgia Melagraki, Hristina Sbirkova-Dimitrova, Boris Shivachev, Michaela Schmidtke, Yavor Mitrev, and et al. 2020. "Synthetic Analogues of Aminoadamantane as Influenza Viral Inhibitors—In Vitro, In Silico and QSAR Studies" Molecules 25, no. 17: 3989. https://doi.org/10.3390/molecules25173989
APA StyleChayrov, R., Parisis, N. A., Chatziathanasiadou, M. V., Vrontaki, E., Moschovou, K., Melagraki, G., Sbirkova-Dimitrova, H., Shivachev, B., Schmidtke, M., Mitrev, Y., Sticha, M., Mavromoustakos, T., Tzakos, A. G., & Stankova, I. (2020). Synthetic Analogues of Aminoadamantane as Influenza Viral Inhibitors—In Vitro, In Silico and QSAR Studies. Molecules, 25(17), 3989. https://doi.org/10.3390/molecules25173989








