Structure-Based Virtual Screening, Molecular Dynamics and Binding Free Energy Calculations of Hit Candidates as ALK-5 Inhibitors
Abstract
1. Introduction
2. Methodology
2.1. Molecular Docking
2.2. Structure-Based Virtual Screening
2.3. Prediction of Pharmacokinetic (PK) Parameters
2.4. Molecular Dynamics (MD) Simulations
2.5. Calculations of Binding Free Energy
3. Results and Discussion
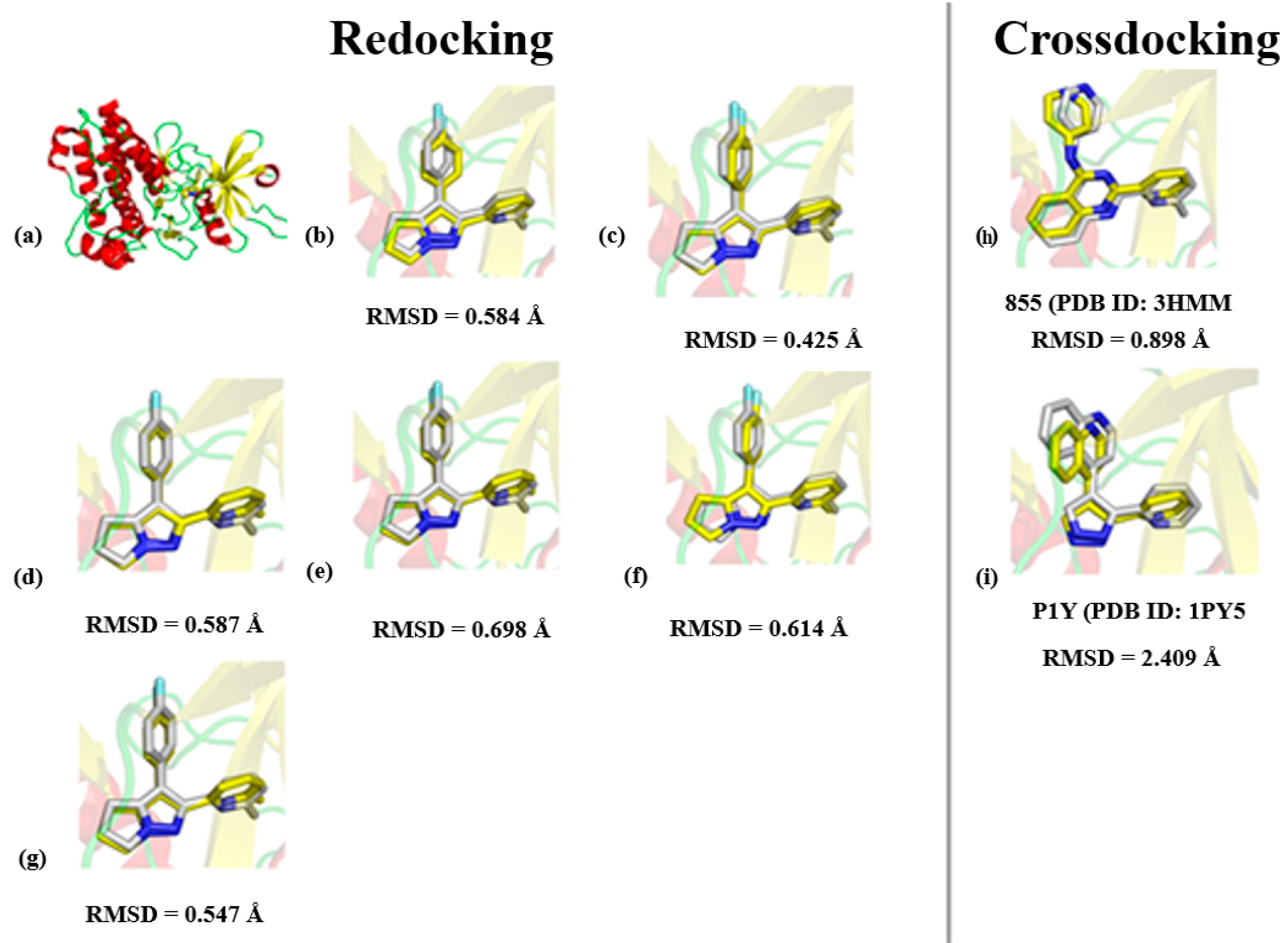
3.1. Molecular Docking
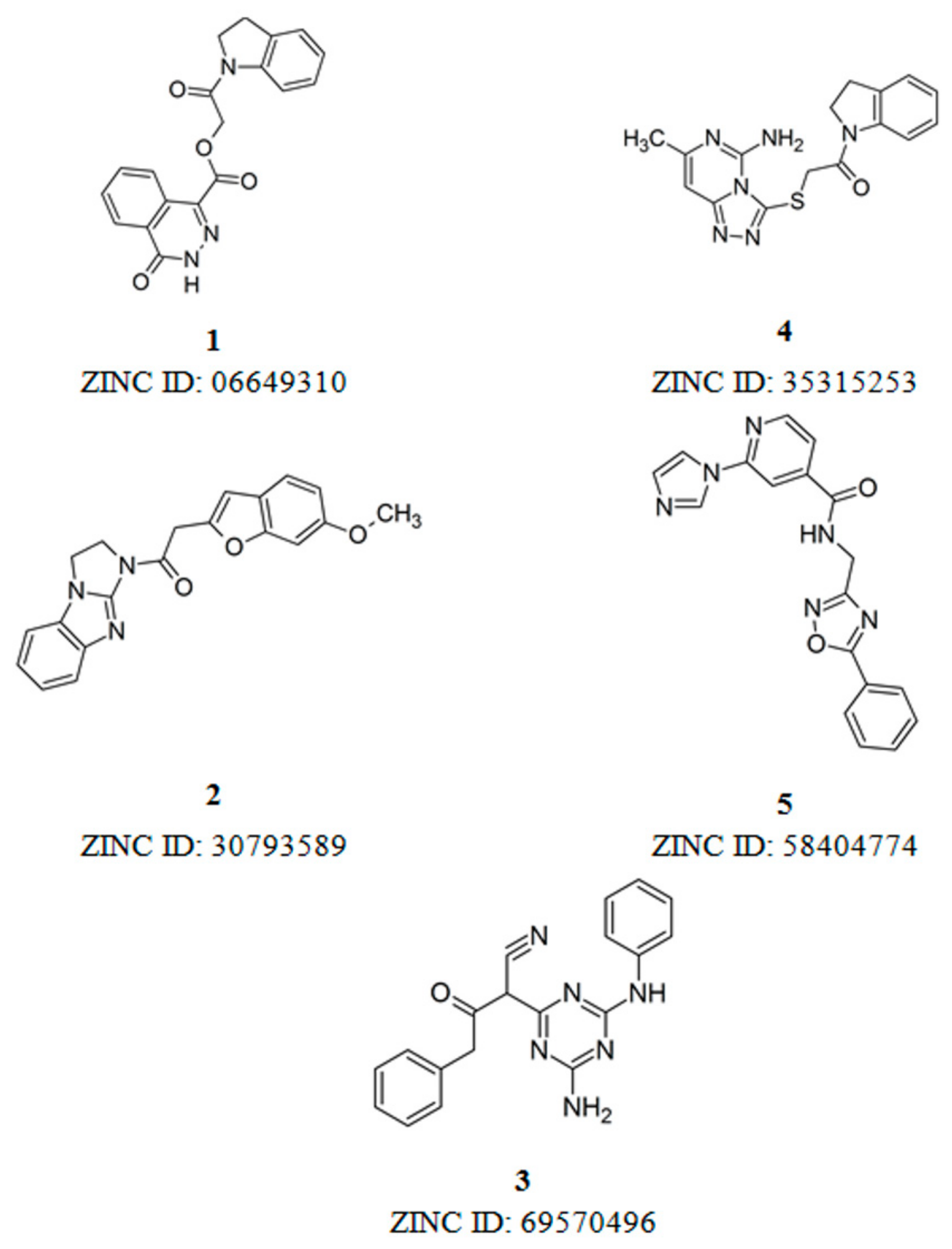
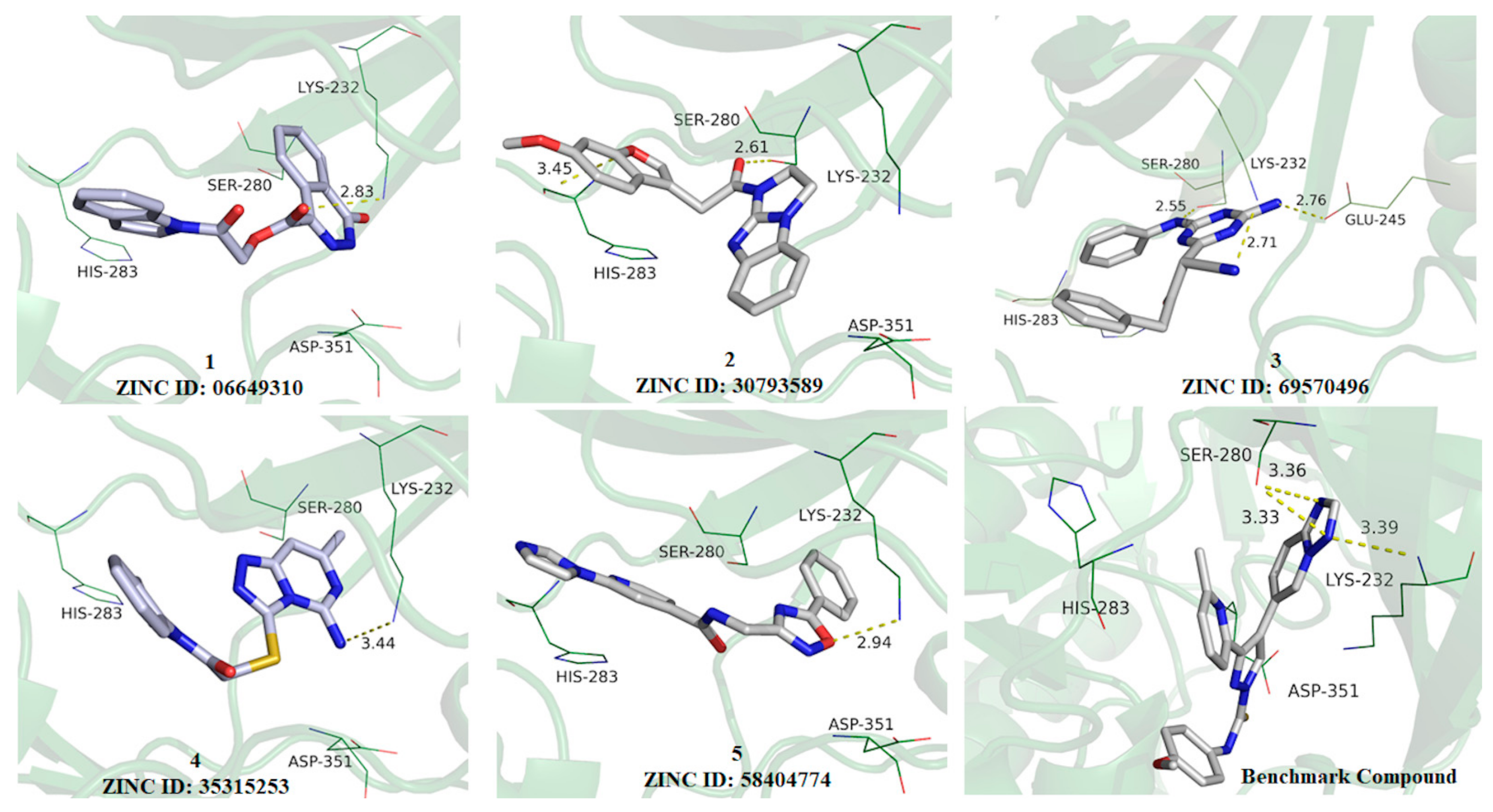
3.2. Structure-Based Virtual Screening and Binding Analysis of the Selected Hits
3.3. ADME-Tox Profile
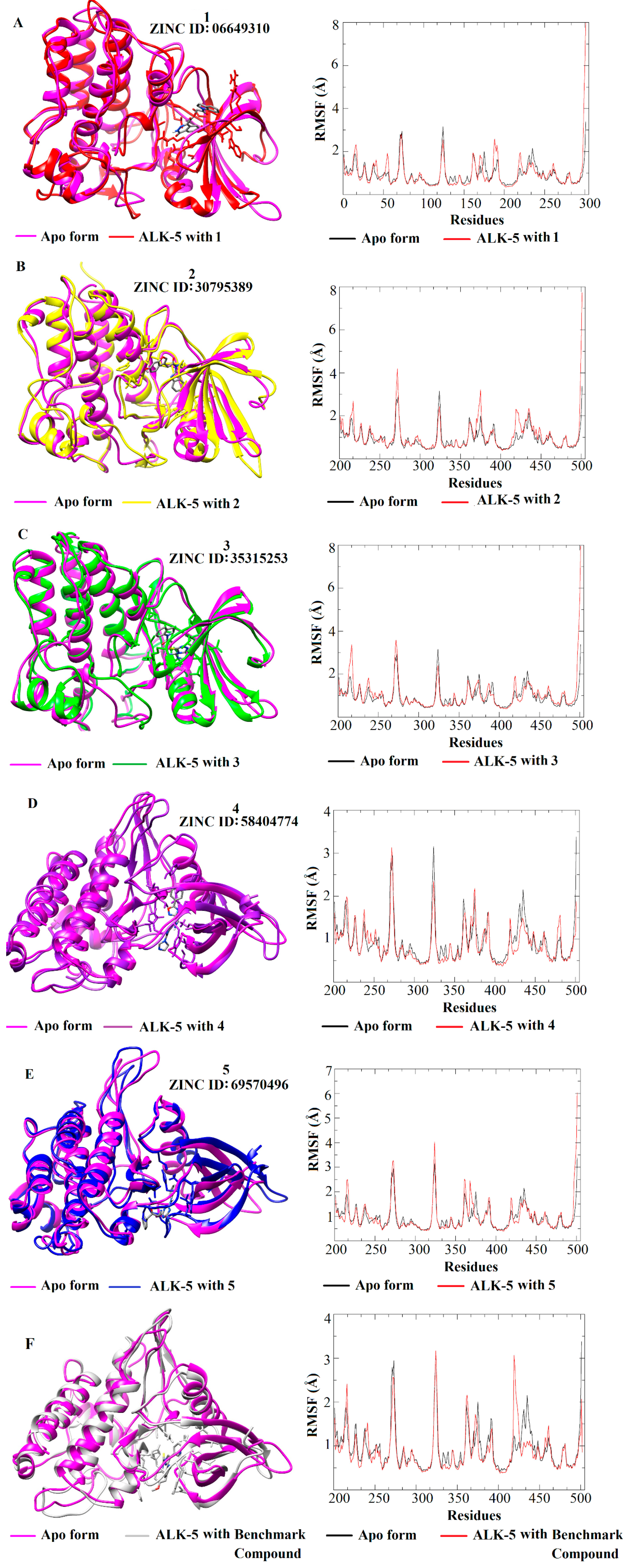
3.4. MD Simulations and Analysis of Binding Mode
3.5. Binding Free Energy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bonafoux, D.; Chuaqui, C.; Boriack-Sjodin, P.A.; Fitch, C.; Hankins, G.; Josiah, S.; Black, C.; Hetu, G.; Ling, L.; Lee, W.C. 2-Aminoimidazoles inhibitors of TGF-β receptor 1. Bioorg. Med. Chem. Lett. 2009, 19, 912–916. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, S.E.; Fiore, A.P.; Siguematu, S.M.; Ebina, K.N.; Friguglietti, C.U.; Ferro, M.C.; Kulcsar, M.A. Kimura ET. Expression of SMAD proteins, TGF-beta/activin signaling mediators, in human thyroid tissues. Arq. Bras. Endocrinol. Metab. 2010, 54, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Neil, J.R.; Schiemann, W.P. Transforming growth factor-β and the hallmarks of cancer. Cell Signal. 2011, 23, 951–962. [Google Scholar] [CrossRef] [PubMed]
- Connolly, E.C.; Freimuth, J.; Akhurst, R.J. Complexities of TGF-beta Targeted Cancer Therapy. Int. J. Biol. Sci. 2012, 8, 964–978. [Google Scholar] [CrossRef] [PubMed]
- Patel, H.M.; Sing, B.; Bhardwaj, V.; Palkar, M.; Shaikh, M.; Rane, R.; Alwan, W.; Gadad, A.K.; Noolvi, M.N.; Karpoormath, R. Design, synthesis and evaluation of small molecule imidazo 2,1-b 1,3,4 thiadiazoles as inhibitors of transforming growth factor-beta type-I receptor kinase (ALK5). Eur. J. Med. Chem. 2015, 93, 599–613. [Google Scholar] [CrossRef] [PubMed]
- Massague, J. TGF-beta signal transduction. Annu. Rev. Biochem. 1998, 67, 753–791. [Google Scholar] [CrossRef] [PubMed]
- Massague, J.; Blain, S.W.; Lo, R.S. TGF beta signaling in growth control, cancer, and heritable disorders. Cell 2000, 103, 295–309. [Google Scholar] [CrossRef]
- Boros, L.G.; Cascante, M.; Lee, W.N. Metabolic profiling of cell growth and death in cancer: Applications in drug discovery. Drug Discov. Today 2002, 7, 364–372. [Google Scholar] [CrossRef]
- Harradine, K.A.; Akhurst, R.J. Mutations of TGF beta signaling molecules in human disease. Ann. Med. 2006, 38, 403–414. [Google Scholar] [CrossRef]
- Cui, X.L.; Shang, S.; Lv, X.; Zhao, J.; Qi, Y.; Liu, Z. Perspectives of small molecule inhibitors of activin receptor-like kinase in anti-tumor treatment and stem cell differentiation. Mol. Med. Rep. 2019, 19, 5053–5062. [Google Scholar] [CrossRef]
- Boldbaatar, A.; Lee, S.; Han, S.; Jeong, A.L.; Ka, H.I.; Buyanravjikh, S.; Lee, J.H.; Lim, J.-S.; Lee, M.S.; Yang, Y. Eupatolide inhibits the TGF-beta 1-induced migration of breast cancer cells via downregulation of SMAD3 phosphorylation and transcriptional repression of ALK5. Oncol. Lett. 2017, 14, 6031–6039. [Google Scholar] [PubMed]
- Chang, H.-H.; Chang, M.C.; Wu, I.H.; Huang, G.F.; Huang, W.L.; Wang, Y.L.; Lee, S.Y.; Yeh, C.Y.; Guo, M.K.; Chan, C.P.; et al. Role of ALK5/Smad2/3 and MEK1/ERK Signaling in Transforming Growth Factor Beta 1-modulated Growth, Collagen Turnover, and Differentiation of Stem Cells from Apical Papilla of Human Tooth. J. Endodont. 2015, 41, 1272–1280. [Google Scholar] [CrossRef] [PubMed]
- Araujo, S.C.; Maltarollo, V.G.; Honorio, K.M. Computational studies of TGF-βRI (ALK-5) inhibitors: Analysis of the 4 binding interactions between ligand–receptor using 2D and 3D 5 techniques. Eur. J. Pharm. Sci. 2013, 49, 542–549. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, F.W.; Ward, R.A.; Powell, S.J.; Debreczeni, J.É.; Norman, R.A.; Roberts, N.J.; Dishington, A.P.; Gingell, H.J.; Wickson, K.F.; Roberts, A.L. Rapid Generation of a High Quality Lead for Transforming Growth Factor-β (TGF-β) Type I Receptor (ALK5). J. Med. Chem. 2009, 52, 7901–7905. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.; Blobe, G.C. Role of transforming growth factor-beta in hematologic malignancies. Blood 2006, 107, 4589–4596. [Google Scholar] [CrossRef]
- Byfield, S.D.; Major, C.; Laping, N.J.; Roberts, A.B. SB-505124 is a selective inhibitor of transforming growth factor-beta type I receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 2004, 65, 744–752. [Google Scholar] [CrossRef]
- De Gouville, A.-C.; Boullay, V.; Krysa, G.; Pilot, J.; Brusq, J.-M.; Loriolle, F.; Gauthier, J.-M.; Papworth, S.A.; Laroze, A.; Gellibert, F.; et al. Inhibition of TGF-beta signaling by an ALK5 inhibitor protects rats from dimethylnitrosamine-induced liver fibrosis. Br. J. Pharmacol. 2005, 145, 166–177. [Google Scholar] [CrossRef]
- Sun, Y.; Ye, P.; Wu, J.; Liu, Z.; Zhang, A.; Ren, L.; Cheng, C.; Huang, X.; Wang, K.; Deng, P.; et al. Inhibition of intimal hyperplasia in murine aortic allografts by the oral administration of the transforming growth factor-beta receptor I kinase inhibitor SD-208. J. Heart Lung Transplant. 2014, 33, 654–661. [Google Scholar] [CrossRef]
- Bueno, L.; de Alwis, D.P.; Pitou, C.; Yingling, J.; Lahn, M.; Glatt, S.; Trocóniz, I.F. Semi-mechanistic modelling of the tumour growth inhibitory effects of LY2157299, a new type I receptor TGF-beta kinase antagonist, in mice. Eur. J. Cancer 2008, 44, 142–150. [Google Scholar] [CrossRef]
- Lin, A.B.; Cai, Z.; Hu, G.; Li, Q. Identification of ALK5 inhibitor via structure-based virtual screening and ADMET prediction. J. Recept. Signal. Transduct. Res. 2015, 35, 559–564. [Google Scholar] [CrossRef]
- Wang, H.; Sessions, R.B.; Prime, S.S.; Shoemark, D.K.; Allen, S.J.; Hong, W.; Narayanan, S.; Paterson, I.C. Identification of novel small molecule TGF-beta antagonists using structure-based drug design. J. Comput. Aided Mol. Des. 2013, 27, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Bierie, B.; Moses, H.L. ; Moses, H.L. TGF-β and cancer. Cytokine Growth F R 2006, 17, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Ciayadi, R.; Potdar, M.; Walton, K.L.; Harrison, C.A.; Kelso, G.F.; Harris, S.J.; Hearn, M.T. 2-Phenyl and 2-heterocyclic-4-(3-(pyridin-2-yl)-1H-pyrazol-4-yl)pyridines as inhibitors of TGF-beta 1 and activin A signalling. Bioorg. Med. Chem. Lett. 2011, 21, 5642–5645. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.S.; Beight, D.W.; Britt, K.S.; Anderson, B.D.; Campbell, R.M.; Goodson, T., Jr.; Herron, D.K.; Li, H.Y.; McMillen, W.T.; Mort, N.; et al. Synthesis and activity of new aryl- and heteroaryl-substituted 5,6-dihydro-4H-pyrrolo[1,2-b]pyrazole inhibitors of the transforming growth factor-β type I receptor kinase domain. Bioorg. Med. Chem. Lett. 2004, 14, 3581–3584. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Sterling, T.; Mysinger, M.M.; Erin, S.; Bolstad, E.S.; Coleman, R.G. ZINC: A Free Tool to Discover Chemistry for Biology. J. Chem. Inf. Model. 2012, 52, 1757–1768. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.H.; Krishnaiah, M.; Sreenu, D.; Subrahmanyam, V.B.; Rao, K.S.; Mohan, A.V.; Park, C.Y.; Son, J.Y.; Sheen, Y.Y.; Kim, D.K. Synthesis and biological evaluation of 1-substituted-3-(6-methylpyridin-2-yl)-4-( 1,2,4 triazolo 1,5-a pyridin-6-yl)pyrazoles as transforming growth factor-beta type 1 receptor kinase inhibitors. Bioorg. Med. Chem. Lett. 2011, 21, 6049–6053. [Google Scholar] [CrossRef]
- Warren, G.L.; Do, T.D.; Kelley, B.P.; Nicholls, A.; Warren, S.D. Essential considerations for using protein-ligand structures in drug discovery. Drug Discov. Today 2012, 17, 1270–1281. [Google Scholar] [CrossRef]
- Krause, L.; Niepötter, B.; Schürmann, C.J.; Stalkea, D.; Herbst-Irmera, R. Validation of experimental charge-density refinement strategies: When do we overfit? Iucrj 2017, 4, 420–430. [Google Scholar] [CrossRef]
- Kleywegt, G.J.; Jones, T.A. Model building and refinement practice. Methods Enzymol. 1997, 277, 208–230. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Makino, S.; Kuntz, I.D. Automated flexible ligand docking method and its application for database search. J. Comput. Chem. 1997, 18, 1812–1825. [Google Scholar] [CrossRef]
- Teramoto, R.; Fukunishi, H. Supervised consensus scoring for docking and virtual screening. J. Chem. Inf. Model. 2007, 47, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Meng, E.C.; Shoichet, B.K.; Kuntz, I.D. Automated docking with grid-based energy evaluation. J. Comput. Chem. 1992, 13, 505–524. [Google Scholar] [CrossRef]
- Kuntz, I.D.; Blaney, J.M.; Oatley, S.J.; Langridge, R.; Ferrin, T.E. A geometric approach to macromolecule-ligand interactions. J. Mol. Biol. 1982, 161, 269–288. [Google Scholar] [CrossRef]
- De Kroon, L.M.G.; Narcisi, R.; Davidson, E.N.B.; Cleary, M.A.; van Beuningen, H.M.; Koevoet, W.J.L.M.; van Osch, G.J.V.M.; van der Kraan, P.M. Activin Receptor-Like Kinase Receptors ALK5 and ALK1 Are Both Required for TGF beta-Induced Chondrogenic Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells. PLoS ONE 2015, 10, 1–18. [Google Scholar] [CrossRef]
- Gellibert, F.; Woolven, J.; Fouchet, M.H.; Mathews, N.; Goodland, H.; Lovegrove, V.; Laroze, A.; Nguyen, V.L.; Sautet, S.; Wang, R.; et al. Identification of 1, 5-naphthyridine derivatives as a novel series of potent and selective TGF-β type I receptor inhibitors. J. Med. Chem. 2004, 47, 4494–4506. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, J.; Hu, C.Q.; Zhang, X.; Ma, B.; Zhang, P. In silico ADME and Toxicity Prediction of Ceftazidime and Its Impurities. Front. Pharmacol. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. PkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Case, D.; Cerutti, D.; Cheatham, T.; Darden, T.; Duke, R.; Giese, T.; Gohlke, H.; Goetz, A.; Greene, D.; Homeyer, N.; et al. AMBER 2018; University of California: San Francisco, CA, USA, 2018. [Google Scholar]
- Wang, J.M.; Cieplak, P.; Kollman, P.A. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000, 21, 1049–1074. [Google Scholar] [CrossRef]
- Ditchfield, R.; Hehre, W.J.; Pople, J.A. Self-consistent molecular-orbital methods. 9. An extended gaussian-type basis for molecular-orbital studies of organic molecules. J. Chem. Phys. 1971, 54, 724–728. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R. Gaussian 09; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [PubMed]
- Naim, M.; Bhat, S.; Rankin, K.N.; Dennis, S.; Chowdhury, S.F.; Siddiqi, I.; Drabik, P.; Sulea, T.; Bayly, C.I.; Jakalian, A.; et al. Solvated interaction energy (SIE) for scoring protein-ligand binding affinities. 1. Exploring the parameter space. J. Chem. Inf. Model. 2007, 47, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Sulea, T.; Cui, Q.; Purisima, E.O. Solvated Interaction Energy (SIE) for Scoring Protein-Ligand Binding Affinities. 2. Benchmark in the CSAR-2010 Scoring Exercise. J. Chem. Inf. Model. 2011, 51, 2066–2081. [Google Scholar] [CrossRef] [PubMed]
- Lill, M.A.; Thompson, J.J. Solvent Interaction Energy Calculations on Molecular Dynamics Trajectories: Increasing the Efficiency Using Systematic Frame Selection. J. Chem. Inf. Model. 2011, 51, 2680–2689. [Google Scholar] [CrossRef] [PubMed]
- Kuntz, I.D. Structure-based strategies for drug design and discovery. Science 1992, 257, 1078–1082. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Lu, A.Y.H. Metabolic bioactivation and drug-related adverse effects: Current status and future directions from a pharmaceutical research perspective. Drug Metab. Rev. 2010, 42, 225–249. [Google Scholar] [CrossRef]
- Fredlund, L.; Winiwarter, S.; Hilgendorf, C. In Vitro Intrinsic Permeability: A Transporter-Independent Measure of Caco-2 Cell Permeability in Drug Design and Development. Mol. Pharm. 2017, 14, 1601–1609. [Google Scholar] [CrossRef]
- Teague, S.J. Implications of protein flexibility for drug discovery. Nat. Rev. Drug Discov. 2003, 2, 527–541. [Google Scholar] [CrossRef]
- Feixas, F.; Lindert, S.; Sinko, W.; McCammon, J.A. Exploring the role of receptor flexibility in structure-based drug discovery. Biophysical Chemistry 2014, 186, 31–45. [Google Scholar] [CrossRef]
- Antunes, D.A.; Devaurs, D.; Kavraki, L.E. Understanding the challenges of protein flexibility in drug design. Expert Opin. Drug Discov. 2015, 10, 1301–1313. [Google Scholar] [CrossRef]
- Almeida, M.O.; Costa, C.H.S.; Gomes, G.C.; Lameira, J.; Alves, C.N.; Honorio, K.M. Computational analyses of interactions between ALK-5 and bioactive ligands: Insights for the design of potential anticancer agents. J. Biomol. Struct. Dyn. 2018, 36, 4010–4022. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |


| ALK-5-Ligand | SIE (kcal/mol) | ChemGrid Score (kcal/mol) |
|---|---|---|
| 1 | –9.49 | –50 |
| 2 | –8.24 | –51 |
| 3 | –7.78 | –49 |
| 4 | –8.00 | –49 |
| 5 | –7.50 | –45 |
| Benchmark compound | –9.60 | –52 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Araujo, S.C.; Maltarollo, V.G.; Almeida, M.O.; Ferreira, L.L.G.; Andricopulo, A.D.; Honorio, K.M. Structure-Based Virtual Screening, Molecular Dynamics and Binding Free Energy Calculations of Hit Candidates as ALK-5 Inhibitors. Molecules 2020, 25, 264. https://doi.org/10.3390/molecules25020264
Araujo SC, Maltarollo VG, Almeida MO, Ferreira LLG, Andricopulo AD, Honorio KM. Structure-Based Virtual Screening, Molecular Dynamics and Binding Free Energy Calculations of Hit Candidates as ALK-5 Inhibitors. Molecules. 2020; 25(2):264. https://doi.org/10.3390/molecules25020264
Chicago/Turabian StyleAraujo, Sheila C., Vinicius G. Maltarollo, Michell O. Almeida, Leonardo L. G. Ferreira, Adriano D. Andricopulo, and Kathia M. Honorio. 2020. "Structure-Based Virtual Screening, Molecular Dynamics and Binding Free Energy Calculations of Hit Candidates as ALK-5 Inhibitors" Molecules 25, no. 2: 264. https://doi.org/10.3390/molecules25020264
APA StyleAraujo, S. C., Maltarollo, V. G., Almeida, M. O., Ferreira, L. L. G., Andricopulo, A. D., & Honorio, K. M. (2020). Structure-Based Virtual Screening, Molecular Dynamics and Binding Free Energy Calculations of Hit Candidates as ALK-5 Inhibitors. Molecules, 25(2), 264. https://doi.org/10.3390/molecules25020264







