Semisynthetic Cardenolides Acting as Antiviral Inhibitors of Influenza A Virus Replication by Preventing Polymerase Complex Formation
Abstract
:1. Introduction
2. Results
2.1. Anti-Influenza Virus Activity of Cardenolide Derivatives
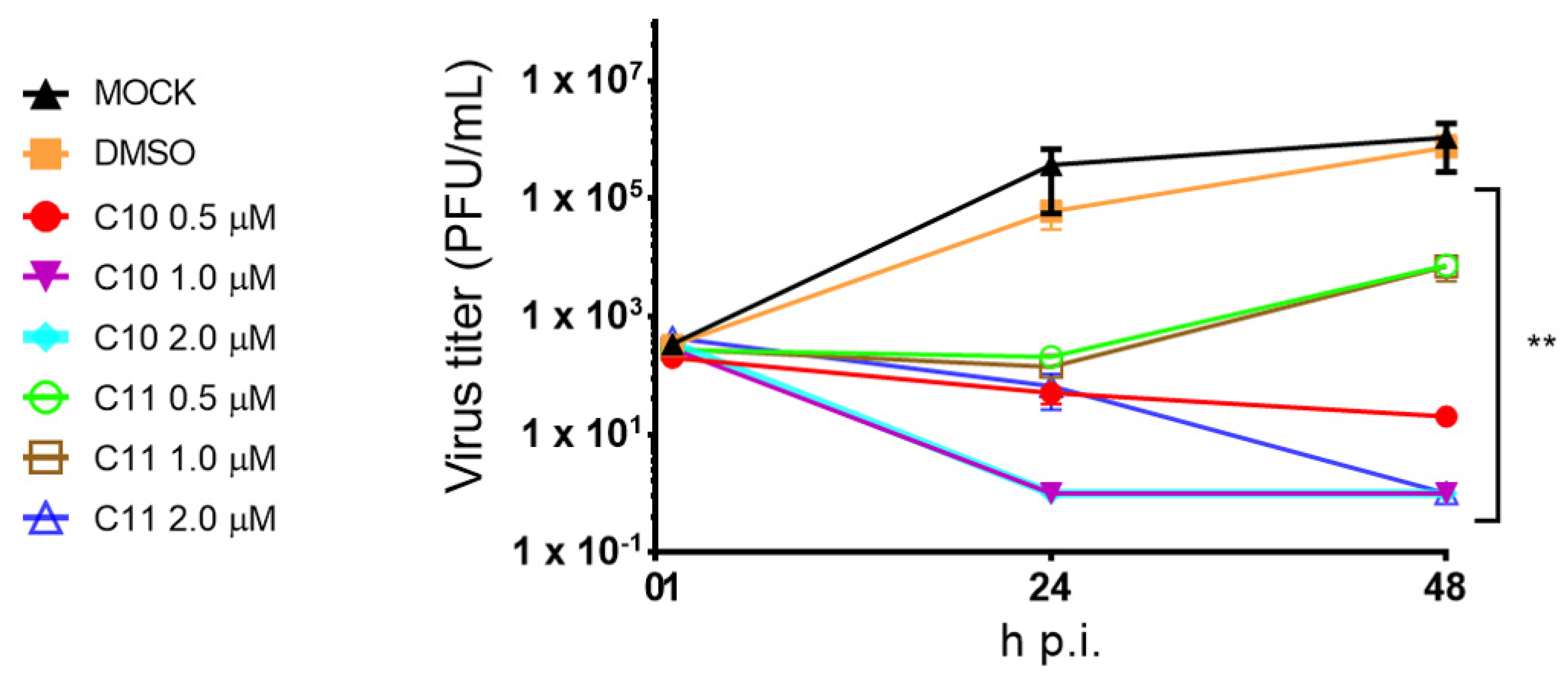
2.2. Cardenolide Derivatives Effects on Cell Viability and Influenza Replication
2.3. Anti-Influenza Virus Mechanism of Action of Compounds C10 and C11
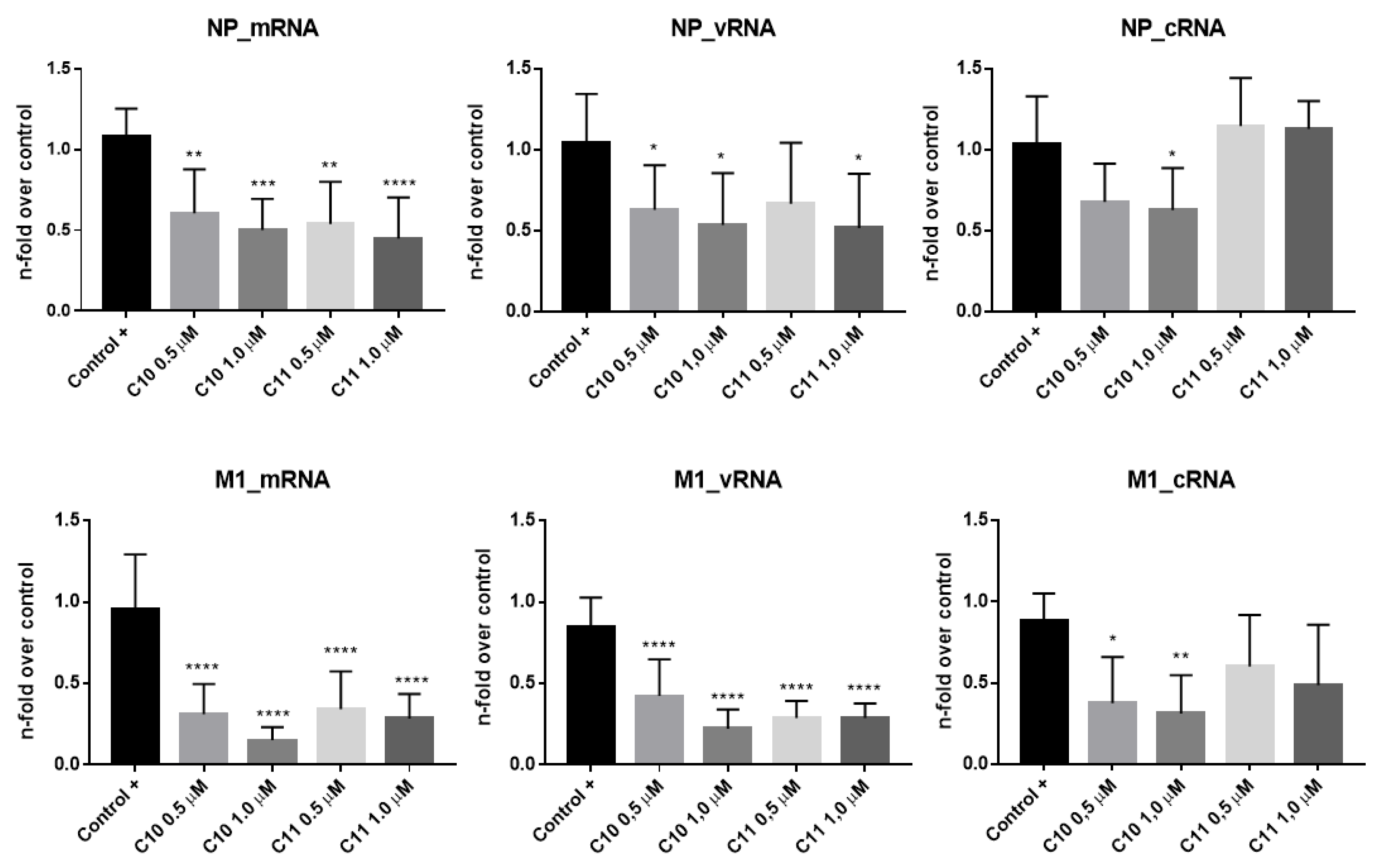
2.3.1. C10 and C11 Affect the Expression of Viral Proteins at the Earlier Stages of Influenza A Virus Replication Cycle
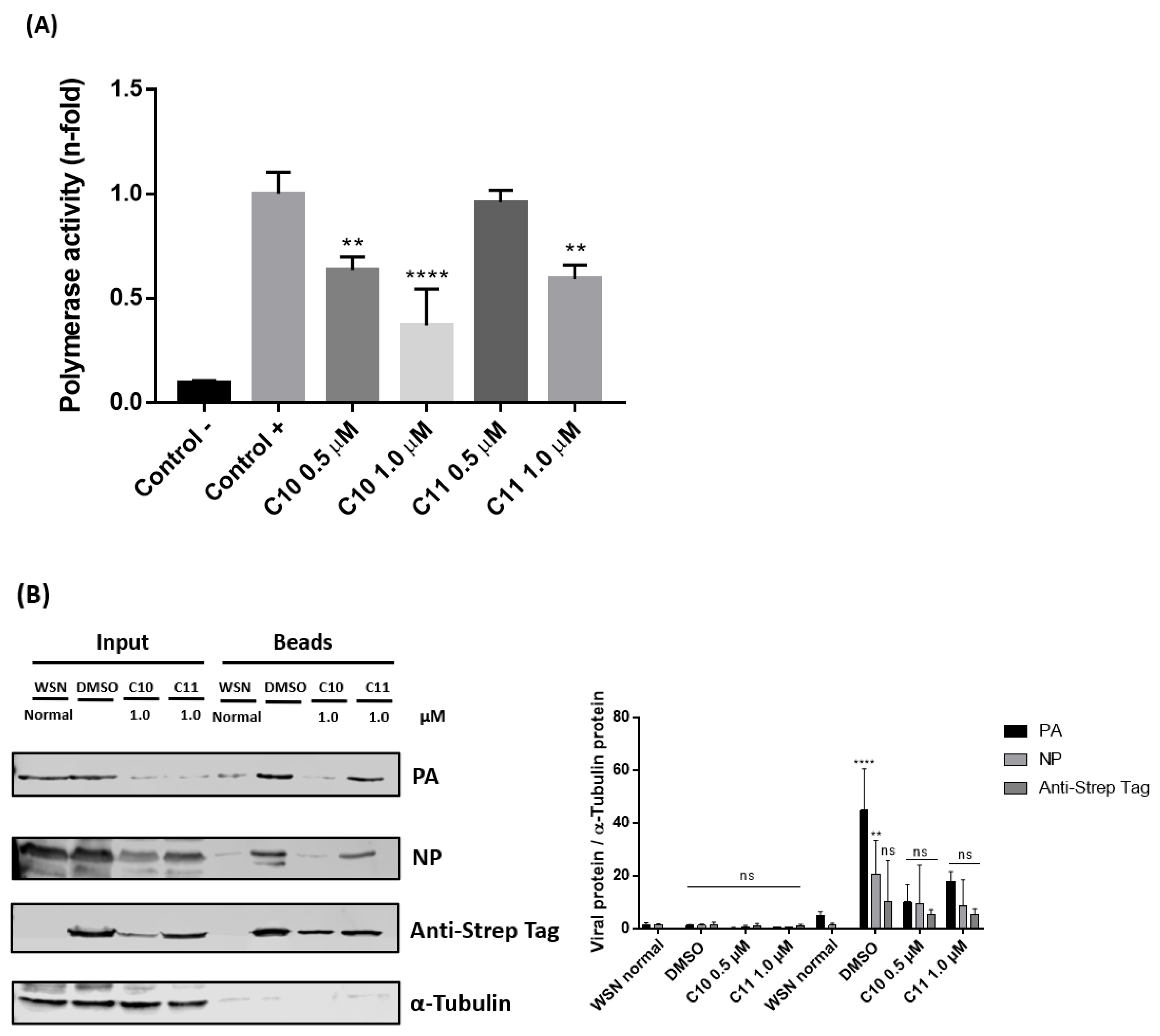
2.3.2. C10 and C11 Affect the Polymerase Complex
2.3.3. C10 and C11 Reduce Viral Replication in an Ex Vivo Model of Human Lung Tissue
3. Discussion
4. Materials and Methods
4.1. Compounds, Cell Lines and Viruses
4.2. Anti-Influenza Screening
4.3. Cytotoxicity Evaluation
4.4. Anti-Influenza Viral Mechanism of Action
4.4.1. Western Blot Analyses
4.4.2. Quantitative Real-Time RT-PCR
4.4.3. Reporter Gene Assay
4.4.4. Virus Infection of Human Lung Tissue Explants
4.5. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| A549 | human alveolar epithelial cells |
| CC50 | 50% cytotoxic concentration |
| DMEM | Dulbecco’s modified eagle’s medium |
| DMSO | dimethyl sulfoxide |
| FBS | fetal bovine serum |
| HA | hemagglutinin |
| IC50 | concentration required for 50% inhibition of viral replication |
| IAV | Influenza A virus |
| IBV | Influenza B virus |
| MDCK-II | Madin-Darby canine kidney type II cells |
| MEM | eagle’s minimum essential medium |
| MOI | multiplicity of infection |
| NA | neuraminidase |
| PBS | phosphate-buffered saline |
| PFU | plaque-forming units |
| p.i. | post-infection |
| p.t. | post-transfection |
| RIPA | radioimmunoprecipitation |
| RLU | relative light units |
| RPMI-1640 | Roswell Park Memorial Institute-1640 |
| SI | selectivity index |
| WB | western blotting |
References
- World Health Organization (WHO). Influenza (Seasonal). Available online: https://www.who.int/en/news-room/fact-sheets/detail/influenza-(seasonal) (accessed on 7 September 2020).
- Bush, R.M. Influenza Evolution. In Encyclopedia of Infectious Diseases; Tibayrenc, M., Ed.; Wiley: Hoboken, NJ, USA, 2007; pp. 199–214. [Google Scholar]
- Su, S.; Fu, X.; Li, G.; Kerlin, F.; Veit, M. Novel Influenza D virus: Epidemiology, pathology, evolution and biological characteristics. Virulence 2017, 8, 1580–1591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Behrens, G.; Stoll, M. Pathogenesis and immunology. In Influenza reports 2006; Kamps, B.S., Hoffmann, C., Preiser, W., Eds.; Flying Publisher: Paris, France, 2006; pp. 92–109. [Google Scholar]
- Shaw, M.L.; Palese, P. Orthomyxoviridae. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Cohen, J.I., Griffin, D.E., Lamb, R.A., Martin, M.A., Racaniello, V.R., Roizman, B., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; pp. 1151–1185. [Google Scholar]
- Lo, C.-Y.; Tang, Y.-S.; Shaw, P.-C. Structure and Function of Influenza Virus Ribonucleoprotein. Subcell. Biochem. 2018, 88, 95–128. [Google Scholar] [CrossRef] [PubMed]
- Dou, D.; Revol, R.; Östbye, H.; Wang, H.; Daniels, R. Influenza A Virus Cell Entry, Replication, Virion Assembly and Movement. Front. Immunol. 2018, 9, 1581. [Google Scholar] [CrossRef] [PubMed]
- Carrat, F.; Flahault, A. Influenza vaccine: The challenge of antigenic drift. Vaccine 2007, 25, 6852–6862. [Google Scholar] [CrossRef]
- Soema, P.C.; Kompier, R.; Amorij, J.-P.; Kersten, G.F. Current and next generation influenza vaccines: Formulation and production strategies. Eur. J. Pharm. Biopharm. 2015, 94, 251–263. [Google Scholar] [CrossRef] [Green Version]
- Principi, N.; Camilloni, B.; Alunno, A.; Polinori, I.; Argentiero, A.; Esposito, S. Drugs for Influenza Treatment: Is There Significant News? Front. Med. 2019, 6, 109. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization (WHO). Influenza Vaccine Viruses and Reagents. Available online: https://www.who.int/influenza/vaccines/virus/en/2020 (accessed on 8 September 2020).
- Chua, J.V.; Chen, W.H. Bench-to-bedside review: Vaccine protection strategies during pandemic flu outbreaks. Crit. Care 2010, 14, 218. [Google Scholar] [CrossRef] [Green Version]
- Hurt, A.C.; Hardie, K.; Wilson, N.J.; Edeng, Y.-M.; Osbourn, B.M.; Gehrig, N.; Kelso, A. Community Transmission of Oseltamivir-Resistant A(H1N1)pdm09 Influenza. N. Engl. J. Med. 2011, 365, 2541–2542. [Google Scholar] [CrossRef]
- Takashita, E.; Ejima, M.; Itoh, R.; Miura, M.; Ohnishi, A.; Nishimura, H.; Odagiri, T.; Tashiro, M. A community cluster of influenza A(H1N1)pdm09 virus exhibiting cross-resistance to oseltamivir and peramivir in Japan, November to December 2013. Eurosurveillance 2014, 19, 20666. [Google Scholar] [CrossRef] [Green Version]
- Jacob, A.; Sood, R.; Chanu, K.V.; Bhatia, S.; Khandia, R.; Pateriya, A.; Nagarajan, S.; Dimri, U.; Kulkarni, D. Amantadine resistance among highly pathogenic avian influenza viruses (H5N1) isolated from India. Microb. Pathog. 2016, 91, 35–40. [Google Scholar] [CrossRef]
- Takashita, E.; Ejima, M.; Ogawa, R.; Fujisaki, S.; Neumann, G.; Furuta, Y.; Kawaoka, Y.; Tashiro, M.; Odagiri, T. Antiviral susceptibility of influenza viruses isolated from patients pre- and post-administration of favipiravir. Antivir. Res. 2016, 132, 170–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matos, A.D.R.; Resende, P.C.; Miranda, M.D.; Garcia, C.C.; Caetano, B.C.; Lopes, J.C.; Debur, M.C.; Cury, A.L.; Vianna, L.A.; Lima, M.C.; et al. Susceptibility of Brazilian influenza A(H1N1)pdm09 viruses to neuraminidase inhibitors in the 2014–2016 seasons: Identification of strains bearing mutations associated with reduced inhibition profile. Antivir. Res. 2018, 154, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Horwood, P.F.; Karlsson, E.A.; Horm, S.V.; Ly, S.; Heng, S.; Chin, S.; Darapheak, C.; Saunders, D.; Chanthap, L.; Rith, S.; et al. Circulation and characterization of seasonal influenza viruses in Cambodia, 2012–2015. Influ. Other Respir. Viruses 2019, 13, 465–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leneva, I.; Burtseva, E.; Yatsyshina, S.; Fedyakina, I.; Kirillova, E.; Selkova, E.; Osipova, E.; Maлeeв, B. Virus susceptibility and clinical effectiveness of anti-influenza drugs during the 2010–2011 influenza season in Russia. Int. J. Infect. Dis. 2016, 43, 77–84. [Google Scholar] [CrossRef] [Green Version]
- Pshenichnaya, N.Y.; Bulgakova, V.A.; Lvov, N.I.; Poromov, A.A.; Selkova, E.P.; Grekova, A.I.; Shestakova, I.V.; Maleev, V.V.; Leneva, I.A. Clinical efficacy of umifenovir in influenza and ARVI (study ARBITR). Ther. Arch. 2019, 91, 56–63. [Google Scholar] [CrossRef]
- Furuta, Y.; Komeno, T.; Nakamura, T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B 2017, 93, 449–463. [Google Scholar] [CrossRef] [Green Version]
- Noshi, T.; Kitano, M.; Taniguchi, K.; Yamamoto, A.; Omoto, S.; Baba, K.; Hashimoto, T.; Ishida, K.; Kushima, Y.; Hattori, K.; et al. In vitro characterization of baloxavir acid, a first-in-class cap-dependent endonuclease inhibitor of the influenza virus polymerase PA subunit. Antivir. Res. 2018, 160, 109–117. [Google Scholar] [CrossRef]
- Hoffmann-La Roche (Roche). Roche Announces FDA Approval of Xofluza (Baloxavir Marboxil) for Influenza. Available online: https://www.roche.com/investors/updates/inv-update-2018-10-24.htm (accessed on 7 September 2020).
- Kreis, W. The Foxgloves (Digitalis) Revisited. Planta Med. 2017, 83, 962–976. [Google Scholar] [CrossRef] [Green Version]
- Bessen, H.A. Therapeutic and toxic effects of digitalis: William Withering, 1785. J. Emerg. Med. 1986, 4, 243–248. [Google Scholar] [CrossRef]
- Rahimtoola, S.H.; Tak, T. The use of digitalis in heart failure. Curr. Probl. Cardiol. 1996, 21, 781–853. [Google Scholar] [CrossRef]
- Patel, S. Plant-derived cardiac glycosides: Role in heart ailments and cancer management. Biomed. Pharmacother. 2016, 84, 1036–1041. [Google Scholar] [CrossRef] [PubMed]
- Reddy, D.; Kumavath, R.; Barh, D.; Azevedo, V.; Ghosh, P. Anticancer and Antiviral Properties of Cardiac Glycosides: A Review to Explore the Mechanism of Actions. Molecules 2020, 25, 3596. [Google Scholar] [CrossRef] [PubMed]
- Grosso, F.; Stoilov, P.; Lingwood, C.; Brown, M.; Cochrane, A. Suppression of Adenovirus Replication by Cardiotonic Steroids. J. Virol. 2016, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashbrook, A.W.; Lentscher, A.J.; Zamora, P.F.; Silva, L.A.; May, N.A.; Bauer, J.A.; Morrison, T.E.; Dermody, T.S. Antagonism of the Sodium-Potassium ATPase Impairs Chikungunya Virus Infection. mBio 2016, 7, e00693-16. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.-W.; Chang, H.-Y.; Hsu, H.-Y.; Lee, Y.-Z.; Chang, H.-S.; Chen, I.-S.; Lee, S.-J. Identification of anti-viral activity of the cardenolides, Na+/K+-ATPase inhibitors, against porcine transmissible gastroenteritis virus. Toxicol. Appl. Pharmacol. 2017, 332, 129–137. [Google Scholar] [CrossRef]
- Yang, C.-W.; Chang, H.-Y.; Lee, Y.-Z.; Hsu, H.-Y.; Lee, S.-J. The cardenolide ouabain suppresses coronaviral replication via augmenting a Na+/K+-ATPase-dependent PI3K_PDK1 axis signaling. Toxicol. Appl. Pharmacol. 2018, 356, 90–97. [Google Scholar] [CrossRef]
- Yang, C.-W.; Hsu, H.-Y.; Chang, H.-Y.; Lee, Y.-Z.; Lee, S.-J. Natural cardenolides suppress coronaviral replication by downregulating JAK1 via a Na+/K+-ATPase independent proteolysise. Biochem. Pharmacol. 2020, 180, 114122. [Google Scholar] [CrossRef]
- Cai, H.; Wang, H.-Y.L.; Venkatadri, R.; Fu, D.-X.; Forman, M.; Bajaj, S.O.; Li, H.; O’Doherty, G.A.; Arav-Boger, R. Digitoxin Analogues with Improved Anticytomegalovirus Activity. ACS Med. Chem. Lett. 2014, 5, 395–399. [Google Scholar] [CrossRef]
- Cohen, T.; Williams, J.D.; Opperman, T.J.; Sanchez, R.; Lurain, N.S.; Tortorella, D. Convallatoxin-Induced Reduction of Methionine Import Effectively Inhibits Human Cytomegalovirus Infection and Replication. J. Virol. 2016, 90, 10715–10727. [Google Scholar] [CrossRef] [Green Version]
- Gardner, T.J.; Cohen, T.; Redmann, V.; Lau, Z.; Felsenfeld, D.; Tortorella, D. Development of a high-content screen for the identification of inhibitors directed against the early steps of the cytomegalovirus infectious cycle. Antivir. Res. 2015, 113, 49–61. [Google Scholar] [CrossRef] [Green Version]
- Cheung, Y.Y.; Chen, K.C.; Chen, H.; Seng, E.K.; Chu, J.J.H. Antiviral activity of lanatoside C against dengue virus infection. Antivir. Res. 2014, 111, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Bertol, J.W.; Rigotto, C.; De Pádua, R.M.; Kreis, W.; Barardi, C.R.M.; Braga, F.C.; Simões, C.M.O. Antiherpes activity of glucoevatromonoside, a cardenolide isolated from a Brazilian cultivar of Digitalis lanata. Antivir. Res. 2011, 92, 73–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dodson, A.W.; Taylor, T.J.; Knipe, D.M.; Coen, D.M. Inhibitors of the sodium potassium ATPase that impair herpes simplex virus replication identified via a chemical screening approach. Virology 2007, 366, 340–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, C.-T.; Hsu, J.T.-A.; Hsieh, H.-P.; Lin, P.-H.; Chen, T.-C.; Kao, C.-L.; Lee, C.-N.; Chen, Y.-C. Anti-HSV activity of digitoxin and its possible mechanisms. Antivir. Res. 2008, 79, 62–70. [Google Scholar] [CrossRef]
- Boff, L.; Schneider, N.F.Z.; Munkert, J.; Ottoni, F.M.; Ramos, G.S.; Kreis, W.; Braga, F.C.; Alves, R.J.; De Pádua, R.M.; Simões, C.M.O. Elucidation of the mechanism of anti-herpes action of two novel semisynthetic cardenolide derivatives. Arch. Virol. 2020, 165, 1385–1396. [Google Scholar] [CrossRef]
- Singh, S.; Shenoy, S.; Nehete, P.N.; Yang, P.; Nehete, B.; Fontenot, D.; Yang, G.; Newman, R.A.; Sastry, K.J. Nerium oleander derived cardiac glycoside oleandrin is a novel inhibitor of HIV infectivity. Fitoterapia 2013, 84, 32–39. [Google Scholar] [CrossRef]
- Wong, R.W.; Balachandran, A.; Ostrowski, M.A.; Cochrane, A. Digoxin Suppresses HIV-1 Replication by Altering Viral RNA Processing. PLoS Pathog. 2013, 9, e1003241. [Google Scholar] [CrossRef]
- Wong, R.W.; Lingwood, C.A.; Ostrowski, M.A.; Cabral, T.; Cochrane, A. Cardiac glycoside/aglycones inhibit HIV-1 gene expression by a mechanism requiring MEK1/2-ERK1/2 signaling. Sci. Rep. 2018, 8, 850. [Google Scholar] [CrossRef] [Green Version]
- Van Der Kolk, T.; Dillingh, M.; Rijneveld, R.; Klaassen, E.; De Koning, M.; Kouwenhoven, S.; Genders, R.; Bavinck, J.N.B.; Feiss, G.; Rissmann, R.; et al. Topical ionic contra-viral therapy comprised of digoxin and furosemide as a potential novel treatment approach for common warts. J. Eur. Acad. Dermatol. Venereol. 2017, 31, 2088–2090. [Google Scholar] [CrossRef]
- Hoffmann, H.-H.; Palese, P.; Shaw, M.L. Modulation of influenza virus replication by alteration of sodium ion transport and protein kinase C activity. Antivir. Res. 2008, 80, 124–134. [Google Scholar] [CrossRef]
- Kiyohara, H.; Ichino, C.; Kawamura, Y.; Nagai, T.; Sato, N.; Yamada, H.; Salama, M.M.; Abdel-Sattar, E.A. In vitro anti-influenza virus activity of a cardiotonic glycoside from Adenium obesum (Forssk.). Phytomedicine 2012, 19, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Amarelle, L.; Katzen, J.; Shigemura, M.; Welch, L.C.; Cajigas, H.; Peteranderl, C.; Celli, D.; Herold, S.; Lecuona, E.; Sznajder, J.I. Cardiac glycosides decrease influenza virus replication by inhibiting cell protein translational machinery. Am. J. Physiol. Cell. Mol. Physiol. 2019, 316, L1094–L1106. [Google Scholar] [CrossRef] [PubMed]
- Norris, M.J.; Malhi, M.; Duan, W.; Ouyang, H.; Granados, A.; Cen, Y.; Tseng, Y.-C.; Gubbay, J.; Maynes, J.; Moraes, T. Targeting Intracellular Ion Homeostasis for the Control of Respiratory Syncytial Virus. Am. J. Respir. Cell Mol. Biol. 2018, 59, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Boff, L.; Munkert, J.; Ottoni, F.M.; Schneider, N.F.Z.; Ramos, G.S.; Kreis, W.; Andrade, S.F.; Souza Filho, J.D.; Braga, F.C.; Alves, R.J.; et al. Potential anti-herpes and cytotoxic action of novel semisynthetic digitoxigenin-derivatives. Eur. J. Med. Chem. 2019, 167, 546–561. [Google Scholar] [CrossRef]
- Boff, L.; Persich, L.; Brambila, P.; Ottoni, F.M.; Munkert, J.; Ramos, G.S.; Viana, A.R.S.; Kreis, W.; Braga, F.C.; Alves, R.J.; et al. Investigation of the cytotoxic activity of two novel digitoxigenin analogues on H460 lung cancer cells. Anti-Cancer Drugs 2020, 31, 452–462. [Google Scholar] [CrossRef]
- Rameix-Welti, M.-A.; Tomoiu, A.; Afonso, E.D.S.; Van Der Werf, S.; Naffakh, N. Avian Influenza A Virus Polymerase Association with Nucleoprotein, but Not Polymerase Assembly, Is Impaired in Human Cells during the Course of Infection. J. Virol. 2008, 83, 1320–1331. [Google Scholar] [CrossRef] [Green Version]
- Matos, A.D.R.; Wunderlich, K.; Schloer, S.; Schughart, K.; Geffers, R.; Seders, M.; De Witt, M.; Christersson, A.; Wiewrodt, R.; Wiebe, K.; et al. Antiviral potential of human IFN-α subtypes against influenza A H3N2 infection in human lung explants reveals subtype-specific activities. Emerg. Microbes Infect. 2019, 8, 1763–1776. [Google Scholar] [CrossRef] [Green Version]
- Prassas, I.; Diamandis, E.P. Novel therapeutic applications of cardiac glycosides. Nat. Rev. Drug Discov. 2008, 7, 926–935. [Google Scholar] [CrossRef]
- Amarelle, L.; Lecuona, E. The Antiviral Effects of Na,K-ATPase Inhibition: A Minireview. Int. J. Mol. Sci. 2018, 19, 2154. [Google Scholar] [CrossRef] [Green Version]
- Hui, E.K.-W.; Nayak, D.P. Role of ATP in Influenza Virus Budding. Virology 2001, 290, 329–341. [Google Scholar] [CrossRef]
- Ishihama, A. A multi-functional enzyme with RNA polymerase and RNase activities: Molecular anatomy of influenza virus RNA polymerase. Biochimie 1996, 78, 1097–1102. [Google Scholar] [CrossRef]
- Honda, A.; Ishihama, A. The molecular anatomy of influenza virus RNA polymerase. Biol. Chem. 1997, 378, 483–488. [Google Scholar] [PubMed]
- Kilkenny, C.; Browne, W.; Cuthill, I.C.; Emerson, M.; Altman, D.G. Animal research: Reporting in vivo experiments: The ARRIVE guidelines. Br. J. Pharmacol. 2010, 160, 1577–1579. [Google Scholar] [CrossRef] [PubMed]
- Perne, A.; Muellner, M.K.; Steinrueck, M.; Craig-Mueller, N.; Mayerhofer, J.; Schwarzinger, I.; Sloane, M.; Uras, I.Z.; Hoermann, G.; Nijman, S.M.B.; et al. Cardiac Glycosides Induce Cell Death in Human Cells by Inhibiting General Protein Synthesis. PLoS ONE 2009, 4, e8292. [Google Scholar] [CrossRef] [PubMed]
- Calderón-Montaño, J.M.; Burgos-Morón, E.; Orta, M.L.; Maldonado-Navas, D.; García-Domínguez, I.; Lopez-Lazaro, M. Evaluating the Cancer Therapeutic Potential of Cardiac Glycosides. BioMed Res. Int. 2014, 2014, 1–9. [Google Scholar] [CrossRef]
- Weinheimer, V.K.; Becher, A.; Tönnies, M.; Holland, G.; Knepper, J.; Bauer, T.T.; Schneider, P.; Neudecker, J.; Rückert, J.C.; Szymanski, K.; et al. Influenza A Viruses Target Type II Pneumocytes in the Human Lung. J. Infect. Dis. 2012, 206, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Du, J.; Huang, C.; Wang, T.; Huang, L.; Yang, S.; Li, L. Discovery of 5-(5-fluoro-1H-pyrrolo [2,3-b]pyridin-3-yl)pyrazin-2(1H)-one derivatives as new potent PB2 inhibitors. Bioorganic Med. Chem. Lett. 2019, 29, 1609–1613. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, Y.; Cui, Q.; Li, P.; Wang, L.; Chen, Z.; Rong, L.; Du, R. A Parallel Phenotypic Versus Target-Based Screening Strategy for RNA-Dependent RNA Polymerase Inhibitors of the Influenza A Virus. Viruses 2019, 11, 826. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Hu, Y.; Wu, N.; Wang, J. Discovery of Influenza Polymerase PA–PB1 Interaction Inhibitors Using an In Vitro Split-Luciferase Complementation-Based Assay. ACS Chem. Biol. 2019, 15, 74–82. [Google Scholar] [CrossRef]
- Hoffmann, E.; Neumann, G.; Kawaoka, Y.; Hobom, G.; Webster, R.G. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 2000, 97, 6108–6113. [Google Scholar] [CrossRef] [Green Version]
- Burleson, F.G.; Chamberts, T.M.; Wiedbrauk, D.L. Virology: A Laboratory Manual; Academic Press: San Diego, CA, USA, 1992; p. 250. [Google Scholar]
- Dudek, S.E.; Luig, C.; Pauli, E.-K.; Schubert, U.; Ludwig, S. The Clinically Approved Proteasome Inhibitor PS-341 Efficiently Blocks Influenza A Virus and Vesicular Stomatitis Virus Propagation by Establishing an Antiviral State. J. Virol. 2010, 84, 9439–9451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schreiber, A.; Liedmann, S.; Brunotte, L.; Anhlan, D.; Ehrhardt, C.; Ludwig, S. Type I interferon antagonistic properties of influenza B virus polymerase proteins. Cell. Microbiol. 2019, 22, e13143. [Google Scholar] [CrossRef] [Green Version]
- Kawakami, E.; Watanabe, T.; Fujii, K.; Goto, H.; Watanabe, S.; Noda, T.; Kawaoka, Y. Strand-specific real-time RT-PCR for distinguishing influenza vRNA, cRNA, and mRNA. J. Virol. Methods 2011, 173, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Börgeling, Y.; Schmolke, M.; Viemann, D.; Nordhoff, C.; Roth, J.; Ludwig, S. Inhibition of p38 Mitogen-activated Protein Kinase Impairs Influenza Virus-induced Primary and Secondary Host Gene Responses and Protects Mice from Lethal H5N1 Infection. J. Biol. Chem. 2013, 289, 13–27. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Saint, D.A. A New Quantitative Method of Real Time Reverse Transcription Polymerase Chain Reaction Assay Based on Simulation of Polymerase Chain Reaction Kinetics. Anal. Biochem. 2002, 302, 52–59. [Google Scholar] [CrossRef] [Green Version]
- Schräder, T.; Dudek, S.E.; Schreiber, A.; Ehrhardt, C.; Planz, O.; Ludwig, S. The clinically approved MEK inhibitor Trametinib efficiently blocks influenza A virus propagation and cytokine expression. Antivir. Res. 2018, 157, 80–92. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-Dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
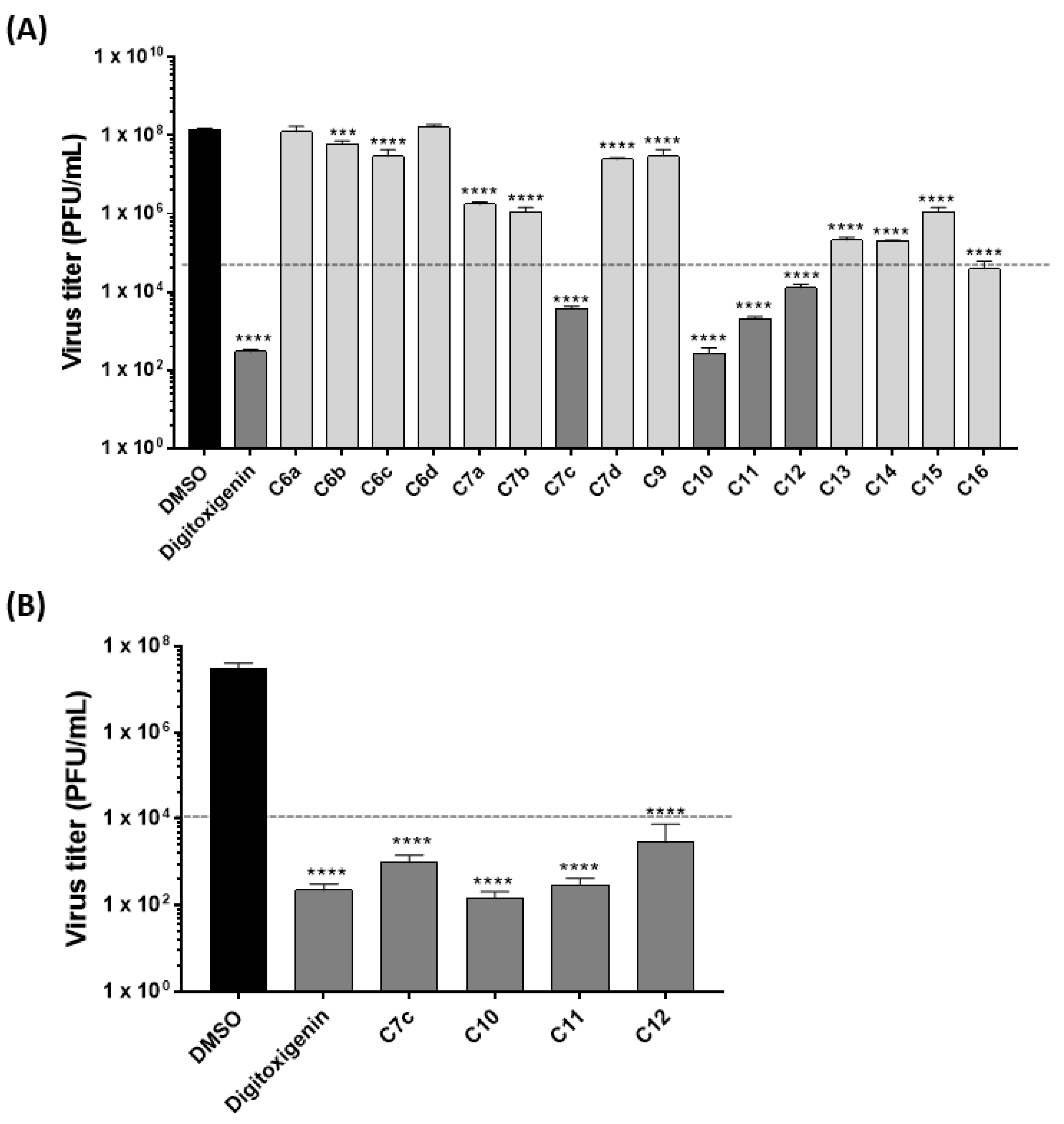
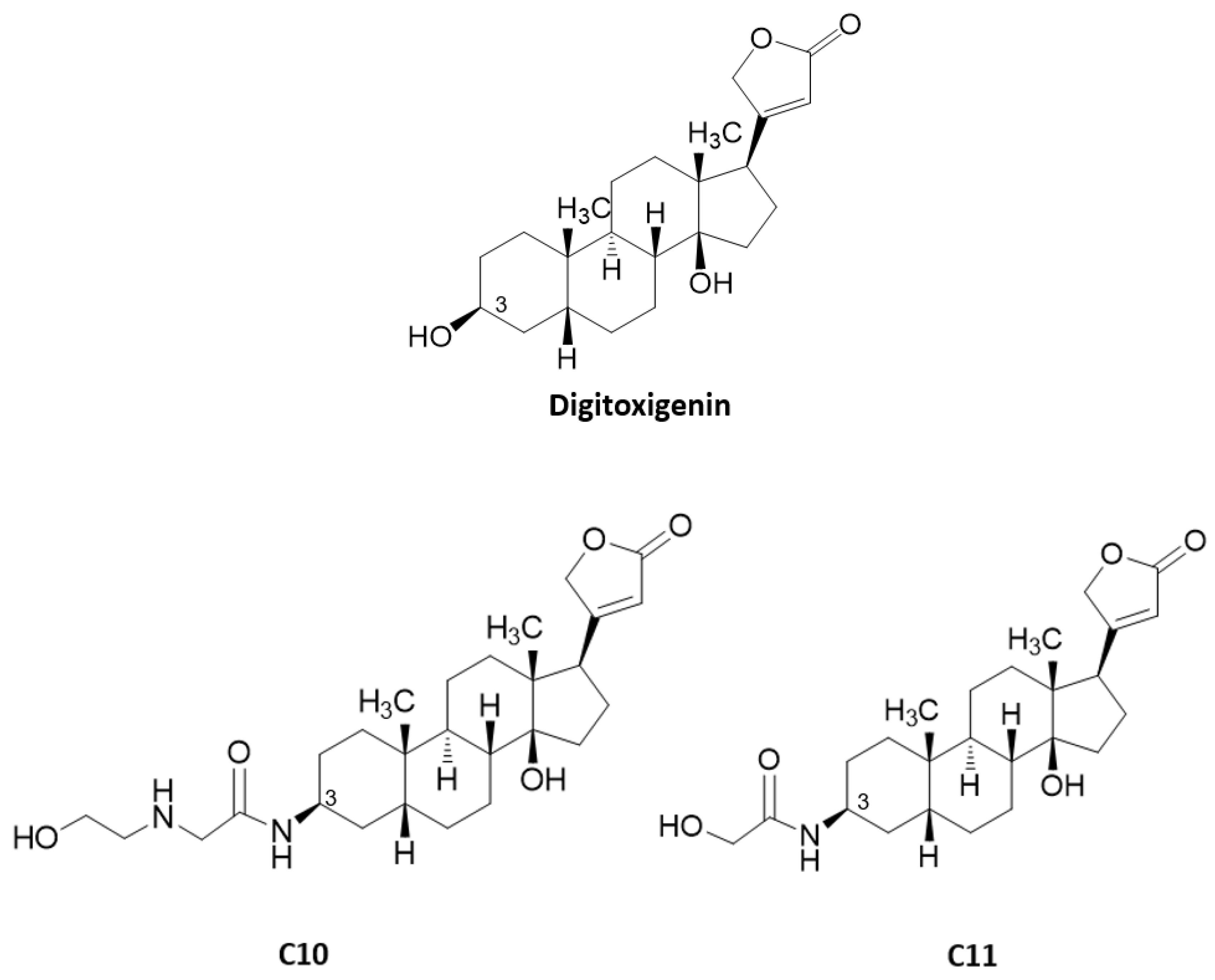

| Compounds | A549 Cells | IAV (Strain WSN/33 (H1N1)) | MDCK Cells | IAV (Strain WSN/33 (H1N1)) | ||
|---|---|---|---|---|---|---|
| CC50 (µM) | IC50 (µM) | SI | CC50 (µM) | IC50 (µM) | SI | |
| C7c | 9.398 ± 0.741 | 0.125 ± 0.010 | 75.282 | 22.170 ± 2.393 | 0.103 ± 0.009 | 216.166 |
| C10 | 12.790 ± 0.754 | 0.057 ± 0.002 | 226.052 | 18.343 ± 0.915 | 0.060 ± 0.004 | 306.164 |
| C11 | 10.025 ± 0.962 | 0.062 ± 0.000 | 161.645 | 15.983 ± 0.235 | 0.066 ± 0.006 | 241.379 |
| C12 | 13.280 ± 1.753 | 0.123 ± 0.013 | 107.647 | 23.340 ± 1.184 | 0.091 ± 0.008 | 257.483 |
| Digitoxigenin | 6.465 ± 1.004 | 0.070 ± 0.010 | 92.366 | 6.360 ± 0.427 | 0.072 ± 0.005 | 88.052 |
Sample Availability: Samples of the compounds C6a, C6b, C6c, C6d, C7a, C7b, C7c, C7d, C9, C10, C11, C12, C13, C14, C15, and C16 are available from the authors. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boff, L.; Schreiber, A.; da Rocha Matos, A.; Del Sarto, J.; Brunotte, L.; Munkert, J.; Melo Ottoni, F.; Silva Ramos, G.; Kreis, W.; Castro Braga, F.; et al. Semisynthetic Cardenolides Acting as Antiviral Inhibitors of Influenza A Virus Replication by Preventing Polymerase Complex Formation. Molecules 2020, 25, 4853. https://doi.org/10.3390/molecules25204853
Boff L, Schreiber A, da Rocha Matos A, Del Sarto J, Brunotte L, Munkert J, Melo Ottoni F, Silva Ramos G, Kreis W, Castro Braga F, et al. Semisynthetic Cardenolides Acting as Antiviral Inhibitors of Influenza A Virus Replication by Preventing Polymerase Complex Formation. Molecules. 2020; 25(20):4853. https://doi.org/10.3390/molecules25204853
Chicago/Turabian StyleBoff, Laurita, André Schreiber, Aline da Rocha Matos, Juliana Del Sarto, Linda Brunotte, Jennifer Munkert, Flaviano Melo Ottoni, Gabriela Silva Ramos, Wolfgang Kreis, Fernão Castro Braga, and et al. 2020. "Semisynthetic Cardenolides Acting as Antiviral Inhibitors of Influenza A Virus Replication by Preventing Polymerase Complex Formation" Molecules 25, no. 20: 4853. https://doi.org/10.3390/molecules25204853








