Branched Ubiquitination: Detection Methods, Biological Functions and Chemical Synthesis
Abstract
1. Introduction
2. Detection of Branched Ubiquitin Chains
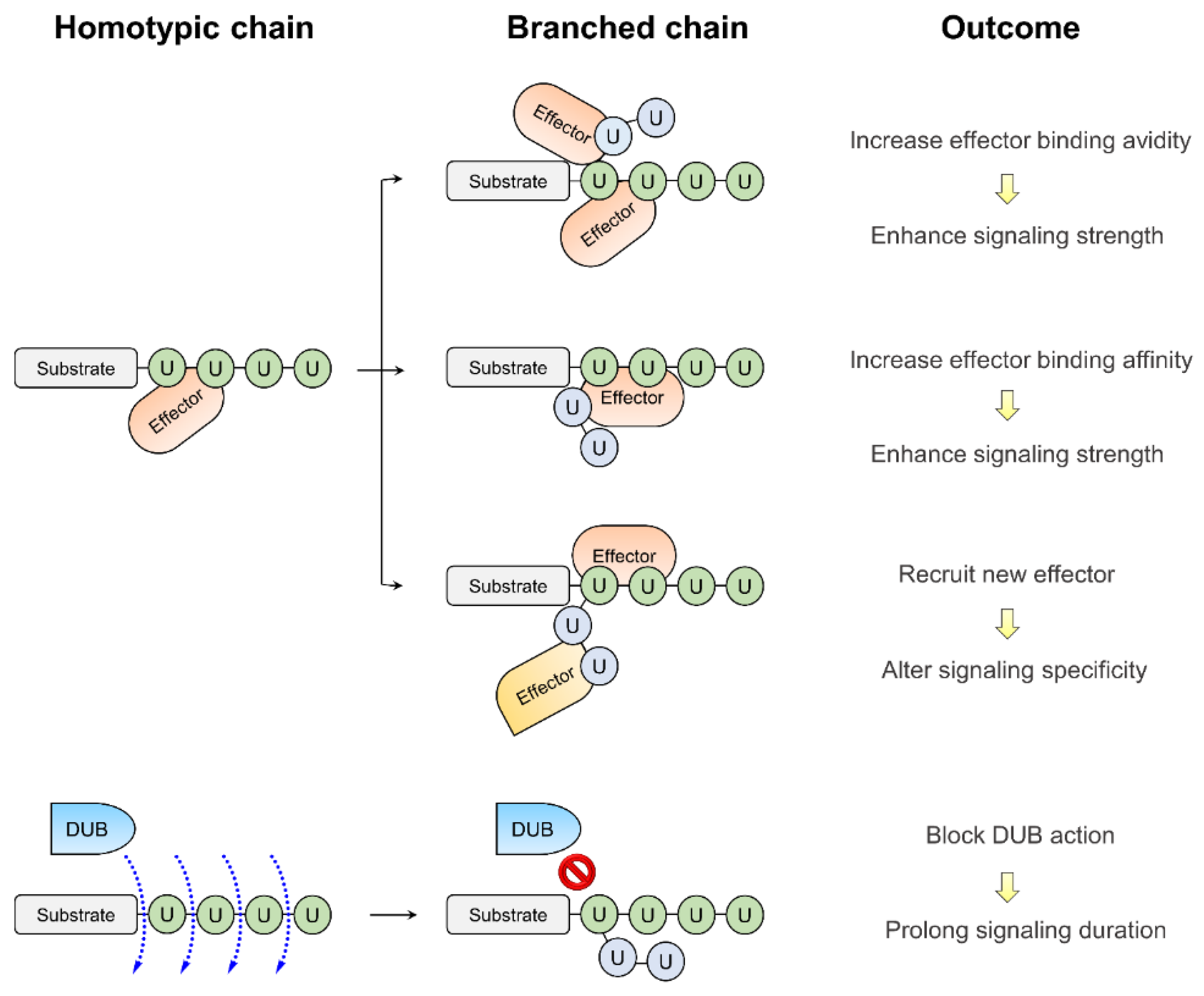
3. The Functional Outcomes of Branched Ubiquitination
3.1. K11/K48 Chain: A High-Priority Proteasomal Degradation Signal in Mitosis and Protein Quality Control
3.2. K63/M1 Chain: A Non-Proteolytic and Combinatory Signal in Regulating NF-κB Pathway
3.3. K48/K63 Chain: A Versatile Signal for Functional Switch or Output Extension
3.4. K29/K48 Chain: An Emerging Proteolytic Signal
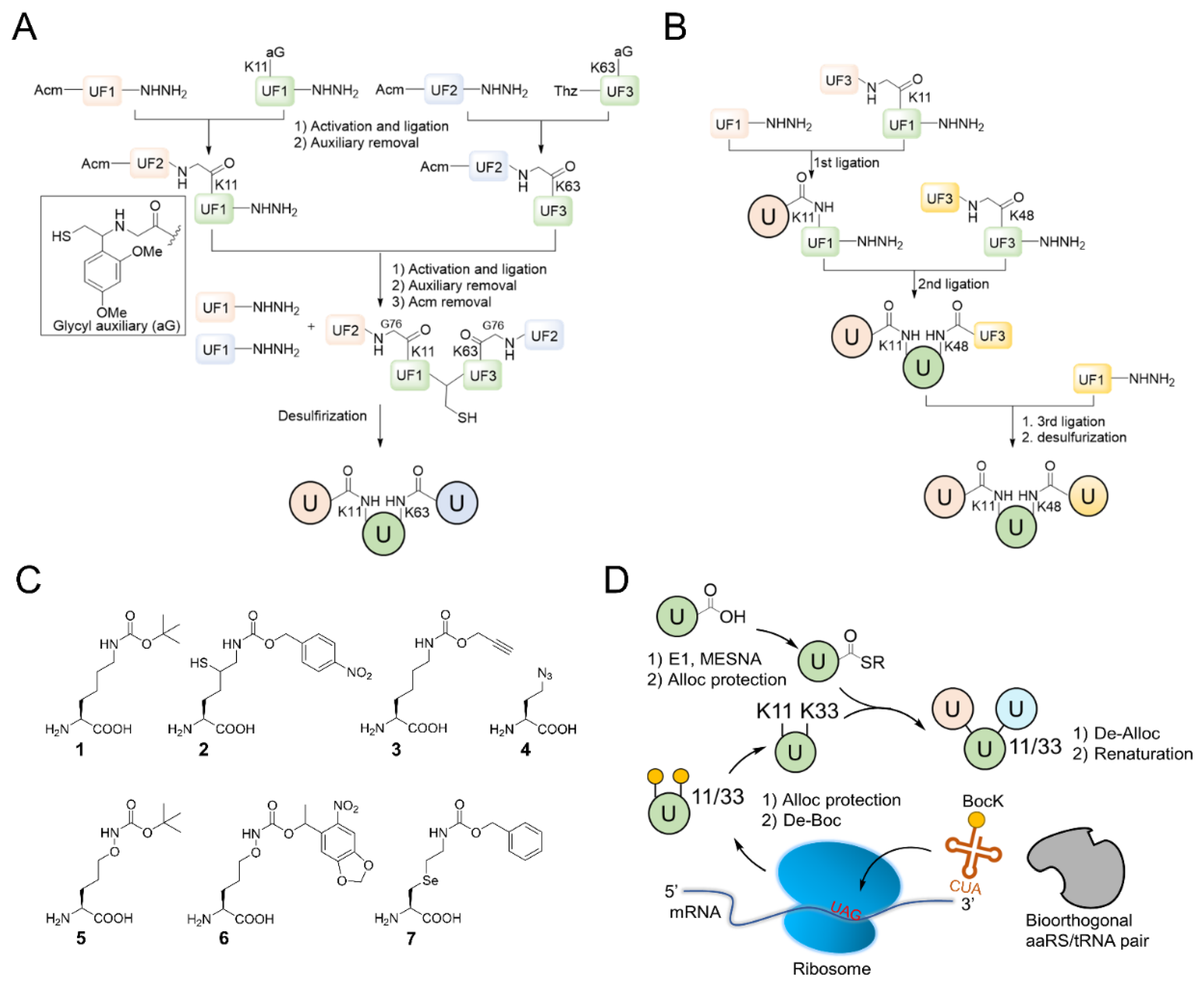
4. In Vitro Synthesis of Branched Ubiquitin Chains
4.1. In Vitro Enzymatic Synthesis of Branched Ubiquitin Chains
4.2. Chemical Synthesis of Branched Ubiquitin Chains
4.3. Semi-Synthesis of Branched Ubiquitin Chains
4.4. Non-Canonical Amino Acids-Assisted Synthesis of Branched Ubiquitin Chains
5. Conclusions and Perspectives
6. Outstanding Questions
Author Contributions
Funding
Conflicts of Interest
References
- Kulathu, Y.; Komander, D. Atypical ubiquitylation—The unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 2012, 13, 508–523. [Google Scholar] [CrossRef] [PubMed]
- Yau, R.; Rape, M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 2016, 18, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Dammer, E.B.; Na, C.H.; Xu, P.; Seyfried, N.T.; Duong, D.M.; Cheng, D.; Gearing, M.; Rees, H.; Lah, J.J.; Levey, A.I.; et al. Polyubiquitin Linkage Profiles in Three Models of Proteolytic Stress Suggest the Etiology of Alzheimer Disease. J. Biol. Chem. 2011, 286, 10457–10465. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Bennett, E.J.; Huttlin, E.L.; Guo, A.; Li, J.; Possemato, A.; Sowa, M.E.; Rad, R.; Rush, J.; Comb, M.J.; et al. Systematic and Quantitative Assessment of the Ubiquitin-Modified Proteome. Mol. Cell 2011, 44, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.A.; Beli, P.; Weinert, B.T.; Nielsen, M.L.; Cox, J.; Mann, M.; Choudhary, C. A Proteome-wide, Quantitative Survey of in vivo Ubiquitylation Sites Reveals Widespread Regulatory Roles. Mol. Cell. Proteom. 2011, 10, M111.013284. [Google Scholar] [CrossRef]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef] [PubMed]
- Crowe, S.O.; Rana, A.S.J.B.; Deol, K.K.; Ge, Y.; Strieter, E.R. Ubiquitin Chain Enrichment Middle-Down Mass Spectrometry Enables Characterization of Branched Ubiquitin Chains in Cellulo. Anal. Chem. 2017, 89, 4428–4434. [Google Scholar] [CrossRef]
- Meyer, H.-J.; Rape, M. Enhanced Protein Degradation by Branched Ubiquitin Chains. Cell 2014, 157, 910–921. [Google Scholar] [CrossRef] [PubMed]
- Swatek, K.N.; Usher, J.L.; Kueck, A.F.; Gladkova, C.; Mevissen, T.E.T.; Pruneda, J.N.; Skern, T.; Komander, D. Insights into ubiquitin chain architecture using Ub-clipping. Nat. Cell Biol. 2019, 572, 533–537. [Google Scholar] [CrossRef]
- Yau, R.G.; Doerner, K.; Castellanos, E.R.; Haakonsen, D.L.; Werner, A.; Wang, N.; Yang, X.W.; Martinez-Martin, N.; Matsumoto, M.L.; Dixit, V.M.; et al. Assembly and Function of Heterotypic Ubiquitin Chains in Cell-Cycle and Protein Quality Control. Cell 2017, 171, 918–933.e20. [Google Scholar] [CrossRef]
- Haakonsen, D.L.; Rape, M. Branching Out: Improved Signaling by Heterotypic Ubiquitin Chains. Trends Cell Biol. 2019, 29, 704–716. [Google Scholar] [CrossRef]
- Scott, D.C.; Schulman, B.A. Dual-color pulse-chase ubiquitination assays to simultaneously monitor substrate priming and extension. Methods Enzym. 2019, 618, 29–48. [Google Scholar] [CrossRef]
- Ohtake, F.; Saeki, Y.; Ishido, S.; Kanno, J.; Tanaka, K. The K48–K63 Branched Ubiquitin Chain Regulates NF-kappaB Signaling. Mol. Cell 2016, 64, 251–266. [Google Scholar] [CrossRef]
- Roscoe, B.P.; Thayer, K.M.; Zeldovich, K.B.; Fushman, D.; Bolon, D.N.A. Analyses of the Effects of All Ubiquitin Point Mutants on Yeast Growth Rate. J. Mol. Biol. 2013, 425, 1363–1377. [Google Scholar] [CrossRef]
- Hospenthal, M.K.; Freund, S.M.; Komander, D. Assembly, analysis and architecture of atypical ubiquitin chains. Nat. Struct. Mol. Biol. 2013, 20, 555–565. [Google Scholar] [CrossRef]
- Hospenthal, M.K.; Mevissen, T.E.; Komander, D. Deubiquitinase-based analysis of ubiquitin chain architecture using Ubiquitin Chain Restriction (UbiCRest). Nat. Protoc. 2015, 10, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.; Jiang, H.; Gekara, N.O. TUBE and UbiCRest assays for elucidating polyubiquitin modifications in protein complexes. Methods Enzymol. 2019, 625, 339–350. [Google Scholar] [CrossRef]
- Mevissen, T.E.; Hospenthal, M.K.; Geurink, P.P.; Elliott, P.R.; Akutsu, M.; Arnaudo, N.; Ekkebus, R.; Kulathu, Y.; Wauer, T.; El Oualid, F.; et al. OTU Deubiquitinases Reveal Mechanisms of Linkage Specificity and Enable Ubiquitin Chain Restriction Analysis. Cell 2013, 154, 169–184. [Google Scholar] [CrossRef]
- Wertz, I.E.; Newton, K.; Seshasayee, D.; Kusam, S.; Lam, C.; Zhang, J.; Popovych, N.; Helgason, E.; Schoeffler, A.; Jeet, S.; et al. Phosphorylation and linear ubiquitin direct A20 inhibition of inflammation. Nat. Cell Biol. 2015, 528, 370–375. [Google Scholar] [CrossRef]
- Valkevich, E.M.; Sanchez, N.A.; Ge, Y.; Strieter, E.R. Middle-Down Mass Spectrometry Enables Characterization of Branched Ubiquitin Chains. Biochemistry 2014, 53, 4979–4989. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, D.S.; Hathaway, N.A.; Hanna, J.; Elsasser, S.; Rush, J.; Finley, D.; King, R.W.; Gygi, S.P. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 2006, 8, 700–710. [Google Scholar] [CrossRef]
- Rana, A.; Ge, Y.; Strieter, E.R. Ubiquitin Chain Enrichment Middle-Down Mass Spectrometry (UbiChEM-MS) Reveals Cell-Cycle Dependent Formation of Lys11/Lys48 Branched Ubiquitin Chains. J. Proteome Res. 2017, 16, 3363–3369. [Google Scholar] [CrossRef]
- Deol, K.K.; Eyles, S.J.; Strieter, E.R. Quantitative Middle-Down MS Analysis of Parkin-Mediated Ubiquitin Chain Assembly. J. Am. Soc. Mass Spectrom. 2020, 31, 1132–1139. [Google Scholar] [CrossRef]
- Swatek, K.N.; Aumayr, M.; Pruneda, J.N.; Visser, L.J.; Berryman, S.; Kueck, A.F.; Geurink, P.P.; Ovaa, H.; Van Kuppeveld, F.J.M.; Tuthill, T.J.; et al. Irreversible inactivation of ISG15 by a viral leader protease enables alternative infection detection strategies. Proc. Natl. Acad. Sci. USA 2018, 115, 2371–2376. [Google Scholar] [CrossRef]
- Hjerpe, R.; Aillet, F.; Lopitz-Otsoa, F.; Lang, V.; England, P.; Rodriguez, M.S. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 2009, 10, 1250–1258. [Google Scholar] [CrossRef]
- Yoshida, Y.; Saeki, Y.; Murakami, A.; Kawawaki, J.; Tsuchiya, H.; Yoshihara, H.; Shindo, M.; Tanaka, K. A comprehensive method for detecting ubiquitinated substrates using TR-TUBE. Proc. Natl. Acad. Sci. USA 2015, 112, 4630–4635. [Google Scholar] [CrossRef]
- Michel, M.A.; Swatek, K.N.; Hospenthal, M.K.; Komander, D. Ubiquitin Linkage-Specific Affimers Reveal Insights into K6-Linked Ubiquitin Signaling. Mol. Cell 2017, 68, 233–246.e5. [Google Scholar] [CrossRef]
- Mark, K.G.; Loveless, T.B.; Toczyski, D.P. Isolation of ubiquitinated substrates by tandem affinity purification of E3 ligase–polyubiquitin-binding domain fusions (ligase traps). Nat. Protoc. 2016, 11, 291–301. [Google Scholar] [CrossRef]
- Hoppe, T. Multiubiquitylation by E4 enzymes: ‘one size’ doesn’t fit all. Trends Biochem. Sci. 2005, 30, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Ohtake, F.; Tsuchiya, H.; Saeki, Y.; Tanaka, K. K63 ubiquitylation triggers proteasomal degradation by seeding branched ubiquitin chains. Proc. Natl. Acad. Sci. USA 2018, 115, E1401–E1408. [Google Scholar] [CrossRef]
- Liu, C.; Liu, W.; Ye, Y.; Li, W. Ufd2p synthesizes branched ubiquitin chains to promote the degradation of substrates modified with atypical chains. Nat. Commun. 2017, 8, 14274. [Google Scholar] [CrossRef]
- Hua, X.; Chu, G.; Li, Y. The Ubiquitin Enigma: Progress in the Detection and Chemical Synthesis of Branched Ubiquitin Chains. ChemBioChem 2020. [Google Scholar] [CrossRef]
- Blythe, E.E.; Olson, K.C.; Chau, V.; Deshaies, R.J. Ubiquitin- and ATP-dependent unfoldase activity of P97/VCP*NPLOC4*UFD1L is enhanced by a mutation that causes multisystem proteinopathy. Proc. Natl. Acad. Sci. USA 2017, 114, E4380–E4388. [Google Scholar] [CrossRef]
- Boughton, A.J.; Krueger, S.; Fushman, D. Branching via K11 and K48 Bestows Ubiquitin Chains with a Unique Interdomain Interface and Enhanced Affinity for Proteasomal Subunit Rpn1. Structure 2020, 28, 29–43.e6. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Liu, S.; Sagum, C.; Chen, J.; Singh, R.; Chaturvedi, A.; Horton, J.R.; Kashyap, T.R.; Fushman, D.; Cheng, X.; et al. Crosstalk between Lys63- and Lys11-polyubiquitin signaling at DNA damage sites is driven by Cezanne. Genes Dev. 2019, 33, 1702–1717. [Google Scholar] [CrossRef]
- Emmerich, C.H.; Ordureau, A.; Strickson, S.; Arthur, J.S.; Pedrioli, P.G.; Komander, D.; Cohen, P. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc. Natl. Acad. Sci. USA 2013, 110, 15247–15252. [Google Scholar] [CrossRef]
- Kim, H.T.; Kim, K.; Lledias, F.; Kisselev, A.F.; Scaglione, K.M.; Skowyra, D.; Gygi, S.P.; Goldberg, A.L. Certain Pairs of Ubiquitin-conjugating Enzymes (E2s) and Ubiquitin-Protein Ligases (E3s) Synthesize Nondegradable Forked Ubiquitin Chains Containing All Possible Isopeptide Linkages. J. Biol. Chem. 2007, 282, 17375–17386. [Google Scholar] [CrossRef]
- Peng, J.; Schwartz, D.T.; Elias, J.; Thoreen, C.C.; Cheng, D.; Marsischky, G.; Roelofs, J.; Finley, D.; Gygi, S.P. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003, 21, 921–926. [Google Scholar] [CrossRef]
- Primorac, I.; Musacchio, A. Panta rhei: The APC/C at steady state. J. Cell Biol. 2013, 201, 177–189. [Google Scholar] [CrossRef]
- Garnett, M.J.; Mansfeld, J.; Godwin, C.; Matsusaka, T.; Wu, J.; Russell, P.; Pines, J.; Venkitaraman, A.R. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat. Cell Biol. 2009, 11, 1363–1369. [Google Scholar] [CrossRef]
- Williamson, A.; Wickliffe, K.E.; Mellone, B.G.; Song, L.; Karpen, G.H.; Rape, M. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc. Natl. Acad. Sci. USA 2009, 106, 18213–18218. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Merbl, Y.; Huo, Y.; Gallop, J.L.; Tzur, A.; Kirschner, M.W. UBE2S drives elongation of K11-linked ubiquitin chains by the Anaphase-Promoting Complex. Proc. Natl. Acad. Sci. USA 2010, 107, 1355–1360. [Google Scholar] [CrossRef]
- Oh, E.; Mark, K.G.; Mocciaro, A.; Watson, E.R.; Prabu, J.R.; Cha, D.D.; Kampmann, M.; Gamarra, N.; Zhou, C.Y.; Rape, M. Gene expression and cell identity controlled by anaphase-promoting complex. Nat. Cell Biol. 2020, 579, 136–140. [Google Scholar] [CrossRef]
- Samant, R.S.; Livingston, C.M.; Sontag, E.M.; Frydman, J. Distinct proteostasis circuits cooperate in nuclear and cytoplasmic protein quality control. Nat. Cell Biol. 2018, 563, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Leto, D.E.; Morgens, D.W.; Zhang, L.; Walczak, C.P.; Elias, J.E.; Bassik, M.C.; Kopito, R.R. Genome-wide CRISPR Analysis Identifies Substrate-Specific Conjugation Modules in ER-Associated Degradation. Mol. Cell 2019, 73, 377–389.e11. [Google Scholar] [CrossRef]
- Emmerich, C.H.; Bakshi, S.; Kelsall, I.R.; Ortiz-Guerrero, J.; Shpiro, N.; Cohen, P. Lys63/Met1-hybrid ubiquitin chains are commonly formed during the activation of innate immune signalling. Biochem. Biophys. Res. Commun. 2016, 474, 452–461. [Google Scholar] [CrossRef]
- Hrdinka, M.; Fiil, B.K.; Zucca, M.; Leske, D.; Bagola, K.; Yabal, M.; Elliott, P.R.; Damgaard, R.B.; Komander, D.; Jost, P.J.; et al. CYLD Limits Lys63- and Met1-Linked Ubiquitin at Receptor Complexes to Regulate Innate Immune Signaling. Cell Rep. 2016, 14, 2846–2858. [Google Scholar] [CrossRef]
- Kulathu, Y.; Akutsu, M.; Bremm, A.; Hofmann, K.; Komander, D. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat. Struct. Mol. Biol. 2009, 16, 1328–1330. [Google Scholar] [CrossRef]
- Rahighi, S.; Ikeda, F.; Kawasaki, M.; Akutsu, M.; Suzuki, N.; Kato, R.; Kensche, T.; Uejima, T.; Bloor, S.; Komander, D.; et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell 2009, 136, 1098–1109. [Google Scholar] [CrossRef]
- Kristariyanto, Y.A.; Rehman, S.A.A.; Campbell, D.G.; Morrice, N.A.; Johnson, C.; Toth, R.; Kulathu, Y. K29-Selective Ubiquitin Binding Domain Reveals Structural Basis of Specificity and Heterotypic Nature of K29 Polyubiquitin. Mol. Cell 2015, 58, 83–94. [Google Scholar] [CrossRef]
- Johnson, E.S.; Ma, P.C.M.; Ota, I.M.; Varshavsky, A. A Proteolytic Pathway That Recognizes Ubiquitin as a Degradation Signal. J. Biol. Chem. 1995, 270, 17442–17456. [Google Scholar] [CrossRef]
- You, J.; Pickart, C.M. A HECT Domain E3 Enzyme Assembles Novel Polyubiquitin Chains. J. Biol. Chem. 2001, 276, 19871–19878. [Google Scholar] [CrossRef]
- Chu, B.W.; Kovary, K.M.; Guillaume, J.; Chen, L.-C.; Teruel, M.N.; Wandless, T.J. The E3 Ubiquitin Ligase UBE3C Enhances Proteasome Processivity by Ubiquitinating Partially Proteolyzed Substrates. J. Biol. Chem. 2013, 288, 34575–34587. [Google Scholar] [CrossRef]
- Gottlieb, C.D.; Thompson, A.C.S.; Ordureau, A.; Harper, J.W.; Kopito, R.R. Acute unfolding of a single protein immediately stimulates recruitment of ubiquitin protein ligase E3C (UBE3C) to 26S proteasomes. J. Biol. Chem. 2019, 294, 16511–16524. [Google Scholar] [CrossRef]
- Kuo, C.-L.; Goldberg, A.L. Ubiquitinated proteins promote the association of proteasomes with the deubiquitinating enzyme Usp14 and the ubiquitin ligase Ube3c. Proc. Natl. Acad. Sci. USA 2017, 114, E3404–E3413. [Google Scholar] [CrossRef]
- Nakasone, M.A.; Livnat-Levanon, N.; Glickman, M.H.; Cohen, R.E.; Fushman, D. Mixed-Linkage Ubiquitin Chains Send Mixed Messages. Structure 2013, 21, 727–740. [Google Scholar] [CrossRef]
- Michel, M.A.; Komander, D.; Elliott, P.R. Enzymatic Assembly of Ubiquitin Chains. In The Ubiquitin Proteasome System; Springer: New York, NY, USA, 2018; Volume 1844, pp. 73–84. [Google Scholar]
- Yan, L.Z.; Dawson, P.E. Synthesis of Peptides and Proteins without Cysteine Residues by Native Chemical Ligation Combined with Desulfurization. J. Am. Chem. Soc. 2001, 123, 526–533. [Google Scholar] [CrossRef]
- Kumar, K.S.A.; Haj-Yahya, M.; Olschewski, D.; Lashuel, H.A.; Brik, A. Highly Efficient and Chemoselective Peptide Ubiquitylation. Angew. Chem. 2009, 121, 8234–8238. [Google Scholar] [CrossRef]
- Yang, R.; Pasunooti, K.K.; Li, F.; Liu, X.-W.; Lui, C.F. Dual Native Chemical Ligation at Lysine. J. Am. Chem. Soc. 2009, 131, 13592–13593. [Google Scholar] [CrossRef]
- Zheng, J.-S.; Tang, S.; Huang, Y.; Liu, L. Development of New Thioester Equivalents for Protein Chemical Synthesis. Accounts Chem. Res. 2013, 46, 2475–2484. [Google Scholar] [CrossRef]
- Fang, G.-M.; Li, Y.-M.; Shen, F.; Huang, Y.; Li, J.; Lin, Y.; Cui, H.-K.; Liu, L. Protein Chemical Synthesis by Ligation of Peptide Hydrazides. Angew. Chem. 2011, 123, 7787–7791. [Google Scholar] [CrossRef]
- Gao, S.; Pan, M.; Zheng, Y.; Huang, Y.; Zheng, Q.; Sun, D.; Lu, L.; Tan, X.; Tan, X.; Lan, H.; et al. Monomer/Oligomer Quasi-Racemic Protein Crystallography. J. Am. Chem. Soc. 2016, 138, 14497–14502. [Google Scholar] [CrossRef]
- Tang, S.; Liang, L.-J.; Si, Y.-Y.; Gao, S.; Wang, J.-X.; Liang, J.; Mei, Z.; Zheng, J.-S.; Liu, L. Practical Chemical Synthesis of Atypical Ubiquitin Chains by Using an Isopeptide-Linked Ub Isomer. Angew. Chem. 2017, 129, 13518–13522. [Google Scholar] [CrossRef]
- Virdee, S.; Ye, Y.; Nguyen, D.P.; Komander, D.; Chin, J.W. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat. Chem. Biol. 2010, 6, 750–757. [Google Scholar] [CrossRef] [PubMed]
- Dixon, E.K.; Castañeda, C.A.; Kashyap, T.R.; Wang, Y.; Fushman, D. Nonenzymatic assembly of branched polyubiquitin chains for structural and biochemical studies. Bioorg. Med. Chem. 2013, 21, 3421–3429. [Google Scholar] [CrossRef]
- Virdee, S.; Kapadnis, P.B.; Elliott, T.; Lang, K.; Madrzak, J.; Nguyen, D.P.; Riechmann, L.; Chin, J.W. Traceless and Site-Specific Ubiquitination of Recombinant Proteins. J. Am. Chem. Soc. 2011, 133, 10708–10711. [Google Scholar] [CrossRef]
- Valkevich, E.M.; Guenette, R.G.; Sanchez, N.A.; Chen, Y.-C.; Ge, Y.; Strieter, E.R. Forging Isopeptide Bonds Using Thiol–Ene Chemistry: Site-Specific Coupling of Ubiquitin Molecules for Studying the Activity of Isopeptidases. J. Am. Chem. Soc. 2012, 134, 6916–6919. [Google Scholar] [CrossRef]
- Chu, G.; Hua, X.; Zuo, C.; Chen, C.; Meng, X.; Zhang, Z.; Fu, Y.; Shi, J.; Li, Y.-M. Efficient Semi-Synthesis of Atypical Ubiquitin Chains and Ubiquitin-Based Probes Forged by Thioether Isopeptide Bonds. Chem. Eur. J. 2019, 25, 16668–16675. [Google Scholar] [CrossRef]
- Si, Y.; Liang, L.; Tang, S.; Qi, Y.; Huang, Y.; Liu, L. Semi-synthesis of disulfide-linked branched tri-ubiquitin mimics. Sci. China Ser. B Chem. 2018, 61, 412–417. [Google Scholar] [CrossRef]
- Schneider, T.; Schneider, D.; Rösner, D.; Malhotra, S.; Mortensen, F.; Mayer, T.U.; Scheffner, M.; Marx, A. Dissecting Ubiquitin Signaling with Linkage-Defined and Protease Resistant Ubiquitin Chains. Angew. Chem. Int. Ed. 2014, 53, 12925–12929. [Google Scholar] [CrossRef]
- Rösner, D.; Schneider, T.; Schneider, D.; Scheffner, M.; Marx, A. Click chemistry for targeted protein ubiquitylation and ubiquitin chain formation. Nat. Protoc. 2015, 10, 1594–1611. [Google Scholar] [CrossRef]
- Zhao, X.; Mißun, M.; Schneider, T.; Müller, F.; Lutz, J.; Scheffner, M.; Marx, A.; Kovermann, M. Artificially Linked Ubiquitin Dimers Characterised Structurally and Dynamically by NMR Spectroscopy. ChemBioChem 2019, 20, 1772–1777. [Google Scholar] [CrossRef]
- Zhao, X.; Lutz, J.; Höllmüller, E.; Scheffner, M.; Marx, A.; Stengel, F. Identification of Proteins Interacting with Ubiquitin Chains. Angew. Chem. Int. Ed. 2017, 56, 15764–15768. [Google Scholar] [CrossRef]
- Zhao, X.; Scheffner, M.; Marx, A. Assembly of branched ubiquitin oligomers by click chemistry. Chem. Commun. 2019, 55, 13093–13095. [Google Scholar] [CrossRef]
- Stanley, M.; Virdee, S. Genetically Directed Production of Recombinant, Isosteric and Nonhydrolysable Ubiquitin Conjugates. ChemBioChem 2016, 17, 1472–1480. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.-K.; Kurkute, P.; Li, C.-L.; Wang, Y.-H.; Chen, P.-J.; Lin, S.-Y.; Wang, Y.-S. Revealing USP7 Deubiquitinase Substrate Specificity by Unbiased Synthesis of Ubiquitin Tagged SUMO2. Biochemistry 2020, 59, 3796–3801. [Google Scholar] [CrossRef]
- Meledin, R.; Mali, S.M.; Singh, S.K.; Brik, A. Protein ubiquitination via dehydroalanine: Development and insights into the diastereoselective 1,4-addition step. Org. Biomol. Chem. 2016, 14, 4817–4823. [Google Scholar] [CrossRef] [PubMed]
- Chu, G.-C.; Pan, M.; Li, J.; Liu, S.; Zuo, C.; Tong, Z.-B.; Bai, J.-S.; Gong, Q.; Ai, H.; Fan, J.; et al. Cysteine-Aminoethylation-Assisted Chemical Ubiquitination of Recombinant Histones. J. Am. Chem. Soc. 2019, 141, 3654–3663. [Google Scholar] [CrossRef]
- Chatterjee, C.; McGinty, R.K.; Fierz, B.; Muir, T.W. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat. Chem. Biol. 2010, 6, 267–269. [Google Scholar] [CrossRef]
- Guo, L.-T.; Wang, Y.-S.; Nakamura, A.; Eiler, D.; Kavran, J.M.; Wong, M.; Kiessling, L.L.; Steitz, T.A.; O’Donoghue, P.; Söll, D. Polyspecific pyrrolysyl-tRNA synthetases from directed evolution. Proc. Natl. Acad. Sci. USA 2014, 111, 16724–16729. [Google Scholar] [CrossRef]
- Bett, J.S.; Ritorto, M.S.; Ewan, R.; Jaffray, E.G.; Virdee, S.; Chin, J.W.; Knebel, A.; Kurz, T.; Trost, M.; Tatham, M.H.; et al. Ubiquitin C-terminal hydrolases cleave isopeptide- and peptide-linked ubiquitin from structured proteins but do not edit ubiquitin homopolymers. Biochem. J. 2015, 466, 489–498. [Google Scholar] [CrossRef]
- Wang, Z.U.; Wang, Y.-S.; Pai, P.-J.; Russell, W.K.; Russell, D.H.; Liu, W.R. A Facile Method to Synthesize Histones with Posttranslational Modification Mimics. Biochemistry 2012, 51, 5232–5234. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, H.; Burana, D.; Ohtake, F.; Arai, N.; Kaiho, A.; Komada, M.; Tanaka, K.; Saeki, Y. Ub-ProT reveals global length and composition of protein ubiquitylation in cells. Nat. Commun. 2018, 9, 524. [Google Scholar] [CrossRef]
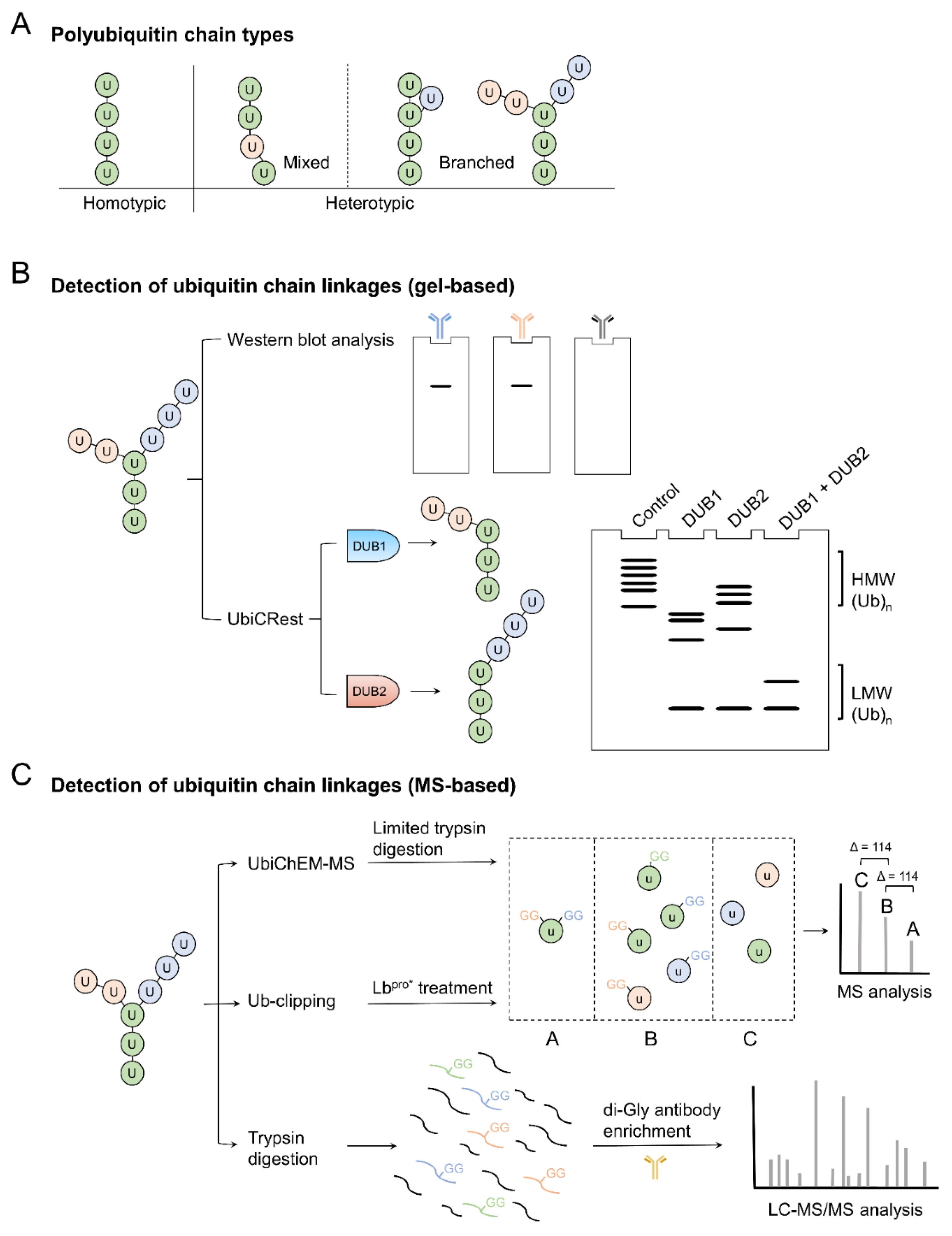
| DUBs (Favored Ub Linkages) |
|---|
| USP21 (non-specific) |
| vOTU (non-specific, except M1) |
| OTUD3 (K6, K11) |
| Cezanne (K11) |
| OTUD2 (K11, K27, K29, K33) |
| TRABID (K29, K33, K63) |
| OTUB1 (K48) |
| OTUD1, AMSH (K63) |
| OTULIN (M1) |
| Enzyme | Linkage | |
|---|---|---|
| E2-E3 pairs | UBE2S-APC/C + UBE2C-APC/C | K11/K48 branched |
| E3s (or E3 + E4) | UBR4 + UBR5 | K11/K48 branched |
| Doa10 or Hrd1 + Ubr1 or San1 | ||
| UBR4 + KCMF1 | ||
| TRAF6 or Pellino 1/2 + LUBAC | K63/M1 heterotypic | |
| ITCH or WWP1 + UBR5 or HUWE1 | K48/K63 branched | |
| TRAF6 + HUWE1 | ||
| Ufd4p + Ufd2p | K29/K48 branched | |
| UBE3C | ||
| DUBs | A20 (inhibited) | K63/M1 heterotypic |
| CYLD (inhibited) | K48/K63 branched | |
| UBDs | Rpn10 | K11/K48 branched |
| Rpn1 | ||
| p97/VCP | ||
| HHR23A | ||
| TAK1 (K63), IKK (M1) | K63/M1 heterotypic |
| Entry | Linkage Structure 1 | Hydrolysable | Topology | Position | Refs. |
|---|---|---|---|---|---|
| 1 |  | O | homogeneous | 6, 29 | [63,64,65,66,67] |
| branched | 11/33, 11/48, 11/63 | ||||
| 2 | 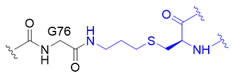 | O | homogeneous | 48, 63 | [68] |
| branched | 6/48, 11/48, 48/63 | ||||
| 3 |  | O | homogeneous | 6, 27, 33 | [69] |
| branched | 11/48 | ||||
| 4 | 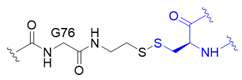 | O | branched | 11/48 | [70] |
| 5 | 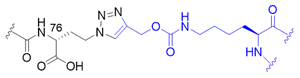 | X | homogeneous | 11, 27, 29, 48 | [71,72] |
| 6 | 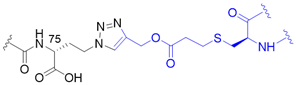 | X | homogeneous | 6, 11, 27, 29, 33, 48, 63 | [73,74,75] |
| branched | 6/11, 11/48, 11/63, 6/11/48 | ||||
| 7 | 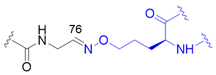 | X | homogeneous | 6, 48 | [76] |
| 8 | 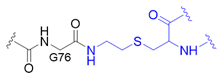 | O | homogeneous | 11 | [77,78] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.-S.; Wu, K.-P.; Jiang, H.-K.; Kurkute, P.; Chen, R.-H. Branched Ubiquitination: Detection Methods, Biological Functions and Chemical Synthesis. Molecules 2020, 25, 5200. https://doi.org/10.3390/molecules25215200
Wang Y-S, Wu K-P, Jiang H-K, Kurkute P, Chen R-H. Branched Ubiquitination: Detection Methods, Biological Functions and Chemical Synthesis. Molecules. 2020; 25(21):5200. https://doi.org/10.3390/molecules25215200
Chicago/Turabian StyleWang, Yane-Shih, Kuen-Phon Wu, Han-Kai Jiang, Prashant Kurkute, and Ruey-Hwa Chen. 2020. "Branched Ubiquitination: Detection Methods, Biological Functions and Chemical Synthesis" Molecules 25, no. 21: 5200. https://doi.org/10.3390/molecules25215200
APA StyleWang, Y.-S., Wu, K.-P., Jiang, H.-K., Kurkute, P., & Chen, R.-H. (2020). Branched Ubiquitination: Detection Methods, Biological Functions and Chemical Synthesis. Molecules, 25(21), 5200. https://doi.org/10.3390/molecules25215200







