P-TEFb as A Promising Therapeutic Target
Abstract
:1. Introduction
2. Discovery of P-TEFb
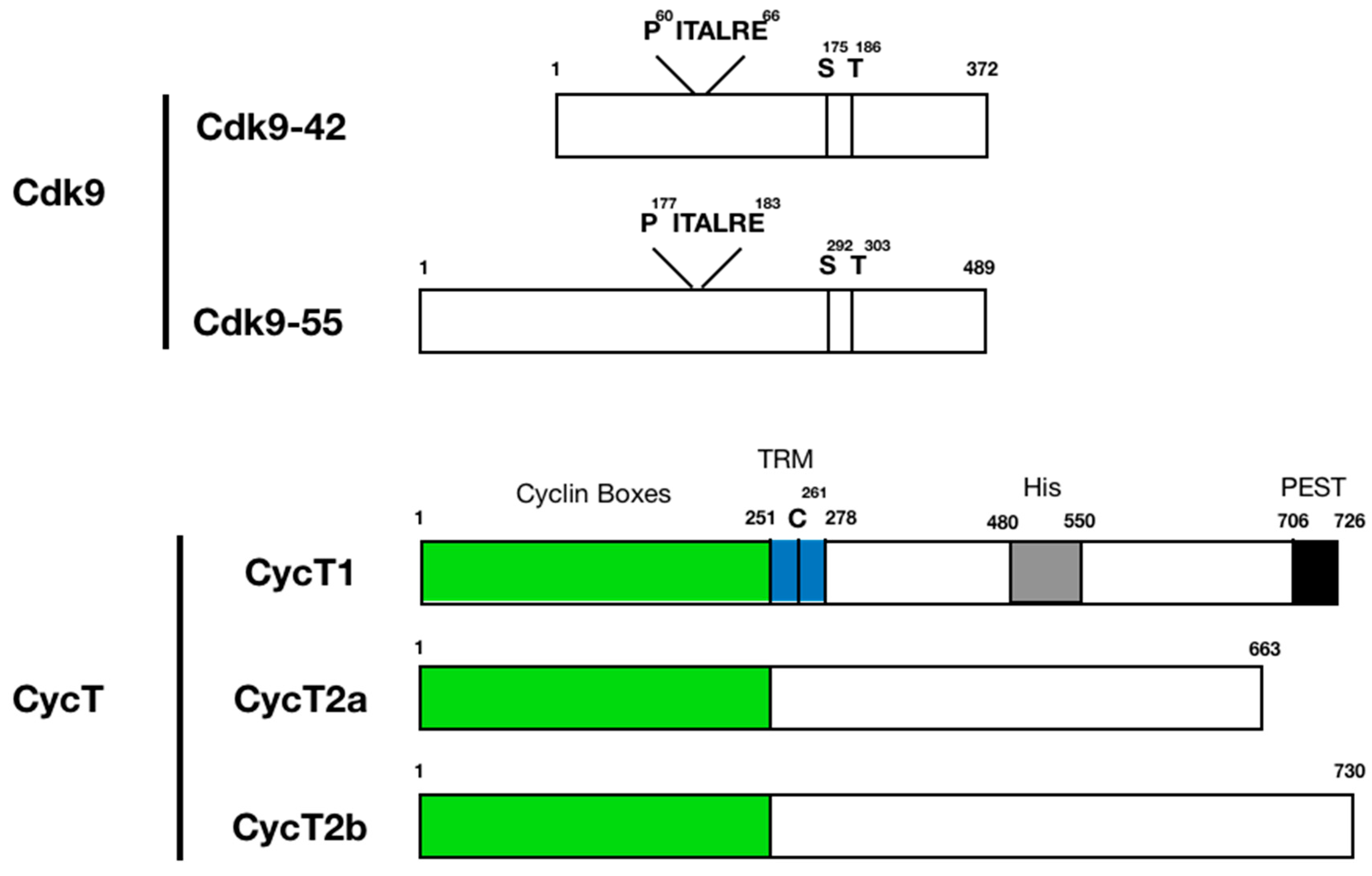
3. Protein Structure and Function of P-TEFb Subunits
4. Substrates of CDK9
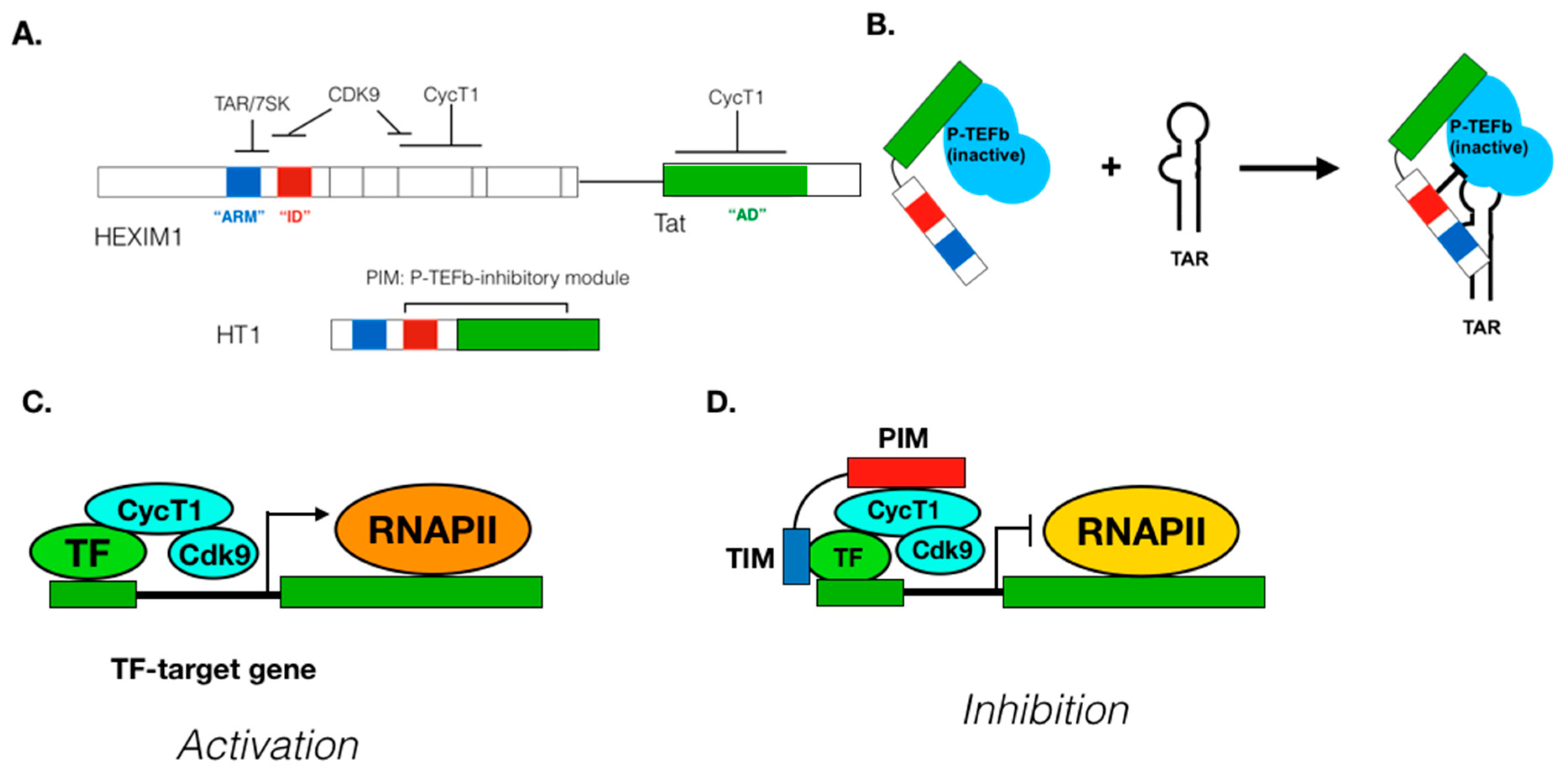
5. Regulation of P-TEFb Functions in Cells
6. Mechanism of P-TEFb Recruitment to Its Target Genes
7. P-TEFb and Human Diseases
8. Approaches to Manipulate P-TEFb Functions
9. Suppressing P-TEFb Activity
10. Increasing P-TEFb Activity
11. Potential Problems/Side Effects
12. Perspectives and Future Directions
Funding
Acknowledgments
Conflicts of Interest
References
- Cramer, P. Eukaryotic Transcription Turns 50. Cell 2019, 179, 808–812. [Google Scholar] [CrossRef]
- Kornberg, R.D. The molecular basis of eukaryotic transcription. Proc. Natl. Acad. Sci. USA 2007, 104, 12955–12961. [Google Scholar] [CrossRef] [Green Version]
- Lis, J.T. A 50 year history of technologies that drove discovery in eukaryotic transcription regulation. Nat. Struct. Mol. Biol. 2019, 26, 777–782. [Google Scholar] [CrossRef]
- Roeder, R.G. 50+ years of eukaryotic transcription: An expanding universe of factors and mechanisms. Nat. Struct. Mol. Biol. 2019, 26, 783–791. [Google Scholar] [CrossRef]
- Jonkers, I.; Lis, J.T. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015, 16, 167–177. [Google Scholar] [CrossRef] [Green Version]
- Rahl, P.B.; Young, R.A. MYC and transcription elongation. Cold Spring Harb. Perspect. Med. 2014, 4, a020990. [Google Scholar] [CrossRef] [Green Version]
- Saldi, T.; Cortazar, M.A.; Sheridan, R.M.; Bentley, D.L. Coupling of RNA Polymerase II Transcription Elongation with Pre-mRNA Splicing. J. Mol. Biol. 2016, 428, 2623–2635. [Google Scholar] [CrossRef] [Green Version]
- Nechaev, S.; Adelman, K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim. Biophys. Acta 2011, 1809, 34–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Q.; Li, T.; Price, D.H. RNA polymerase II elongation control. Annu. Rev. Biochem. 2012, 81, 119–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilchrist, D.A.; Fargo, D.C.; Adelman, K. Using ChIP-chip and ChIP-seq to study the regulation of gene expression: Genome-wide localization studies reveal widespread regulation of transcription elongation. Methods 2009, 48, 398–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AJ, C.Q.; Bugai, A.; Barboric, M. Cracking the control of RNA polymerase II elongation by 7SK snRNP and P-TEFb. Nucleic Acids Res. 2016, 44, 7527–7539. [Google Scholar] [CrossRef] [Green Version]
- Bres, V.; Yoh, S.M.; Jones, K.A. The multi-tasking P-TEFb complex. Curr. Opin. Cell Biol. 2008, 20, 334–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.; Yik, J.H.; Lew, Q.J.; Chao, S.H. Brd4 and HEXIM1: Multiple roles in P-TEFb regulation and cancer. Biomed. Res. Int. 2014, 2014, 232870. [Google Scholar] [CrossRef] [PubMed]
- Peterlin, B.M.; Price, D.H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 2006, 23, 297–305. [Google Scholar] [CrossRef]
- Zhou, Q.; Yik, J.H. The Yin and Yang of P-TEFb regulation: Implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol. Mol. Biol. Rev. 2006, 70, 646–659. [Google Scholar] [CrossRef] [Green Version]
- Eick, D.; Geyer, M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 2013, 113, 8456–8490. [Google Scholar] [CrossRef]
- Conaway, J.W.; Conaway, R.C. Transcription elongation and human disease. Annu. Rev. Biochem. 1999, 68, 301–319. [Google Scholar] [CrossRef]
- Bacon, C.W.; D’Orso, I. CDK9: A signaling hub for transcriptional control. Transcription 2019, 10, 57–75. [Google Scholar] [CrossRef]
- Romano, G.; Giordano, A. Role of the cyclin-dependent kinase 9-related pathway in mammalian gene expression and human diseases. Cell Cycle 2008, 7, 3664–3668. [Google Scholar] [CrossRef] [Green Version]
- Bensaude, O. Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription 2011, 2, 103–108. [Google Scholar] [CrossRef] [Green Version]
- Boffo, S.; Damato, A.; Alfano, L.; Giordano, A. CDK9 inhibitors in acute myeloid leukemia. J. Exp. Clin. Cancer Res. 2018, 37, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bose, P.; Simmons, G.L.; Grant, S. Cyclin-dependent kinase inhibitor therapy for hematologic malignancies. Expert Opin. Investig. Drugs 2013, 22, 723–738. [Google Scholar] [CrossRef] [PubMed]
- Echalier, A.; Endicott, J.A.; Noble, M.E. Recent developments in cyclin-dependent kinase biochemical and structural studies. Biochim. Biophys. Acta 2010, 1804, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, G.I. Cyclin-dependent kinase pathways as targets for cancer treatment. J. Clin. Oncol. 2006, 24, 1770–1783. [Google Scholar] [CrossRef]
- Marshall, N.F.; Price, D.H. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol. Cell Biol. 1992, 12, 2078–2090. [Google Scholar] [CrossRef] [Green Version]
- Marshall, N.F.; Price, D.H. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 1995, 270, 12335–12338. [Google Scholar] [CrossRef] [Green Version]
- Kao, S.Y.; Calman, A.F.; Luciw, P.A.; Peterlin, B.M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 1987, 330, 489–493. [Google Scholar] [CrossRef]
- Feinberg, M.B.; Baltimore, D.; Frankel, A.D. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc. Natl. Acad. Sci. USA 1991, 88, 4045–4049. [Google Scholar] [CrossRef] [Green Version]
- Kato, H.; Sumimoto, H.; Pognonec, P.; Chen, C.H.; Rosen, C.A.; Roeder, R.G. HIV-1 Tat acts as a processivity factor in vitro in conjunction with cellular elongation factors. Genes Dev. 1992, 6, 655–666. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Welsh, T.M.; Peterlin, B.M. The human immunodeficiency virus type 1 long terminal repeat specifies two different transcription complexes, only one of which is regulated by Tat. J. Virol. 1993, 67, 1752–1760. [Google Scholar] [CrossRef] [Green Version]
- Herrmann, C.H.; Rice, A.P. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: Candidate for a Tat cofactor. J. Virol. 1995, 69, 1612–1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grana, X.; De Luca, A.; Sang, N.; Fu, Y.; Claudio, P.P.; Rosenblatt, J.; Morgan, D.O.; Giordano, A. PITALRE, a nuclear CDC2-related protein kinase that phosphorylates the retinoblastoma protein in vitro. Proc. Natl. Acad. Sci. USA 1994, 91, 3834–3838. [Google Scholar] [CrossRef] [Green Version]
- Mancebo, H.S.; Lee, G.; Flygare, J.; Tomassini, J.; Luu, P.; Zhu, Y.; Peng, J.; Blau, C.; Hazuda, D.; Price, D.; et al. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997, 11, 2633–2644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Pe’ery, T.; Peng, J.; Ramanathan, Y.; Marshall, N.; Marshall, T.; Amendt, B.; Mathews, M.B.; Price, D.H. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997, 11, 2622–2632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, P.; Garber, M.E.; Fang, S.M.; Fischer, W.H.; Jones, K.A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 1998, 92, 451–462. [Google Scholar] [CrossRef] [Green Version]
- Garber, M.E.; Wei, P.; KewalRamani, V.N.; Mayall, T.P.; Herrmann, C.H.; Rice, A.P.; Littman, D.R.; Jones, K.A. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998, 12, 3512–3527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramanathan, Y.; Reza, S.M.; Young, T.M.; Mathews, M.B.; Pe’ery, T. Human and rodent transcription elongation factor P-TEFb: Interactions with human immunodeficiency virus type 1 tat and carboxy-terminal domain substrate. J. Virol. 1999, 73, 5448–5458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bieniasz, P.D.; Grdina, T.A.; Bogerd, H.P.; Cullen, B.R. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 1998, 17, 7056–7065. [Google Scholar] [CrossRef] [Green Version]
- Fujinaga, K.; Taube, R.; Wimmer, J.; Cujec, T.P.; Peterlin, B.M. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc. Natl. Acad. Sci. USA 1999, 96, 1285–1290. [Google Scholar] [CrossRef] [Green Version]
- Kwak, Y.T.; Ivanov, D.; Guo, J.; Nee, E.; Gaynor, R.B. Role of the human and murine cyclin T proteins in regulating HIV-1 tat-activation. J. Mol. Biol. 1999, 288, 57–69. [Google Scholar] [CrossRef]
- Hart, C.E.; Ou, C.Y.; Galphin, J.C.; Moore, J.; Bacheler, L.T.; Wasmuth, J.J.; Petteway, S.R., Jr.; Schochetman, G. Human chromosome 12 is required for elevated HIV-1 expression in human-hamster hybrid cells. Science 1989, 246, 488–491. [Google Scholar] [CrossRef] [PubMed]
- Winslow, B.J.; Trono, D. The blocks to human immunodeficiency virus type 1 Tat and Rev functions in mouse cell lines are independent. J. Virol. 1993, 67, 2349–2354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karn, J. Tackling Tat. J. Mol. Biol. 1999, 293, 235–254. [Google Scholar] [CrossRef]
- Sano, M.; Abdellatif, M.; Oh, H.; Xie, M.; Bagella, L.; Giordano, A.; Michael, L.H.; DeMayo, F.J.; Schneider, M.D. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat. Med. 2002, 8, 1310–1317. [Google Scholar] [CrossRef]
- Barboric, M.; Nissen, R.M.; Kanazawa, S.; Jabrane-Ferrat, N.; Peterlin, B.M. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 2001, 8, 327–337. [Google Scholar] [CrossRef]
- Eberhardy, S.R.; Farnham, P.J. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J. Biol. Chem. 2002, 277, 40156–40162. [Google Scholar] [CrossRef] [Green Version]
- Kanazawa, S.; Soucek, L.; Evan, G.; Okamoto, T.; Peterlin, B.M. c-Myc recruits P-TEFb for transcription, cellular proliferation and apoptosis. Oncogene 2003, 22, 5707–5711. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.K.; Duan, H.O.; Chang, C. Androgen receptor interacts with the positive elongation factor P-TEFb and enhances the efficiency of transcriptional elongation. J. Biol. Chem. 2001, 276, 9978–9984. [Google Scholar] [CrossRef] [Green Version]
- Wittmann, B.M.; Fujinaga, K.; Deng, H.; Ogba, N.; Montano, M.M. The breast cell growth inhibitor, estrogen down regulated gene 1, modulates a novel functional interaction between estrogen receptor alpha and transcriptional elongation factor cyclin T1. Oncogene 2005, 24, 5576–5588. [Google Scholar] [CrossRef] [Green Version]
- Mitra, P.; Pereira, L.A.; Drabsch, Y.; Ramsay, R.G.; Gonda, T.J. Estrogen receptor-alpha recruits P-TEFb to overcome transcriptional pausing in intron 1 of the MYB gene. Nucleic Acids Res. 2012, 40, 5988–6000. [Google Scholar] [CrossRef] [Green Version]
- Franco, L.C.; Morales, F.; Boffo, S.; Giordano, A. CDK9: A key player in cancer and other diseases. J. Cell Biochem 2018, 119, 1273–1284. [Google Scholar] [CrossRef] [PubMed]
- Paparidis, N.F.; Durvale, M.C.; Canduri, F. The emerging picture of CDK9/P-TEFb: More than 20 years of advances since PITALRE. Mol. Biosyst. 2017, 13, 246–276. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.K.; Mochizuki, K.; Zhou, M.; Jeong, H.S.; Brady, J.N.; Ozato, K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 2005, 19, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Yik, J.H.; Chen, R.; He, N.; Jang, M.K.; Ozato, K.; Zhou, Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 2005, 19, 535–545. [Google Scholar] [CrossRef]
- Takahashi, H.; Parmely, T.J.; Sato, S.; Tomomori-Sato, C.; Banks, C.A.; Kong, S.E.; Szutorisz, H.; Swanson, S.K.; Martin-Brown, S.; Washburn, M.P.; et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 2011, 146, 92–104. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.; Smith, E.R.; Takahashi, H.; Lai, K.C.; Martin-Brown, S.; Florens, L.; Washburn, M.P.; Conaway, J.W.; Conaway, R.C.; Shilatifard, A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell 2010, 37, 429–437. [Google Scholar] [CrossRef] [Green Version]
- Lis, J.T.; Mason, P.; Peng, J.; Price, D.H.; Werner, J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000, 14, 792–803. [Google Scholar]
- Hargreaves, D.C.; Horng, T.; Medzhitov, R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 2009, 138, 129–145. [Google Scholar] [CrossRef] [Green Version]
- Rahl, P.B.; Lin, C.Y.; Seila, A.C.; Flynn, R.A.; McCuine, S.; Burge, C.B.; Sharp, P.A.; Young, R.A. c-Myc regulates transcriptional pause release. Cell 2010, 141, 432–445. [Google Scholar] [CrossRef] [Green Version]
- Kohoutek, J. P-TEFb—The final frontier. Cell Div. 2009, 4, 19. [Google Scholar] [CrossRef] [Green Version]
- Taube, R.; Peterlin, M. Lost in transcription: Molecular mechanisms that control HIV latency. Viruses 2013, 5, 902–927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giacinti, C.; Bagella, L.; Puri, P.L.; Giordano, A.; Simone, C. MyoD recruits the cdk9/cyclin T2 complex on myogenic-genes regulatory regions. J. Cell Physiol. 2006, 206, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Ray, S.; Brasier, A.R. The functional role of an interleukin 6-inducible CDK9.STAT3 complex in human gamma-fibrinogen gene expression. J. Biol. Chem. 2007, 282, 37091–37102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eberhardy, S.R.; Farnham, P.J. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J. Biol. Chem. 2001, 276, 48562–48571. [Google Scholar] [CrossRef] [Green Version]
- Nojima, M.; Huang, Y.; Tyagi, M.; Kao, H.Y.; Fujinaga, K. The positive transcription elongation factor b is an essential cofactor for the activation of transcription by myocyte enhancer factor 2. J. Mol. Biol. 2008, 382, 275–287. [Google Scholar] [CrossRef] [Green Version]
- Kanazawa, S.; Okamoto, T.; Peterlin, B.M. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity 2000, 12, 61–70. [Google Scholar] [CrossRef] [Green Version]
- Oven, I.; Brdickova, N.; Kohoutek, J.; Vaupotic, T.; Narat, M.; Peterlin, B.M. AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol. Cell Biol. 2007, 27, 8815–8823. [Google Scholar] [CrossRef] [Green Version]
- Arter, J.; Wegner, M. Transcription factors Sox10 and Sox2 functionally interact with positive transcription elongation factor b in Schwann cells. J. Neurochem. 2015, 132, 384–393. [Google Scholar] [CrossRef]
- Bottardi, S.; Mavoungou, L.; Pak, H.; Daou, S.; Bourgoin, V.; Lakehal, Y.A.; Affar el, B.; Milot, E. The IKAROS interaction with a complex including chromatin remodeling and transcription elongation activities is required for hematopoiesis. Plos Genet. 2014, 10, e1004827. [Google Scholar] [CrossRef]
- Iankova, I.; Petersen, R.K.; Annicotte, J.S.; Chavey, C.; Hansen, J.B.; Kratchmarova, I.; Sarruf, D.; Benkirane, M.; Kristiansen, K.; Fajas, L. Peroxisome proliferator-activated receptor gamma recruits the positive transcription elongation factor b complex to activate transcription and promote adipogenesis. Mol. Endocrinol. 2006, 20, 1494–1505. [Google Scholar] [CrossRef]
- Meier, N.; Krpic, S.; Rodriguez, P.; Strouboulis, J.; Monti, M.; Krijgsveld, J.; Gering, M.; Patient, R.; Hostert, A.; Grosveld, F. Novel binding partners of Ldb1 are required for haematopoietic development. Development 2006, 133, 4913–4923. [Google Scholar] [CrossRef] [Green Version]
- Shore, S.M.; Byers, S.A.; Dent, P.; Price, D.H. Characterization of Cdk9(55) and differential regulation of two Cdk9 isoforms. Gene 2005, 350, 51–58. [Google Scholar] [CrossRef]
- Giacinti, C.; Musaro, A.; De Falco, G.; Jourdan, I.; Molinaro, M.; Bagella, L.; Simone, C.; Giordano, A. Cdk9-55: A new player in muscle regeneration. J. Cell Physiol. 2008, 216, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Morales, F.; Giordano, A. Overview of CDK9 as a target in cancer research. Cell Cycle 2016, 15, 519–527. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Herrmann, C.H. Differential localization and expression of the Cdk9 42k and 55k isoforms. J. Cell Physiol. 2005, 203, 251–260. [Google Scholar] [CrossRef]
- Wang, S.; Fischer, P.M. Cyclin-dependent kinase 9: A key transcriptional regulator and potential drug target in oncology, virology and cardiology. Trends Pharm. Sci. 2008, 29, 302–313. [Google Scholar] [CrossRef]
- Sonawane, Y.A.; Taylor, M.A.; Napoleon, J.V.; Rana, S.; Contreras, J.I.; Natarajan, A. Cyclin Dependent Kinase 9 Inhibitors for Cancer Therapy. J. Med. Chem. 2016, 59, 8667–8684. [Google Scholar] [CrossRef]
- Marshall, R.M.; Grana, X. Mechanisms controlling CDK9 activity. Front. Biosci. 2006, 11, 2598–2613. [Google Scholar] [CrossRef]
- Li, Q.; Price, J.P.; Byers, S.A.; Cheng, D.; Peng, J.; Price, D.H. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J. Biol. Chem. 2005, 280, 28819–28826. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Dow, E.C.; Liang, Y.Y.; Ramakrishnan, R.; Liu, H.; Sung, T.L.; Lin, X.; Rice, A.P. Phosphatase PPM1A regulates phosphorylation of Thr-186 in the Cdk9 T-loop. J. Biol. Chem. 2008, 283, 33578–33584. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Yang, Z.; Zhou, Q. Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J. Biol. Chem. 2004, 279, 4153–4160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramakrishnan, R.; Dow, E.C.; Rice, A.P. Characterization of Cdk9 T-loop phosphorylation in resting and activated CD4(+) T lymphocytes. J. Leukoc Biol. 2009, 86, 1345–1350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramakrishnan, R.; Rice, A.P. Cdk9 T-loop phosphorylation is regulated by the calcium signaling pathway. J. Cell Physiol. 2012, 227, 609–617. [Google Scholar] [CrossRef] [Green Version]
- Mbonye, U.R.; Wang, B.; Gokulrangan, G.; Chance, M.R.; Karn, J. Phosphorylation of HEXIM1 at Tyr271 and Tyr274 Promotes Release of P-TEFb from the 7SK snRNP Complex and Enhances Proviral HIV Gene Expression. Proteomics 2015, 15, 2078–2086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mbonye, U.R.; Gokulrangan, G.; Datt, M.; Dobrowolski, C.; Cooper, M.; Chance, M.R.; Karn, J. Phosphorylation of CDK9 at Ser175 enhances HIV transcription and is a marker of activated P-TEFb in CD4(+) T lymphocytes. PLoS Pathog 2013, 9, e1003338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ammosova, T.; Obukhov, Y.; Kotelkin, A.; Breuer, D.; Beullens, M.; Gordeuk, V.R.; Bollen, M.; Nekhai, S. Protein phosphatase-1 activates CDK9 by dephosphorylating Ser175. PLoS ONE 2011, 6, e18985. [Google Scholar] [CrossRef]
- Anand, K.; Schulte, A.; Fujinaga, K.; Scheffzek, K.; Geyer, M. Cyclin box structure of the P-TEFb subunit cyclin T1 derived from a fusion complex with EIAV tat. J. Mol. Biol. 2007, 370, 826–836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baumli, S.; Lolli, G.; Lowe, E.D.; Troiani, S.; Rusconi, L.; Bullock, A.N.; Debreczeni, J.E.; Knapp, S.; Johnson, L.N. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J. 2008, 27, 1907–1918. [Google Scholar] [CrossRef]
- Jadlowsky, J.K.; Nojima, M.; Okamoto, T.; Fujinaga, K. Dominant negative mutant cyclin T1 proteins that inhibit HIV transcription by forming a kinase inactive complex with Tat. J. Gen. Virol. 2008, 89, 2783–2787. [Google Scholar] [CrossRef]
- Jadlowsky, J.K.; Nojima, M.; Schulte, A.; Geyer, M.; Okamoto, T.; Fujinaga, K. Dominant negative mutant cyclin T1 proteins inhibit HIV transcription by specifically degrading Tat. Retrovirology 2008, 5, 63. [Google Scholar] [CrossRef] [Green Version]
- Kuzmina, A.; Verstraete, N.; Galker, S.; Maatook, M.; Bensaude, O.; Taube, R. A single point mutation in cyclin T1 eliminates binding to Hexim1, Cdk9 and RNA but not to AFF4 and enforces repression of HIV transcription. Retrovirology 2014, 11, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verstraete, N.; Kuzmina, A.; Diribarne, G.; Nguyen, V.T.; Kobbi, L.; Ludanyi, M.; Taube, R.; Bensaude, O. A Cyclin T1 point mutation that abolishes positive transcription elongation factor (P-TEFb) binding to Hexim1 and HIV tat. Retrovirology 2014, 11, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, J.; Zhu, Y.; Milton, J.T.; Price, D.H. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998, 12, 755–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taube, R.; Lin, X.; Irwin, D.; Fujinaga, K.; Peterlin, B.M. Interaction between P-TEFb and the C-terminal domain of RNA polymerase II activates transcriptional elongation from sites upstream or downstream of target genes. Mol. Cell Biol. 2002, 22, 321–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, H.; Yu, D.; Hansen, A.S.; Ganguly, S.; Liu, R.; Heckert, A.; Darzacq, X.; Zhou, Q. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 2018, 558, 318–323. [Google Scholar] [CrossRef] [PubMed]
- De Luca, A.; Esposito, V.; Baldi, A.; Claudio, P.P.; Fu, Y.; Caputi, M.; Pisano, M.M.; Baldi, F.; Giordano, A. CDC2-related kinase PITALRE phosphorylates pRb exclusively on serine and is widely expressed in human tissues. J. Cell Physiol. 1997, 172, 265–273. [Google Scholar] [CrossRef]
- Ni, Z.; Saunders, A.; Fuda, N.J.; Yao, J.; Suarez, J.R.; Webb, W.W.; Lis, J.T. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol. Cell Biol. 2008, 28, 1161–1170. [Google Scholar] [CrossRef] [Green Version]
- Schuller, R.; Forne, I.; Straub, T.; Schreieck, A.; Texier, Y.; Shah, N.; Decker, T.M.; Cramer, P.; Imhof, A.; Eick, D. Heptad-Specific Phosphorylation of RNA Polymerase II CTD. Mol. Cell 2016, 61, 305–314. [Google Scholar] [CrossRef] [Green Version]
- Fujita, T.; Ryser, S.; Piuz, I.; Schlegel, W. Up-regulation of P-TEFb by the MEK1-extracellular signal-regulated kinase signaling pathway contributes to stimulated transcription elongation of immediate early genes in neuroendocrine cells. Mol. Cell Biol. 2008, 28, 1630–1643. [Google Scholar] [CrossRef] [Green Version]
- Mayer, A.; Lidschreiber, M.; Siebert, M.; Leike, K.; Soding, J.; Cramer, P. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 2010, 17, 1272–1278. [Google Scholar] [CrossRef] [Green Version]
- Ramanathan, Y.; Rajpara, S.M.; Reza, S.M.; Lees, E.; Shuman, S.; Mathews, M.B.; Pe’ery, T. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. J. Biol. Chem. 2001, 276, 10913–10920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, M.; Halanski, M.A.; Radonovich, M.F.; Kashanchi, F.; Peng, J.; Price, D.H.; Brady, J.N. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell Biol. 2000, 20, 5077–5086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czudnochowski, N.; Bosken, C.A.; Geyer, M. Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat. Commun. 2012, 3, 842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujinaga, K.; Irwin, D.; Huang, Y.; Taube, R.; Kurosu, T.; Peterlin, B.M. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol. Cell Biol. 2004, 24, 787–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narita, T.; Yamaguchi, Y.; Yano, K.; Sugimoto, S.; Chanarat, S.; Wada, T.; Kim, D.K.; Hasegawa, J.; Omori, M.; Inukai, N.; et al. Human transcription elongation factor NELF: Identification of novel subunits and reconstitution of the functionally active complex. Mol. Cell Biol. 2003, 23, 1863–1873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jadlowsky, J.K.; Wong, J.Y.; Graham, A.C.; Dobrowolski, C.; Devor, R.L.; Adams, M.D.; Fujinaga, K.; Karn, J. Negative elongation factor is required for the maintenance of proviral latency but does not induce promoter-proximal pausing of RNA polymerase II on the HIV long terminal repeat. Mol. Cell Biol. 2014, 34, 1911–1928. [Google Scholar] [CrossRef] [Green Version]
- Bourgeois, C.F.; Kim, Y.K.; Churcher, M.J.; West, M.J.; Karn, J. Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Mol. Cell Biol. 2002, 22, 1079–1093. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, D.; Kwak, Y.T.; Guo, J.; Gaynor, R.B. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell Biol. 2000, 20, 2970–2983. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.B.; Sharp, P.A. Positive transcription elongation factor B phosphorylates hSPT5 and RNA polymerase II carboxyl-terminal domain independently of cyclin-dependent kinase-activating kinase. J. Biol. Chem. 2001, 276, 12317–12323. [Google Scholar] [CrossRef] [Green Version]
- Wada, T.; Takagi, T.; Yamaguchi, Y.; Watanabe, D.; Handa, H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998, 17, 7395–7403. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Yamaguchi, Y.; Tsugeno, Y.; Yamamoto, J.; Yamada, T.; Nakamura, M.; Hisatake, K.; Handa, H. DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev. 2009, 23, 2765–2777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.; Zhu, X.; Li, Y.; Liu, M.; Yu, B.; Wang, Y.; Rao, M.; Yang, H.; Zhou, K.; Wang, Y.; et al. Multiple P-TEFbs cooperatively regulate the release of promoter-proximally paused RNA polymerase II. Nucleic Acids Res. 2016, 44, 6853–6867. [Google Scholar] [CrossRef] [PubMed]
- Sanso, M.; Levin, R.S.; Lipp, J.J.; Wang, V.Y.; Greifenberg, A.K.; Quezada, E.M.; Ali, A.; Ghosh, A.; Larochelle, S.; Rana, T.M.; et al. P-TEFb regulation of transcription termination factor Xrn2 revealed by a chemical genetic screen for Cdk9 substrates. Genes Dev. 2016, 30, 117–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castelo-Branco, G.; Amaral, P.P.; Engstrom, P.G.; Robson, S.C.; Marques, S.C.; Bertone, P.; Kouzarides, T. The non-coding snRNA 7SK controls transcriptional termination, poising, and bidirectionality in embryonic stem cells. Genome Biol. 2013, 14, R98. [Google Scholar] [CrossRef]
- Decker, T.M.; Forne, I.; Straub, T.; Elsaman, H.; Ma, G.; Shah, N.; Imhof, A.; Eick, D. Analog-sensitive cell line identifies cellular substrates of CDK9. Oncotarget 2019, 10, 6934–6943. [Google Scholar] [CrossRef] [Green Version]
- Tsai, W.H.; Wang, P.W.; Lin, S.Y.; Wu, I.L.; Ko, Y.C.; Chen, Y.L.; Li, M.; Lin, S.F. Ser-634 and Ser-636 of Kaposi’s Sarcoma-Associated Herpesvirus RTA are Involved in Transactivation and are Potential Cdk9 Phosphorylation Sites. Front. Microbiol. 2012, 3, 60. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Zhu, Q.; Luo, K.; Zhou, Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 2001, 414, 317–322. [Google Scholar] [CrossRef]
- Nguyen, V.T.; Kiss, T.; Michels, A.A.; Bensaude, O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 2001, 414, 322–325. [Google Scholar] [CrossRef]
- Barboric, M.; Lenasi, T.; Chen, H.; Johansen, E.B.; Guo, S.; Peterlin, B.M. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc. Natl. Acad. Sci. USA 2009, 106, 7798–7803. [Google Scholar] [CrossRef] [Green Version]
- Diribarne, G.; Bensaude, O. 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNA Biol. 2009, 6, 122–128. [Google Scholar] [CrossRef] [Green Version]
- He, N.; Jahchan, N.S.; Hong, E.; Li, Q.; Bayfield, M.A.; Maraia, R.J.; Luo, K.; Zhou, Q. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol. Cell 2008, 29, 588–599. [Google Scholar] [CrossRef] [PubMed]
- Krueger, B.J.; Jeronimo, C.; Roy, B.B.; Bouchard, A.; Barrandon, C.; Byers, S.A.; Searcey, C.E.; Cooper, J.J.; Bensaude, O.; Cohen, E.A.; et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008, 36, 2219–2229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michels, A.A.; Fraldi, A.; Li, Q.; Adamson, T.E.; Bonnet, F.; Nguyen, V.T.; Sedore, S.C.; Price, J.P.; Price, D.H.; Lania, L.; et al. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004, 23, 2608–2619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yik, J.H.; Chen, R.; Nishimura, R.; Jennings, J.L.; Link, A.J.; Zhou, Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 2003, 12, 971–982. [Google Scholar] [CrossRef]
- Yik, J.H.; Chen, R.; Pezda, A.C.; Samford, C.S.; Zhou, Q. A human immunodeficiency virus type 1 Tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb. Mol. Cell Biol. 2004, 24, 5094–5105. [Google Scholar] [CrossRef] [Green Version]
- Peterlin, B.M.; Brogie, J.E.; Price, D.H. 7SK snRNA: A noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip. Rev. RNA 2012, 3, 92–103. [Google Scholar] [CrossRef] [Green Version]
- Bugai, A.; Quaresma, A.J.C.; Friedel, C.C.; Lenasi, T.; Duster, R.; Sibley, C.R.; Fujinaga, K.; Kukanja, P.; Hennig, T.; Blasius, M.; et al. P-TEFb Activation by RBM7 Shapes a Pro-survival Transcriptional Response to Genotoxic Stress. Mol. Cell 2019, 74, 254–267 e210. [Google Scholar] [CrossRef] [Green Version]
- Bartholomeeusen, K.; Fujinaga, K.; Xiang, Y.; Peterlin, B.M. Histone deacetylase inhibitors (HDACis) that release the positive transcription elongation factor b (P-TEFb) from its inhibitory complex also activate HIV transcription. J. Biol. Chem. 2013, 288, 14400–14407. [Google Scholar] [CrossRef] [Green Version]
- Bartholomeeusen, K.; Xiang, Y.; Fujinaga, K.; Peterlin, B.M. Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein. J. Biol. Chem. 2012, 287, 36609–36616. [Google Scholar] [CrossRef] [Green Version]
- Contreras, X.; Barboric, M.; Lenasi, T.; Peterlin, B.M. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog 2007, 3, 1459–1469. [Google Scholar] [CrossRef]
- Kim, Y.K.; Mbonye, U.; Hokello, J.; Karn, J. T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J. Mol. Biol. 2011, 410, 896–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Contreras, X.; Schweneker, M.; Chen, C.S.; McCune, J.M.; Deeks, S.G.; Martin, J.; Peterlin, B.M. Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J. Biol. Chem. 2009, 284, 6782–6789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gargano, B.; Amente, S.; Majello, B.; Lania, L. P-TEFb is a crucial co-factor for Myc transactivation. Cell Cycle 2007, 6, 2031–2037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michels, A.A.; Bensaude, O. RNA-driven cyclin-dependent kinase regulation: When CDK9/cyclin T subunits of P-TEFb meet their ribonucleoprotein partners. Biotechnol. J. 2008, 3, 1022–1032. [Google Scholar] [CrossRef] [PubMed]
- Navone, N.M. P-TEFb joins the family of cdks in oncology, promotes cell growth of cancer cells. Cell Cycle 2010, 9, 2935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.; Xiang, Y.; Fujinaga, K.; Bartholomeeusen, K.; Nilson, K.A.; Price, D.H.; Peterlin, B.M. Release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein (snRNP) activates hexamethylene bisacetamide-inducible protein (HEXIM1) transcription. J. Biol. Chem. 2014, 289, 9918–9925. [Google Scholar] [CrossRef] [Green Version]
- Tan, J.L.; Fogley, R.D.; Flynn, R.A.; Ablain, J.; Yang, S.; Saint-Andre, V.; Fan, Z.P.; Do, B.T.; Laga, A.C.; Fujinaga, K.; et al. Stress from Nucleotide Depletion Activates the Transcriptional Regulator HEXIM1 to Suppress Melanoma. Mol. Cell 2016, 62, 34–46. [Google Scholar] [CrossRef] [Green Version]
- Bandukwala, H.S.; Gagnon, J.; Togher, S.; Greenbaum, J.A.; Lamperti, E.D.; Parr, N.J.; Molesworth, A.M.; Smithers, N.; Lee, K.; Witherington, J.; et al. Selective inhibition of CD4+ T-cell cytokine production and autoimmunity by BET protein and c-Myc inhibitors. Proc. Natl. Acad. Sci. USA 2012, 109, 14532–14537. [Google Scholar] [CrossRef] [Green Version]
- Bowry, A.; Piberger, A.L.; Rojas, P.; Saponaro, M.; Petermann, E. BET Inhibition Induces HEXIM1- and RAD51-Dependent Conflicts between Transcription and Replication. Cell Rep. 2018, 25, 2061–2069 e2064. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishnan, R.; Liu, H.; Rice, A.P. Short communication: SAHA (vorinostat) induces CDK9 Thr-186 (T-loop) phosphorylation in resting CD4+ T cells: Implications for reactivation of latent HIV. Aids Res. Hum. Retrovir. 2015, 31, 137–141. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Yang, W.; Ni, T.; Tang, Z.; Nakadai, T.; Zhu, J.; Roeder, R.G. RNA polymerase II-associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II. Science 2015, 350, 1383–1386. [Google Scholar] [CrossRef] [Green Version]
- He, N.; Chan, C.K.; Sobhian, B.; Chou, S.; Xue, Y.; Liu, M.; Alber, T.; Benkirane, M.; Zhou, Q. Human Polymerase-Associated Factor complex (PAFc) connects the Super Elongation Complex (SEC) to RNA polymerase II on chromatin. Proc. Natl. Acad. Sci. USA 2011, 108, E636–645. [Google Scholar] [CrossRef] [Green Version]
- Lu, H.; Li, Z.; Xue, Y.; Schulze-Gahmen, U.; Johnson, J.R.; Krogan, N.J.; Alber, T.; Zhou, Q. AFF1 is a ubiquitous P-TEFb partner to enable Tat extraction of P-TEFb from 7SK snRNP and formation of SECs for HIV transactivation. Proc. Natl. Acad. Sci. USA 2014, 111, E15–24. [Google Scholar] [CrossRef] [Green Version]
- Luo, Z.; Lin, C.; Shilatifard, A. The super elongation complex (SEC) family in transcriptional control. Nat. Rev. Mol. Cell Biol. 2012, 13, 543–547. [Google Scholar] [CrossRef]
- Smith, E.; Lin, C.; Shilatifard, A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011, 25, 661–672. [Google Scholar] [CrossRef] [Green Version]
- Brasier, A.R. Expanding role of cyclin dependent kinases in cytokine inducible gene expression. Cell Cycle 2008, 7, 2661–2666. [Google Scholar] [CrossRef] [Green Version]
- Giraud, S.; Hurlstone, A.; Avril, S.; Coqueret, O. Implication of BRG1 and cdk9 in the STAT3-mediated activation of the p21waf1 gene. Oncogene 2004, 23, 7391–7398. [Google Scholar] [CrossRef]
- Simone, C.; Stiegler, P.; Bagella, L.; Pucci, B.; Bellan, C.; De Falco, G.; De Luca, A.; Guanti, G.; Puri, P.L.; Giordano, A. Activation of MyoD-dependent transcription by cdk9/cyclin T2. Oncogene 2002, 21, 4137–4148. [Google Scholar] [CrossRef] [Green Version]
- Zumer, K.; Plemenitas, A.; Saksela, K.; Peterlin, B.M. Patient mutation in AIRE disrupts P-TEFb binding and target gene transcription. Nucleic Acids Res. 2011, 39, 7908–7919. [Google Scholar] [CrossRef] [Green Version]
- Asamitsu, K.; Fujinaga, K.; Okamoto, T. HIV Tat/P-TEFb Interaction: A Potential Target for Novel Anti-HIV Therapies. Molecules 2018, 23, 933. [Google Scholar] [CrossRef] [Green Version]
- Jeang, K.T. Tat, Tat-associated kinase, and transcription. J. Biomed. Sci. 1998, 5, 24–27. [Google Scholar] [CrossRef]
- Jones, K.A. Taking a new TAK on tat transactivation. Genes Dev. 1997, 11, 2593–2599. [Google Scholar] [CrossRef] [Green Version]
- Karn, J. The molecular biology of HIV latency: Breaking and restoring the Tat-dependent transcriptional circuit. Curr. Opin. Hiv. Aids 2011, 6, 4–11. [Google Scholar] [CrossRef]
- Ott, M.; Geyer, M.; Zhou, Q. The control of HIV transcription: Keeping RNA polymerase II on track. Cell Host Microbe 2011, 10, 426–435. [Google Scholar] [CrossRef] [Green Version]
- Rice, A.P. Roles of CDKs in RNA polymerase II transcription of the HIV-1 genome. Transcription 2019, 10, 111–117. [Google Scholar] [CrossRef]
- Dawson, M.A.; Prinjha, R.K.; Dittmann, A.; Giotopoulos, G.; Bantscheff, M.; Chan, W.I.; Robson, S.C.; Chung, C.W.; Hopf, C.; Savitski, M.M.; et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 2011, 478, 529–533. [Google Scholar] [CrossRef] [Green Version]
- Mertz, J.A.; Conery, A.R.; Bryant, B.M.; Sandy, P.; Balasubramanian, S.; Mele, D.A.; Bergeron, L.; Sims, R.J., 3rd. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl. Acad. Sci. USA 2011, 108, 16669–16674. [Google Scholar] [CrossRef] [Green Version]
- Delmore, J.E.; Issa, G.C.; Lemieux, M.E.; Rahl, P.B.; Shi, J.; Jacobs, H.M.; Kastritis, E.; Gilpatrick, T.; Paranal, R.M.; Qi, J.; et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011, 146, 904–917. [Google Scholar] [CrossRef] [Green Version]
- Prasanth, K.V.; Camiolo, M.; Chan, G.; Tripathi, V.; Denis, L.; Nakamura, T.; Hubner, M.R.; Spector, D.L. Nuclear organization and dynamics of 7SK RNA in regulating gene expression. Mol. Biol. Cell 2010, 21, 4184–4196. [Google Scholar] [CrossRef] [Green Version]
- Rice, A.P. The HIV-1 Tat Protein: Mechanism of Action and Target for HIV-1 Cure Strategies. Curr. Pharm. Des. 2017, 23, 4098–4102. [Google Scholar] [CrossRef] [Green Version]
- Cary, D.C.; Fujinaga, K.; Peterlin, B.M. Molecular mechanisms of HIV latency. J. Clin. Investig. 2016, 126, 448–454. [Google Scholar] [CrossRef]
- Mbonye, U.; Karn, J. The Molecular Basis for Human Immunodeficiency Virus Latency. Annu. Rev. Virol. 2017, 4, 261–285. [Google Scholar] [CrossRef]
- Biglione, S.; Byers, S.A.; Price, J.P.; Nguyen, V.T.; Bensaude, O.; Price, D.H.; Maury, W. Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology 2007, 4, 47. [Google Scholar] [CrossRef] [Green Version]
- Chao, S.H.; Fujinaga, K.; Marion, J.E.; Taube, R.; Sausville, E.A.; Senderowicz, A.M.; Peterlin, B.M.; Price, D.H. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J. Biol. Chem. 2000, 275, 28345–28348. [Google Scholar] [CrossRef] [Green Version]
- de la Fuente, C.; Maddukuri, A.; Kehn, K.; Baylor, S.Y.; Deng, L.; Pumfery, A.; Kashanchi, F. Pharmacological cyclin-dependent kinase inhibitors as HIV-1 antiviral therapeutics. Curr. Hiv. Res. 2003, 1, 131–152. [Google Scholar] [CrossRef]
- Medina-Moreno, S.; Dowling, T.C.; Zapata, J.C.; Le, N.M.; Sausville, E.; Bryant, J.; Redfield, R.R.; Heredia, A. Targeting of CDK9 with indirubin 3’-monoxime safely and durably reduces HIV viremia in chronically infected humanized mice. PLoS ONE 2017, 12, e0183425. [Google Scholar] [CrossRef] [Green Version]
- Nemeth, G.; Varga, Z.; Greff, Z.; Bencze, G.; Sipos, A.; Szantai-Kis, C.; Baska, F.; Gyuris, A.; Kelemenics, K.; Szathmary, Z.; et al. Novel, selective CDK9 inhibitors for the treatment of HIV infection. Curr. Med. Chem. 2011, 18, 342–358. [Google Scholar] [CrossRef]
- Okamoto, M.; Hidaka, A.; Toyama, M.; Hosoya, T.; Yamamoto, M.; Hagiwara, M.; Baba, M. Selective inhibition of HIV-1 replication by the CDK9 inhibitor FIT-039. Antivir. Res. 2015, 123, 1–4. [Google Scholar] [CrossRef]
- Pisell, T.L.; Ho, O.; Lee, G.; Butera, S.T. Spectrum of cdk-9 inhibitor activity against HIV-1 replication among various models of chronic and latent infection. Antivir Chem. Chemother. 2001, 12 (Suppl. 1), 33–41. [Google Scholar]
- Dahabieh, M.S.; Battivelli, E.; Verdin, E. Understanding HIV latency: The road to an HIV cure. Annu. Rev. Med. 2015, 66, 407–421. [Google Scholar] [CrossRef] [Green Version]
- Darcis, G.; Van Driessche, B.; Van Lint, C. HIV Latency: Should We Shock or Lock? Trends Immunol. 2017, 38, 217–228. [Google Scholar] [CrossRef]
- Margolis, D.M. Mechanisms of HIV latency: An emerging picture of complexity. Curr. Hiv. Aids Rep. 2010, 7, 37–43. [Google Scholar] [CrossRef]
- Rasmussen, T.A.; Lewin, S.R. Shocking HIV out of hiding: Where are we with clinical trials of latency reversing agents? Curr. Opin. Hiv. Aids 2016, 11, 394–401. [Google Scholar] [CrossRef] [Green Version]
- Siliciano, R.F.; Greene, W.C. HIV latency. Cold Spring Harb. Perspect. Med. 2011, 1, a007096. [Google Scholar] [CrossRef] [Green Version]
- Xing, S.; Siliciano, R.F. Targeting HIV latency: Pharmacologic strategies toward eradication. Drug Discov. Today 2013, 18, 541–551. [Google Scholar] [CrossRef] [Green Version]
- Kimata, J.T.; Rice, A.P.; Wang, J. Challenges and strategies for the eradication of the HIV reservoir. Curr. Opin. Immunol. 2016, 42, 65–70. [Google Scholar] [CrossRef] [Green Version]
- Sengupta, S.; Siliciano, R.F. Targeting the Latent Reservoir for HIV-1. Immunity 2018, 48, 872–895. [Google Scholar] [CrossRef] [Green Version]
- International AIDS Society Scientific Working Group on HIV Cure; Deeks, S.G.; Autran, B.; Berkhout, B.; Benkirane, M.; Cairns, S.; Chomont, N.; Chun, T.W.; Churchill, M.; Di Mascio, M.; et al. Towards an HIV cure: A global scientific strategy. Nat. Rev. Immunol. 2012, 12, 607–614. [Google Scholar] [CrossRef]
- Churchill, M.J.; Deeks, S.G.; Margolis, D.M.; Siliciano, R.F.; Swanstrom, R. HIV reservoirs: What, where and how to target them. Nat. Rev. Microbiol. 2016, 14, 55–60. [Google Scholar] [CrossRef]
- Martinez-Picado, J.; Deeks, S.G. Persistent HIV-1 replication during antiretroviral therapy. Curr. Opin. Hiv. Aids 2016, 11, 417–423. [Google Scholar] [CrossRef]
- Chomont, N.; DaFonseca, S.; Vandergeeten, C.; Ancuta, P.; Sekaly, R.P. Maintenance of CD4+ T-cell memory and HIV persistence: Keeping memory, keeping HIV. Curr. Opin. Hiv. Aids 2011, 6, 30–36. [Google Scholar] [CrossRef]
- Eisele, E.; Siliciano, R.F. Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity 2012, 37, 377–388. [Google Scholar] [CrossRef] [Green Version]
- Murray, A.J.; Kwon, K.J.; Farber, D.L.; Siliciano, R.F. The Latent Reservoir for HIV-1: How Immunologic Memory and Clonal Expansion Contribute to HIV-1 Persistence. J. Immunol. 2016, 197, 407–417. [Google Scholar] [CrossRef] [Green Version]
- Marshall, R.M.; Salerno, D.; Garriga, J.; Grana, X. Cyclin T1 expression is regulated by multiple signaling pathways and mechanisms during activation of human peripheral blood lymphocytes. J. Immunol. 2005, 175, 6402–6411. [Google Scholar] [CrossRef] [Green Version]
- Liou, L.Y.; Haaland, R.E.; Herrmann, C.H.; Rice, A.P. Cyclin T1 but not cyclin T2a is induced by a post-transcriptional mechanism in PAMP-activated monocyte-derived macrophages. J. Leukoc. Biol. 2006, 79, 388–396. [Google Scholar] [CrossRef]
- Liou, L.Y.; Herrmann, C.H.; Rice, A.P. Transient induction of cyclin T1 during human macrophage differentiation regulates human immunodeficiency virus type 1 Tat transactivation function. J. Virol. 2002, 76, 10579–10587. [Google Scholar] [CrossRef] [Green Version]
- Rice, A.P.; Herrmann, C.H. Regulation of TAK/P-TEFb in CD4+ T lymphocytes and macrophages. Curr. Hiv. Res. 2003, 1, 395–404. [Google Scholar] [CrossRef]
- Garriga, J.; Peng, J.; Parreno, M.; Price, D.H.; Henderson, E.E.; Grana, X. Upregulation of cyclin T1/CDK9 complexes during T cell activation. Oncogene 1998, 17, 3093–3102. [Google Scholar] [CrossRef]
- Krystof, V.; Chamrad, I.; Jorda, R.; Kohoutek, J. Pharmacological targeting of CDK9 in cardiac hypertrophy. Med. Res. Rev. 2010, 30, 646–666. [Google Scholar] [CrossRef]
- Sano, M.; Schneider, M.D. Cyclin-dependent kinase-9: An RNAPII kinase at the nexus of cardiac growth and death cascades. Circ. Res. 2004, 95, 867–876. [Google Scholar] [CrossRef] [Green Version]
- Yoshikawa, N.; Shimizu, N.; Maruyama, T.; Sano, M.; Matsuhashi, T.; Fukuda, K.; Kataoka, M.; Satoh, T.; Ojima, H.; Sawai, T.; et al. Cardiomyocyte-specific overexpression of HEXIM1 prevents right ventricular hypertrophy in hypoxia-induced pulmonary hypertension in mice. PLoS ONE 2012, 7, e52522. [Google Scholar] [CrossRef]
- Kulkarni, P.A.; Sano, M.; Schneider, M.D. Phosphorylation of RNA polymerase II in cardiac hypertrophy: Cell enlargement signals converge on cyclin T/Cdk9. Recent. Prog. Horm. Res. 2004, 59, 125–139. [Google Scholar] [CrossRef]
- Espinoza-Derout, J.; Wagner, M.; Salciccioli, L.; Lazar, J.M.; Bhaduri, S.; Mascareno, E.; Chaqour, B.; Siddiqui, M.A. Positive transcription elongation factor b activity in compensatory myocardial hypertrophy is regulated by cardiac lineage protein-1. Circ. Res. 2009, 104, 1347–1354. [Google Scholar] [CrossRef]
- Espinoza-Derout, J.; Wagner, M.; Shahmiri, K.; Mascareno, E.; Chaqour, B.; Siddiqui, M.A. Pivotal role of cardiac lineage protein-1 (CLP-1) in transcriptional elongation factor P-TEFb complex formation in cardiac hypertrophy. Cardiovasc. Res. 2007, 75, 129–138. [Google Scholar] [CrossRef]
- Montano, M.M.; Doughman, Y.Q.; Deng, H.; Chaplin, L.; Yang, J.; Wang, N.; Zhou, Q.; Ward, N.L.; Watanabe, M. Mutation of the HEXIM1 gene results in defects during heart and vascular development partly through downregulation of vascular endothelial growth factor. Circ. Res. 2008, 102, 415–422. [Google Scholar] [CrossRef] [Green Version]
- Romano, G. Deregulations in the cyclin-dependent kinase-9-related pathway in cancer: Implications for drug discovery and development. ISRN Oncol. 2013, 2013, 305371. [Google Scholar] [CrossRef] [Green Version]
- Baluapuri, A.; Hofstetter, J.; Dudvarski Stankovic, N.; Endres, T.; Bhandare, P.; Vos, S.M.; Adhikari, B.; Schwarz, J.D.; Narain, A.; Vogt, M.; et al. MYC Recruits SPT5 to RNA Polymerase II to Promote Processive Transcription Elongation. Mol. Cell 2019, 74, 674–687 e611. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.; Jin, Z.; Agarwal, R.; Ma, K.; Yang, J.; Ibrahim, S.; Olaru, A.V.; David, S.; Ashktorab, H.; Smoot, D.T.; et al. LARP7 is a potential tumor suppressor gene in gastric cancer. Lab. InvestIG. 2012, 92, 1013–1019. [Google Scholar] [CrossRef] [Green Version]
- Dey, A.; Chao, S.H.; Lane, D.P. HEXIM1 and the control of transcription elongation: From cancer and inflammation to AIDS and cardiac hypertrophy. Cell Cycle 2007, 6, 1856–1863. [Google Scholar] [CrossRef]
- Ji, X.; Lu, H.; Zhou, Q.; Luo, K. LARP7 suppresses P-TEFb activity to inhibit breast cancer progression and metastasis. Elife 2014, 3, e02907. [Google Scholar] [CrossRef]
- Kretz, A.L.; Schaum, M.; Richter, J.; Kitzig, E.F.; Engler, C.C.; Leithauser, F.; Henne-Bruns, D.; Knippschild, U.; Lemke, J. CDK9 is a prognostic marker and therapeutic target in pancreatic cancer. Tumour Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, H.; Seebacher, N.A.; Hornicek, F.J.; Duan, Z. Cyclin-dependent kinase 9 (CDK9) is a novel prognostic marker and therapeutic target in osteosarcoma. EBioMedicine 2019, 39, 182–193. [Google Scholar] [CrossRef] [Green Version]
- Moiola, C.; De Luca, P.; Gardner, K.; Vazquez, E.; De Siervi, A. Cyclin T1 overexpression induces malignant transformation and tumor growth. Cell Cycle 2010, 9, 3119–3126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlafstein, A.J.; Withers, A.E.; Rudra, S.; Danelia, D.; Switchenko, J.M.; Mister, D.; Harari, S.; Zhang, H.; Daddacha, W.; Ehdaivand, S.; et al. CDK9 Expression Shows Role as a Potential Prognostic Biomarker in Breast Cancer Patients Who Fail to Achieve Pathologic Complete Response after Neoadjuvant Chemotherapy. Int. J. Breast Cancer 2018, 2018, 6945129. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Dean, D.C.; Hornicek, F.J.; Shi, H.; Duan, Z. Cyclin-dependent kinase 9 (CDK9) is a novel prognostic marker and therapeutic target in ovarian cancer. FASEB J. 2019, 33, 5990–6000. [Google Scholar] [CrossRef]
- Napolitano, G.; Majello, B.; Lania, L. Role of cyclinT/Cdk9 complex in basal and regulated transcription (review). Int. J. Oncol. 2002, 21, 171–177. [Google Scholar] [CrossRef]
- Claudio, P.P.; Cui, J.; Ghafouri, M.; Mariano, C.; White, M.K.; Safak, M.; Sheffield, J.B.; Giordano, A.; Khalili, K.; Amini, S.; et al. Cdk9 phosphorylates p53 on serine 392 independently of CKII. J. Cell Physiol. 2006, 208, 602–612. [Google Scholar] [CrossRef]
- Simone, C.; Bagella, L.; Bellan, C.; Giordano, A. Physical interaction between pRb and cdk9/cyclinT2 complex. Oncogene 2002, 21, 4158–4165. [Google Scholar] [CrossRef] [Green Version]
- Garriga, J.; Grana, X. CDK9 inhibition strategy defines distinct sets of target genes. BMC Res. Notes 2014, 7, 301. [Google Scholar] [CrossRef] [Green Version]
- Garriga, J.; Xie, H.; Obradovic, Z.; Grana, X. Selective control of gene expression by CDK9 in human cells. J. Cell Physiol. 2010, 222, 200–208. [Google Scholar] [CrossRef] [Green Version]
- Gomes, N.P.; Bjerke, G.; Llorente, B.; Szostek, S.A.; Emerson, B.M.; Espinosa, J.M. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006, 20, 601–612. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Zhan, Y.; Hou, G.; Li, L. Cyclin-dependent kinase 9 may as a novel target in downregulating the atherosclerosis inflammation (Review). Biomed. Rep. 2014, 2, 775–779. [Google Scholar] [CrossRef] [PubMed]
- Hellvard, A.; Zeitlmann, L.; Heiser, U.; Kehlen, A.; Niestroj, A.; Demuth, H.U.; Koziel, J.; Delaleu, N.; Jan, P.; Mydel, P. Inhibition of CDK9 as a therapeutic strategy for inflammatory arthritis. Sci. Rep. 2016, 6, 31441. [Google Scholar] [CrossRef] [PubMed]
- Yik, J.H.; Hu, Z.; Kumari, R.; Christiansen, B.A.; Haudenschild, D.R. Cyclin-dependent kinase 9 inhibition protects cartilage from the catabolic effects of proinflammatory cytokines. Arthritis Rheumatol. 2014, 66, 1537–1546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medzhitov, R.; Horng, T. Transcriptional control of the inflammatory response. Nat. Rev. Immunol. 2009, 9, 692–703. [Google Scholar] [CrossRef]
- Gupte, R.; Muse, G.W.; Chinenov, Y.; Adelman, K.; Rogatsky, I. Glucocorticoid receptor represses proinflammatory genes at distinct steps of the transcription cycle. Proc. Natl. Acad. Sci. USA 2013, 110, 14616–14621. [Google Scholar] [CrossRef] [Green Version]
- Kanazawa, S.; Ota, S.; Sekine, C.; Tada, T.; Otsuka, T.; Okamoto, T.; Sonderstrup, G.; Peterlin, B.M. Aberrant MHC class II expression in mouse joints leads to arthritis with extraarticular manifestations similar to rheumatoid arthritis. Proc. Natl. Acad. Sci. USA 2006, 103, 14465–14470. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Hampson, P.; Hazeldine, J.; Krystof, V.; Strnad, M.; Pechan, P.; Janet, M. Cyclin-dependent kinase 9 activity regulates neutrophil spontaneous apoptosis. PLoS ONE 2012, 7, e30128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Z.; Yik, J.H.; Cissell, D.D.; Michelier, P.V.; Athanasiou, K.A.; Haudenschild, D.R. Inhibition of CDK9 prevents mechanical injury-induced inflammation, apoptosis and matrix degradation in cartilage explants. Eur. Cell Mater. 2016, 30, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Schrecengost, R.S.; Green, C.L.; Zhuang, Y.; Keller, S.N.; Smith, R.A.; Maines, L.W.; Smith, C.D. In Vitro and In Vivo Antitumor and Anti-Inflammatory Capabilities of the Novel GSK3 and CDK9 Inhibitor ABC1183. J. Pharm. Exp. Ther. 2018, 365, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Zaborowska, J.; Isa, N.F.; Murphy, S. P-TEFb goes viral. Inside Cell 2016, 1, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Barboric, M.; Taube, R.; Nekrep, N.; Fujinaga, K.; Peterlin, B.M. Binding of Tat to TAR and recruitment of positive transcription elongation factor b occur independently in bovine immunodeficiency virus. J. Virol. 2000, 74, 6039–6044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bieniasz, P.D.; Grdina, T.A.; Bogerd, H.P.; Cullen, B.R. Highly divergent lentiviral Tat proteins activate viral gene expression by a common mechanism. Mol. Cell Biol. 1999, 19, 4592–4599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taube, R.; Fujinaga, K.; Irwin, D.; Wimmer, J.; Geyer, M.; Peterlin, B.M. Interactions between equine cyclin T1, Tat, and TAR are disrupted by a leucine-to-valine substitution found in human cyclin T1. J. Virol. 2000, 74, 892–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaborowska, J.; Baumli, S.; Laitem, C.; O’Reilly, D.; Thomas, P.H.; O’Hare, P.; Murphy, S. Herpes Simplex Virus 1 (HSV-1) ICP22 protein directly interacts with cyclin-dependent kinase (CDK)9 to inhibit RNA polymerase II transcription elongation. PLoS ONE 2014, 9, e107654. [Google Scholar] [CrossRef]
- Guo, L.; Wu, W.J.; Liu, L.D.; Wang, L.C.; Zhang, Y.; Wu, L.Q.; Guan, Y.; Li, Q.H. Herpes simplex virus 1 ICP22 inhibits the transcription of viral gene promoters by binding to and blocking the recruitment of P-TEFb. PLoS ONE 2012, 7, e45749. [Google Scholar] [CrossRef] [Green Version]
- Kurosu, T.; Peterlin, B.M. VP16 and ubiquitin; binding of P-TEFb via its activation domain and ubiquitin facilitates elongation of transcription of target genes. Curr. Biol. 2004, 14, 1112–1116. [Google Scholar] [CrossRef] [Green Version]
- Chang, P.C.; Li, M. Kaposi’s sarcoma-associated herpesvirus K-cyclin interacts with Cdk9 and stimulates Cdk9-mediated phosphorylation of p53 tumor suppressor. J. Virol. 2008, 82, 278–290. [Google Scholar] [CrossRef] [Green Version]
- Vijayalingam, S.; Chinnadurai, G. Adenovirus L-E1A activates transcription through mediator complex-dependent recruitment of the super elongation complex. J. Virol. 2013, 87, 3425–3434. [Google Scholar] [CrossRef] [Green Version]
- Palermo, R.D.; Webb, H.M.; West, M.J. RNA polymerase II stalling promotes nucleosome occlusion and pTEFb recruitment to drive immortalization by Epstein-Barr virus. PLoS Pathog. 2011, 7, e1002334. [Google Scholar] [CrossRef]
- Choi, K.H. Viral polymerases. Adv. Exp. Med. Biol. 2012, 726, 267–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Li, G.; Ye, X. Cyclin T1/CDK9 interacts with influenza A virus polymerase and facilitates its association with cellular RNA polymerase II. J. Virol. 2010, 84, 12619–12627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, T.; Okuyama-Dobashi, K.; Murakami, S.; Chen, W.; Okamoto, T.; Ueda, K.; Hosoya, T.; Matsuura, Y.; Ryo, A.; Tanaka, Y.; et al. Inhibitory effect of CDK9 inhibitor FIT-039 on hepatitis B virus propagation. Antivir. Res. 2016, 133, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Onogi, H.; Kii, I.; Yoshida, S.; Iida, K.; Sakai, H.; Abe, M.; Tsubota, T.; Ito, N.; Hosoya, T.; et al. CDK9 inhibitor FIT-039 prevents replication of multiple DNA viruses. J. Clin. Investig. 2014, 124, 3479–3488. [Google Scholar] [CrossRef] [PubMed]
- Chao, S.H.; Price, D.H. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 2001, 276, 31793–31799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlson, B.A.; Dubay, M.M.; Sausville, E.A.; Brizuela, L.; Worland, P.J. Flavopiridol induces G1 arrest with inhibition of cyclin-dependent kinase (CDK) 2 and CDK4 in human breast carcinoma cells. Cancer Res. 1996, 56, 2973–2978. [Google Scholar]
- Sedlacek, H.; Czech, J.; Naik, R.; Kaur, G.; Worland, P.; Losiewicz, M.; Parker, B.; Carlson, B.; Smith, A.; Senderowicz, A.; et al. Flavopiridol (L86 8275; NSC 649890), a new kinase inhibitor for tumor therapy. Int. J. Oncol. 1996, 9, 1143–1168. [Google Scholar] [CrossRef]
- Wang, L.M.; Ren, D.M. Flavopiridol, the first cyclin-dependent kinase inhibitor: Recent advances in combination chemotherapy. Mini Rev. Med. Chem. 2010, 10, 1058–1070. [Google Scholar] [CrossRef]
- Christian, B.A.; Grever, M.R.; Byrd, J.C.; Lin, T.S. Flavopiridol in chronic lymphocytic leukemia: A concise review. Clin. Lymphoma Myeloma 2009, 9 (Suppl. 3), S179–S185. [Google Scholar] [CrossRef]
- Wiernik, P.H. Alvocidib (flavopiridol) for the treatment of chronic lymphocytic leukemia. Expert Opin. Investig. Drugs 2016, 25, 729–734. [Google Scholar] [CrossRef]
- Zeidner, J.F.; Karp, J.E. Clinical activity of alvocidib (flavopiridol) in acute myeloid leukemia. Leuk. Res. 2015, 39, 1312–1318. [Google Scholar] [CrossRef] [PubMed]
- Sekine, C.; Sugihara, T.; Miyake, S.; Hirai, H.; Yoshida, M.; Miyasaka, N.; Kohsaka, H. Successful treatment of animal models of rheumatoid arthritis with small-molecule cyclin-dependent kinase inhibitors. J. Immunol. 2008, 180, 1954–1961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.; Zhao, S.; Gong, Y.; Hou, G.; Li, X.; Li, L. Serum cyclin-dependent kinase 9 is a potential biomarker of atherosclerotic inflammation. Oncotarget 2016, 7, 1854–1862. [Google Scholar] [CrossRef] [PubMed]
- Lucking, U.; Scholz, A.; Lienau, P.; Siemeister, G.; Kosemund, D.; Bohlmann, R.; Briem, H.; Terebesi, I.; Meyer, K.; Prelle, K.; et al. Identification of Atuveciclib (BAY 1143572), the First Highly Selective, Clinical PTEFb/CDK9 Inhibitor for the Treatment of Cancer. ChemMedChem 2017, 12, 1776–1793. [Google Scholar] [CrossRef] [PubMed]
- Hashiguchi, T.; Bruss, N.; Best, S.; Lam, V.; Danilova, O.; Paiva, C.J.; Wolf, J.; Gilbert, E.W.; Okada, C.Y.; Kaur, P.; et al. Cyclin-Dependent Kinase-9 Is a Therapeutic Target in MYC-Expressing Diffuse Large B-Cell Lymphoma. Mol. Cancer Ther. 2019, 18, 1520–1532. [Google Scholar] [CrossRef] [PubMed]
- Veeranki, O.L.; Tong, Z.; Dokey, R.; Mejia, A.; Zhang, J.; Qiao, Y.; Singh, P.K.; Katkhuda, R.; Mino, B.; Tailor, R.; et al. Targeting cyclin-dependent kinase 9 by a novel inhibitor enhances radiosensitization and identifies Axl as a novel downstream target in esophageal adenocarcinoma. Oncotarget 2019, 10, 4703–4718. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, S.; Ishida, T.; Ito, A.; Narita, T.; Masaki, A.; Suzuki, S.; Yoshida, T.; Ri, M.; Kusumoto, S.; Komatsu, H.; et al. Cyclin-dependent kinase 9 as a potential specific molecular target in NK-cell leukemia/lymphoma. Haematologica 2018, 103, 2059–2068. [Google Scholar] [CrossRef]
- Natoni, A.; Coyne, M.R.; Jacobsen, A.; Rainey, M.D.; O’Brien, G.; Healy, S.; Montagnoli, A.; Moll, J.; O’Dwyer, M.; Santocanale, C. Characterization of a Dual CDC7/CDK9 Inhibitor in Multiple Myeloma Cellular Models. Cancers 2013, 5, 901–918. [Google Scholar] [CrossRef] [Green Version]
- Yin, T.; Lallena, M.J.; Kreklau, E.L.; Fales, K.R.; Carballares, S.; Torrres, R.; Wishart, G.N.; Ajamie, R.T.; Cronier, D.M.; Iversen, P.W.; et al. A novel CDK9 inhibitor shows potent antitumor efficacy in preclinical hematologic tumor models. Mol. Cancer Ther. 2014, 13, 1442–1456. [Google Scholar] [CrossRef] [Green Version]
- Baker, A.; Gregory, G.P.; Verbrugge, I.; Kats, L.; Hilton, J.J.; Vidacs, E.; Lee, E.M.; Lock, R.B.; Zuber, J.; Shortt, J.; et al. The CDK9 Inhibitor Dinaciclib Exerts Potent Apoptotic and Antitumor Effects in Preclinical Models of MLL-Rearranged Acute Myeloid Leukemia. Cancer Res. 2016, 76, 1158–1169. [Google Scholar] [CrossRef] [Green Version]
- MacCallum, D.E.; Melville, J.; Frame, S.; Watt, K.; Anderson, S.; Gianella-Borradori, A.; Lane, D.P.; Green, S.R. Seliciclib (CYC202, R-Roscovitine) induces cell death in multiple myeloma cells by inhibition of RNA polymerase II-dependent transcription and down-regulation of Mcl-1. Cancer Res. 2005, 65, 5399–5407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dey, J.; Deckwerth, T.L.; Kerwin, W.S.; Casalini, J.R.; Merrell, A.J.; Grenley, M.O.; Burns, C.; Ditzler, S.H.; Dixon, C.P.; Beirne, E.; et al. Voruciclib, a clinical stage oral CDK9 inhibitor, represses MCL-1 and sensitizes high-risk Diffuse Large B-cell Lymphoma to BCL2 inhibition. Sci. Rep. 2017, 7, 18007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, W.G.; Chen, R.; Plunkett, W.; Siegel, D.; Sinha, R.; Harvey, R.D.; Badros, A.Z.; Popplewell, L.; Coutre, S.; Fox, J.A.; et al. Phase I and pharmacologic study of SNS-032, a potent and selective Cdk2, 7, and 9 inhibitor, in patients with advanced chronic lymphocytic leukemia and multiple myeloma. J. Clin. Oncol. 2010, 28, 3015–3022. [Google Scholar] [CrossRef] [PubMed]
- Manohar, S.M.; Rathos, M.J.; Sonawane, V.; Rao, S.V.; Joshi, K.S. Cyclin-dependent kinase inhibitor, P276-00 induces apoptosis in multiple myeloma cells by inhibition of Cdk9-T1 and RNA polymerase II-dependent transcription. Leuk. Res. 2011, 35, 821–830. [Google Scholar] [CrossRef]
- Rahaman, M.H.; Yu, Y.; Zhong, L.; Adams, J.; Lam, F.; Li, P.; Noll, B.; Milne, R.; Peng, J.; Wang, S. CDKI-73: An orally bioavailable and highly efficacious CDK9 inhibitor against acute myeloid leukemia. Investig. New Drugs 2019, 37, 625–635. [Google Scholar] [CrossRef]
- Lu, H.; Xue, Y.; Yu, G.K.; Arias, C.; Lin, J.; Fong, S.; Faure, M.; Weisburd, B.; Ji, X.; Mercier, A.; et al. Compensatory induction of MYC expression by sustained CDK9 inhibition via a BRD4-dependent mechanism. Elife 2015, 4, e06535. [Google Scholar] [CrossRef]
- Polier, G.; Ding, J.; Konkimalla, B.V.; Eick, D.; Ribeiro, N.; Kohler, R.; Giaisi, M.; Efferth, T.; Desaubry, L.; Krammer, P.H.; et al. Wogonin and related natural flavones are inhibitors of CDK9 that induce apoptosis in cancer cells by transcriptional suppression of Mcl-1. Cell Death Dis. 2011, 2, e182. [Google Scholar] [CrossRef] [Green Version]
- Whittaker, S.R.; Barlow, C.; Martin, M.P.; Mancusi, C.; Wagner, S.; Self, A.; Barrie, E.; Te Poele, R.; Sharp, S.; Brown, N.; et al. Molecular profiling and combinatorial activity of CCT068127: A potent CDK2 and CDK9 inhibitor. Mol. Oncol. 2018, 12, 287–304. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Cuellar, M.P.; Fuller, E.; Mathner, E.; Breitinger, C.; Hetzner, K.; Zeitlmann, L.; Borkhardt, A.; Slany, R.K. Efficacy of cyclin-dependent-kinase 9 inhibitors in a murine model of mixed-lineage leukemia. Leukemia 2014, 28, 1427–1435. [Google Scholar] [CrossRef]
- Olson, C.M.; Jiang, B.; Erb, M.A.; Liang, Y.; Doctor, Z.M.; Zhang, Z.; Zhang, T.; Kwiatkowski, N.; Boukhali, M.; Green, J.L.; et al. Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat. Chem. Biol. 2018, 14, 163–170. [Google Scholar] [CrossRef]
- Zhang, H.; Pandey, S.; Travers, M.; Sun, H.; Morton, G.; Madzo, J.; Chung, W.; Khowsathit, J.; Perez-Leal, O.; Barrero, C.A.; et al. Targeting CDK9 Reactivates Epigenetically Silenced Genes in Cancer. Cell 2018, 175, 1244–1258.e1226. [Google Scholar] [CrossRef] [Green Version]
- Goh, K.C.; Novotny-Diermayr, V.; Hart, S.; Ong, L.C.; Loh, Y.K.; Cheong, A.; Tan, Y.C.; Hu, C.; Jayaraman, R.; William, A.D.; et al. TG02, a novel oral multi-kinase inhibitor of CDKs, JAK2 and FLT3 with potent anti-leukemic properties. Leukemia 2012, 26, 236–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cidado, J.; Boiko, S.; Proia, T.; Ferguson, D.; Criscione, S.W.; San Martin, M.; Pop-Damkov, P.; Su, N.; Roamio Franklin, V.N.; Sekhar Reddy Chilamakuri, C.; et al. AZD4573 is a highly selective CDK9 inhibitor that suppresses Mcl-1 and induces apoptosis in hematological cancer cells. Clin. Cancer Res. 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, A.; Giuliano, C.J.; Palladino, A.; John, K.M.; Abramowicz, C.; Yuan, M.L.; Sausville, E.L.; Lukow, D.A.; Liu, L.; Chait, A.R.; et al. Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci. Transl. Med. 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Fujinaga, K.; Barboric, M.; Li, Q.; Luo, Z.; Price, D.H.; Peterlin, B.M. PKC phosphorylates HEXIM1 and regulates P-TEFb activity. Nucleic Acids Res. 2012, 40, 9160–9170. [Google Scholar] [CrossRef] [PubMed]
- Casse, C.; Giannoni, F.; Nguyen, V.T.; Dubois, M.F.; Bensaude, O. The transcriptional inhibitors, actinomycin D and alpha-amanitin, activate the HIV-1 promoter and favor phosphorylation of the RNA polymerase II C-terminal domain. J. Biol. Chem. 1999, 274, 16097–16106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, K.; Smith, E.R.; Aoi, Y.; Stoltz, K.L.; Katagi, H.; Woodfin, A.R.; Rendleman, E.J.; Marshall, S.A.; Murray, D.C.; Wang, L.; et al. Targeting Processive Transcription Elongation via SEC Disruption for MYC-Induced Cancer Therapy. Cell 2018, 175, 766–779.e717. [Google Scholar] [CrossRef] [Green Version]
- Leoz, M.; Kukanja, P.; Luo, Z.; Huang, F.; Cary, D.C.; Peterlin, B.M.; Fujinaga, K. HEXIM1-Tat chimera inhibits HIV-1 replication. PLoS Pathog. 2018, 14, e1007402. [Google Scholar] [CrossRef]
- Neklesa, T.K.; Winkler, J.D.; Crews, C.M. Targeted protein degradation by PROTACs. Pharmacol. Ther. 2017, 174, 138–144. [Google Scholar] [CrossRef]
- Mullard, A. First targeted protein degrader hits the clinic. Nat. Rev. Drug Discov. 2019. [Google Scholar] [CrossRef]
- Bian, J.; Ren, J.; Li, Y.; Wang, J.; Xu, X.; Feng, Y.; Tang, H.; Wang, Y.; Li, Z. Discovery of Wogonin-based PROTACs against CDK9 and capable of achieving antitumor activity. Bioorg Chem. 2018, 81, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Robb, C.M.; Contreras, J.I.; Kour, S.; Taylor, M.A.; Abid, M.; Sonawane, Y.A.; Zahid, M.; Murry, D.J.; Natarajan, A.; Rana, S. Chemically induced degradation of CDK9 by a proteolysis targeting chimera (PROTAC). Chem. Commun. 2017, 53, 7577–7580. [Google Scholar] [CrossRef] [PubMed]
- Minzel, W.; Venkatachalam, A.; Fink, A.; Hung, E.; Brachya, G.; Burstain, I.; Shaham, M.; Rivlin, A.; Omer, I.; Zinger, A.; et al. Small Molecules Co-targeting CKIalpha and the Transcriptional Kinases CDK7/9 Control AML in Preclinical Models. Cell 2018, 175, 171–185.e125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darcis, G.; Kula, A.; Bouchat, S.; Fujinaga, K.; Corazza, F.; Ait-Ammar, A.; Delacourt, N.; Melard, A.; Kabeya, K.; Vanhulle, C.; et al. An In-Depth Comparison of Latency-Reversing Agent Combinations in Various In Vitro and Ex Vivo HIV-1 Latency Models Identified Bryostatin-1+JQ1 and Ingenol-B+JQ1 to Potently Reactivate Viral Gene Expression. PLoS Pathog. 2015, 11, e1005063. [Google Scholar] [CrossRef] [PubMed]
- Fujinaga, K.; Luo, Z.; Schaufele, F.; Peterlin, B.M. Visualization of positive transcription elongation factor b (P-TEFb) activation in living cells. J. Biol. Chem. 2015, 290, 1829–1836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cary, D.C.; Fujinaga, K.; Peterlin, B.M. Euphorbia Kansui Reactivates Latent HIV. PLoS ONE 2016, 11, e0168027. [Google Scholar] [CrossRef] [Green Version]
- Ammosova, T.; Washington, K.; Debebe, Z.; Brady, J.; Nekhai, S. Dephosphorylation of CDK9 by protein phosphatase 2A and protein phosphatase-1 in Tat-activated HIV-1 transcription. Retrovirology 2005, 2, 47. [Google Scholar] [CrossRef] [Green Version]
- Budhiraja, S.; Ramakrishnan, R.; Rice, A.P. Phosphatase PPM1A negatively regulates P-TEFb function in resting CD4(+) T cells and inhibits HIV-1 gene expression. Retrovirology 2012, 9, 52. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Liu, M.; Li, H.; Xue, Y.; Ramey, W.N.; He, N.; Ai, N.; Luo, H.; Zhu, Y.; Zhou, N.; et al. PP2B and PP1alpha cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev. 2008, 22, 1356–1368. [Google Scholar] [CrossRef] [Green Version]
- Gudipaty, S.A.; McNamara, R.P.; Morton, E.L.; D’Orso, I. PPM1G Binds 7SK RNA and Hexim1 To Block P-TEFb Assembly into the 7SK snRNP and Sustain Transcription Elongation. Mol. Cell Biol. 2015, 35, 3810–3828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Gao, Y.; Ye, H.; Gerrin, S.; Ma, F.; Wu, Y.; Zhang, T.; Russo, J.; Cai, C.; Yuan, X.; et al. Positive feedback loop mediated by protein phosphatase 1alpha mobilization of P-TEFb and basal CDK1 drives androgen receptor in prostate cancer. Nucleic Acids Res. 2017, 45, 3738–3751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nekhai, S.; Petukhov, M.; Breuer, D. Regulation of CDK9 activity by phosphorylation and dephosphorylation. Biomed. Res. Int. 2014, 2014, 964964. [Google Scholar] [CrossRef] [PubMed]
- Parua, P.K.; Booth, G.T.; Sanso, M.; Benjamin, B.; Tanny, J.C.; Lis, J.T.; Fisher, R.P. A Cdk9-PP1 switch regulates the elongation-termination transition of RNA polymerase II. Nature 2018, 558, 460–464. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Schroeder, S.; Ott, M. CYCLINg through transcription: Posttranslational modifications of P-TEFb regulate transcription elongation. Cell Cycle 2010, 9, 1697–1705. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Transcription Factor | Category * | Refs |
|---|---|---|
| NFκB | DNA-binding transcription activator activity | [45] |
| Myoblast Determination Protein 1 (MyoD) | DNA-binding transcription activator activity | [62] |
| Estrogen Receptor | nuclear receptor activity | [49] |
| Androgen Receptor | nuclear receptor activity | [48] |
| Signal Transducer and Activator of Transcription 3 (STAT3) | DNA-binding transcription factor activity | [63] |
| cMyc | DNA-binding transcription activator activity | [47,64] |
| Myocyte Enhancer Factor-2 (MEF2) | DNA-binding transcription activator activity | [65] |
| Class II Transactivator (CIITA) | activating transcription factor binding | [66] |
| Autoimmune regulator (AIRE) | DNA-binding transcription activator activity | [67] |
| SRY-related HMG-box 2 and 10 (Sox2/Sox10) | DNA-binding transcription factor | [68] |
| IKAROS | DNA-binding transcription factor activity | [69] |
| Peroxisome Proliferator Activated Receptor gamma (PPARγ) | DNA-binding transcription factor activity | [70] |
| Lim Domain Binding 1 (Ldb1) | enhancer sequence-specific DNA binding | [71] |
| Inhibitor | Alternative Names | IC50 | (nM) | Refs | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cdk1 | cdk2 | cdk3 | cdk4 | cdk5 | cdk6 | cdk7 | cdk9 | Other targets | |||
| Flavopiridol | Alvocidib | 30–400 | 100 | 410 | 20–40 | 110 | 60 | 110–300 | 6 | ** | |
| BAY1143572 | Atuveciclib | 1093 | 997 | 893 | n.d. | n.d. | n.d. | n.i. | 6 | GSK3 | ** |
| PHA-767491 | CAY10572 | 250 | 240 | n.d. | n.d. | 460 | n.d. | n.i. | 34 | cdc7 | ** |
| LY2857785 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 246 | 11 | ** | ||
| Dinaciclib | SCH727965 | 3 to 10 | 1 | 1 | n.d. | 0.8–1 | n.d. | 5 | 1 to 4 | ** | |
| Roscovitine | Seliciclib | 330–650 | 170–700 | 1500 | n.i. | 280 | 5100 | 800 | 230 | ** | |
| Voruciclib | 5.4–9.1 | n.d. | n.d. | 3.9 | n.d. | 2.9 | n.d. | 0.6–1.6 | [252] | ||
| SNS-032 | 480 | 38–48 | 56 | 925 | 740 | n.d. | 62 | 4 | ** | ||
| P276-00 | Riviciclib | 79 | 224–2500 | n.d. | 63 | n.d. | n.d. | 2870 | 40 | ** | |
| CDKI-73 | 4 to 8 | 30–33 | n.d. | 8 | n.d. | 38 | 91–134 | 4 to 6 | ** | ||
| i-CDK9 | HY16462, CDK9-IN-2 | 1700 | 240 | n.d. | 1800 | n.d. | 2 | 0.4 | ** | ||
| Wogonin | Vogonin | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 12300 | 190 | ** | |
| TG02 | Zotiraciclib | 9 | 5 | 8 | n.d. | n.d. | n.d. | 4 | 3 | Lck, Fyn, JAK, FLT3, etc. | [262] |
| FIT-039 | n.i. | n.d. | 30000 | n.i. | n.i. | n.i. | 5.8 | [234] | |||
| CCT068127 | 1100 | 10–110 | n.d. | 4800 | 70 | 6200 | 520 | 90 | [258] | ||
| AZD4573 | 37 | n.i. | n.d. | 1100 | 1100 | 1100 | 14 | [263] | |||
| MC180295 | 138 | 233–367 | 399 | 112 | 186 | 712 | 555 | 5 | [261] |
| Inhibitor | Alternative Names | Conditions or Diseases | Phase |
|---|---|---|---|
| Flavopiridol | Alvocidib | B-cell Chronic Lymphocytic Leukemia | 2 |
| Prostate cancer | 2 | ||
| (26 other phase 2 clinical trials and 33 phase 1 clinical trials) | |||
| BAY1143572 | Atuveciclib | Advanced Cancer | 1 |
| Acute Leukemia | 1 | ||
| Dinaciclib | SCH727965 | Refractory Chronic Lymphocytic Leukemia | 3 |
| Stage IV Melanoma | 2 | ||
| (Three other phase 2 clinical trials and 11 phase 1 clinical trials) | 2 | ||
| Roscovitine | Seliciclib | Cushing Disease | 2 |
| Moderately to Severely Active Ulcerative Colitis | 2 | ||
| Voruciclib | B-cell malignancies or AML | 1 | |
| SNS-032 | Advanced B-lymphoid Malignancies | 1 | |
| Advanced solid tumor | 1 | ||
| P276-00 | Riviciclib | Head and Neck Cancer | 2 |
| Melanoma | 2 | ||
| (Four other phase 2 clinical trials and four phase 1 clinical trials) | |||
| TG02 | Zotiraciclib | Brain Timor | 2 |
| Advanced Hematological Malignancies | 1 | ||
| (Three other phase 1 clinical trials) | |||
| AZD4573 | Relapsed or Refractory Hematological Malignancies | 1 | |
| TP-1287 | Advanced Solid Tumor | 1 |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fujinaga, K. P-TEFb as A Promising Therapeutic Target. Molecules 2020, 25, 838. https://doi.org/10.3390/molecules25040838
Fujinaga K. P-TEFb as A Promising Therapeutic Target. Molecules. 2020; 25(4):838. https://doi.org/10.3390/molecules25040838
Chicago/Turabian StyleFujinaga, Koh. 2020. "P-TEFb as A Promising Therapeutic Target" Molecules 25, no. 4: 838. https://doi.org/10.3390/molecules25040838
APA StyleFujinaga, K. (2020). P-TEFb as A Promising Therapeutic Target. Molecules, 25(4), 838. https://doi.org/10.3390/molecules25040838





