Targeted Isolation of Indole Alkaloids from Streptomyces sp. CT37
Abstract
1. Introduction
2. Results and Discussion
2.1. Structure Elucidation
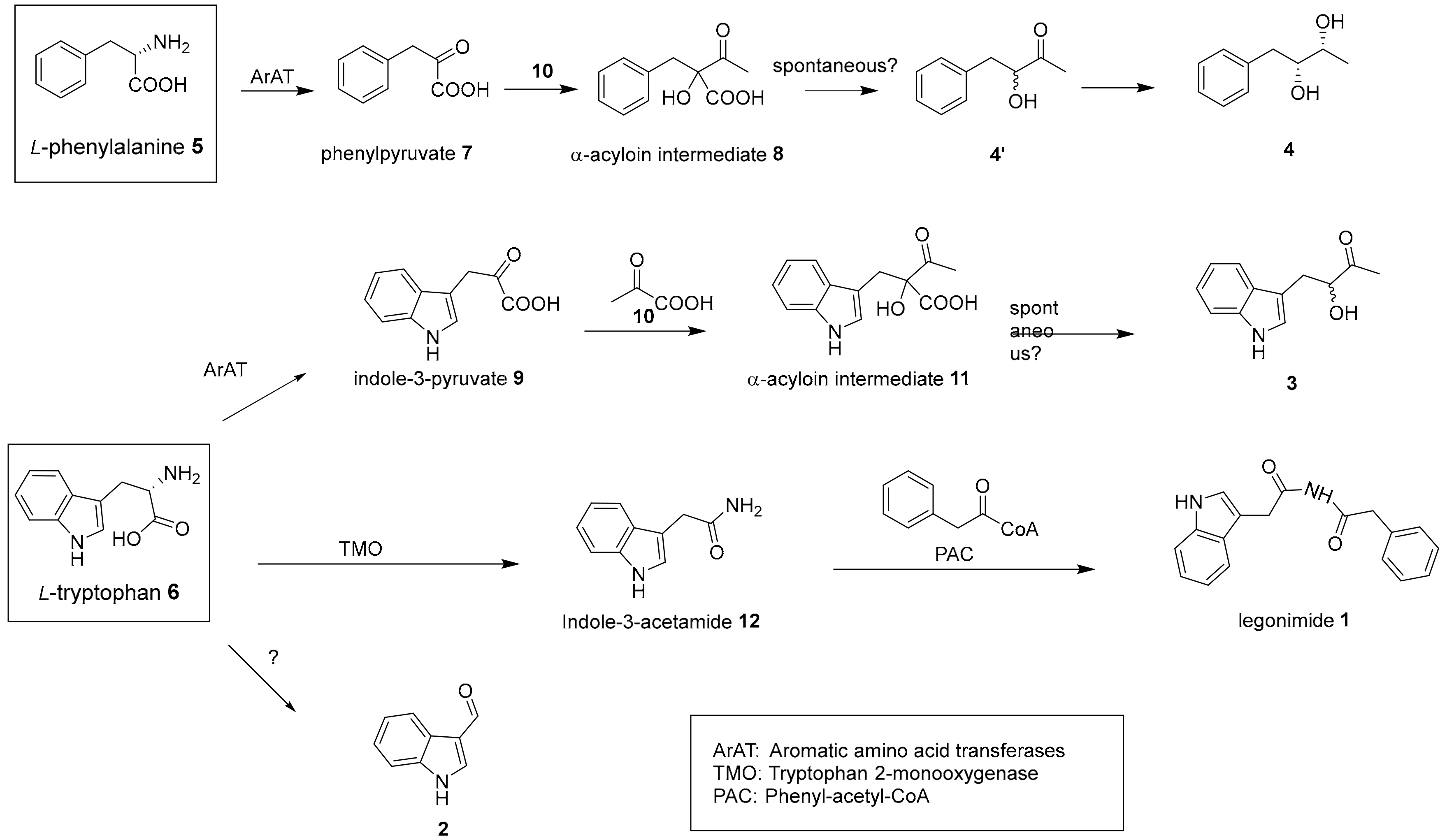
2.2. Proposed Biosynthesis Pathway of 1–4
2.3. Biological Test
3. Experimental
3.1. General Experimental Procedures
3.2. Biological Material Collection and Identification
3.3. Smale Scale Cultivation of Streptomyces sp. CT37
3.4. Disc Diffusion Assay
3.5. Large-Scale Fermentation of Streptomyces sp. CT37
3.6. Feature Based Molecular Networking
3.7. HPLC Isolation
3.8. Minimum Inhibitory Concentration
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ngo, L.T.; Okogun, J.I.; Folk, W.R. 21st Century natural product research and drug development and traditional medicines. Nat. Prod. Rep. 2013, 30, 584–592. [Google Scholar] [CrossRef]
- Mohana, N.C.; Rao, H.C.Y.; Rakshith, D.; Mithun, P.R.; Nuthan, B.R.; Satish, S. Omics based approach for biodiscovery of microbial natural products in antibiotic resistance era. J. Genet. Eng. Biotechnol. 2018, 16, 1–8. [Google Scholar] [CrossRef]
- Hopwood, D.A. How do antibiotic-producing bacteria ensure their self-resistance before antibiotic biosynthesis incapacitates them? Mol. Microbiol. 2007, 63, 937–940. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Maglangit, F.; Wu, L.; Ebel, R.; Kyeremeh, K.; Andersen, J.H.; Annang, F.; Pérez-Moreno, G.; Reyes, F.; Deng, H. Signalling and bioactive metabolites from Streptomyces sp. RK44. Molecules 2020, 25, 460. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Ma, L.; Bandaranayaka, N.; Qin, Z.; Mann, G.; Kyeremeh, K.; Yu, Y.; Shepherd, T.; Naismith, J.H.; O’Hagan, D. Identification of Fluorinases from Streptomyces sp MA37, Norcardia brasiliensis, and Actinoplanes sp N902-109 by genome mining. ChemBioChem 2014, 15, 364–368. [Google Scholar] [CrossRef] [PubMed]
- Maglangit, F.; Tong, M.H.; Jaspars, M.; Kyeremeh, K.; Deng, H. Legonoxamines A-B, two new hydroxamate siderophores from the soil bacterium, Streptomyces sp. MA37. Tetrahedron Lett. 2019, 60, 75–79. [Google Scholar] [CrossRef]
- Maglangit, F.; Fang, Q.; Leman, V.; Soldatou, S.; Ebel, R.; Kyeremeh, K.; Deng, H. Accramycin A, a new aromatic polyketide, from the soil bacterium, Streptomyces sp. MA37. Molecules 2019, 24, 3384. [Google Scholar] [CrossRef]
- Marmann, A.; Aly, A.H.; Lin, W.; Wang, B.; Proksch, P. Co-cultivation—A powerful emerging tool for enhancing the chemical diversity of microorganisms. Mar. Drugs 2014, 12, 1043–1065. [Google Scholar] [CrossRef]
- Tabudravu, J.N.; Pellissier, L.; Smith, A.J.; Subko, K.; Autreáu, C.; Feussner, K.; Hardy, D.; Butler, D.; Kidd, R.; Milton, E.J.; et al. LC-HRMS-database screening metrics for rapid prioritization of samples to accelerate the discovery of structurally new natural products. J. Nat. Prod. 2019, 82, 211–220. [Google Scholar] [CrossRef]
- Su, L.; Zhang, R.; Kyeremeh, K.; Deng, Z.; Deng, H.; Yu, Y. Dissection of the neocarazostatin: A C4 alkyl side chain biosynthesis by in vitro reconstitution. Org. Biomol. Chem. 2017, 3843–3848. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Lv, M.; Kyeremeh, K.; Deng, Z.; Deng, H.; Yu, Y. A ThDP-dependent enzymatic carboligation reaction involved in neocarazostatin a tricyclic carbazole formation. Org. Biomol. Chem. 2016, 14, 8679–8684. [Google Scholar] [CrossRef] [PubMed]
- Rateb, M.E.; Zhai, Y.; Ehrner, E.; Rath, C.M.; Wang, X.; Tabudravu, J.; Ebel, R.; Bibb, M.; Kyeremeh, K.; Dorrestein, P.C.; et al. Legonaridin, a new member of linaridin RiPP from a Ghanaian Streptomyces isolate. Org. Biomol. Chem. 2015, 13, 9585–9592. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Tabudravu, J.; Elsayed, S.S.; Travert, J.; Peace, D.; Tong, M.H.; Kyeremeh, K.; Kelly, S.M.; Trembleau, L.; Ebel, R.; et al. Discovery of a single monooxygenase that catalyzes carbamate formation and ring contraction in the biosynthesis of the legonmycins. Angew. Chem. Int. Ed. 2015, 54, 12697–12701. [Google Scholar] [CrossRef] [PubMed]
- Maglangit, F.; Fang, Q.; Kyeremeh, K.; Sternberg, J.M.; Ebel, R.; Deng, H. Co-Culturing Approach Enables Discovery and Biosynthesis of a Bioactive Indole Alkaloid Metabolite. Molecules 2020, 25, 256. [Google Scholar] [CrossRef]
- Callahan, D.L.; Elliott, C.E.; Walker, J.M. Metabolomics Tools for Natural Product Discovery; Methods in Molecular Biology 1055; HUMANA: Louisville, KY, USA, 2013; Volume 1055, ISBN 978-1-62703-576-7. [Google Scholar]
- Cox, D.G.; Oh, J.; Keasling, A.; Colson, K.; Hamann, M.T. The utility of metabolomics in natural product and biomarker characterization. Biochim. Biophys. Acta 2014, 1840, 3460–3474. [Google Scholar] [CrossRef]
- Networking, S.M.; Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.D.T.; Watrous, J.; Kapono, C.A.; et al. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar]
- Ernst, M.; Kang, K.B.; Caraballo-Rodríguez, A.M.; Nothias, L.-F.; Wandy, J.; Chen, C.; Wang, M.; Rogers, S.; Medema, M.H.; Dorrestein, P.C.; et al. MolNetEnhancer: Enhanced molecular networks by integrating metabolome mining and annotation tools. Metabolites 2019, 9, 144. [Google Scholar] [CrossRef]
- Borges, R.M.; Taujale, R.; de Souza, J.S.; de Andrade Bezerra, T.; Silva, E.L.E.; Herzog, R.; Ponce, F.V.; Wolfender, J.-L.; Edison, A.S. Dereplication of plant phenolics using a mass-spectrometry database independent method. Phytochem. Anal. 2018, 29, 601–612. [Google Scholar] [CrossRef]
- Blaženović, I.; Kind, T.; Torbašinović, H.; Obrenović, S.; Mehta, S.S.; Tsugawa, H.; Wermuth, T.; Schauer, N.; Jahn, M.; Biedendieck, R.; et al. Comprehensive comparison of in silico MS/MS fragmentation tools of the CASMI contest: Database boosting is needed to achieve 93% accuracy. J. Cheminform. 2017, 9, 32. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of streptomyces species. Int. J. Syst. Bacteriol. 1966, 16, 313–340. [Google Scholar] [CrossRef]
- Mohimani, H.; Gurevich, A.; Shlemov, A.; Mikheenko, A.; Korobeynikov, A.; Cao, L.; Shcherbin, E.; Nothias, L.-F.; Dorrestein, P.C.; Pevzner, P.A. Dereplication of microbial metabolites through database search of mass spectra. Nat. Commun. 2018, 9, 4035. [Google Scholar] [CrossRef] [PubMed]
- Santen, J.; Van, A.; Jacob, G.; Singh, A.L.; Aniebok, V.; Balunas, M.J.; Bunsko, D.; Neto, F.C.; Castaño-Espriu, L.; Chang, C.; et al. The natural products atlas: An open access knowledge base for microbial natural products discovery. ACS Cent. Sci. 2019, 5, 1824–1833. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Dong, W.; Li, X.; Zhang, H. Indole alkaloids from the roots of isatis ingigotica and their antiherpes simplex virus type 2 (HSV-2) activity in vitro. Chem. Nat. Compd. 2010, 46, 763–766. [Google Scholar] [CrossRef]
- Awano, K.I.; Yanai, T.; Watanabe, I.; Takagi, Y.; Kitahara, T.; Mori, K. Synthesis of all four possible stereoisomers of 1-phenyl-2,3-butanediol and both enantiomers of 3-hydroxy-4-phenyl-2-butanone to determine the absolute configuration of the natural constituents. Biosci. Biotechnol. Biochem. 1995, 59, 1251–1254. [Google Scholar] [CrossRef][Green Version]
- Huang, S.X.; Powell, E.; Rajski, S.R.; Zhao, L.X.; Jiang, C.L.; Duan, Y.; Xu, W.; Shen, B. Discovery and total synthesis of a new estrogen receptor heterodimerizing actinopolymorphol A from actinopolymorpha rutilus. Org. Lett. 2010, 12, 3525–3527. [Google Scholar] [CrossRef]
- Parra, R.D.; Furukawa, M.; Gong, B.; Zeng, X.C. Energetics and cooperativity in three-center hydrogen bonding interactions. I. diacetamide-X dimers (X=HCN, CH3OH). J. Chem. Phys. 2001, 115, 6030–6035. [Google Scholar] [CrossRef]
- Hvoslef, J.; Tracy, M.L.; Nash, C.P. Interatomic distances and angles in four planar systems with adjacent C–O and C–N bonds: Structures of pivalamide (I), dipivalamide (II),N-pivaloylpivalamidinium pyrosulfate (III) and N-pivaloylpivalamidine (IV). Acta Crystallogr. Sect. C Cryst. Struct. Commun. 1986, 42, 353–360. [Google Scholar] [CrossRef]
- Hinterberger, S.; Hofer, O.; Greger, H. Synthesis and corrected structures of sulphur-containing amides from glycosmis species: Sinharines, penimides, and illukumbins. Tetrahedron 1994, 50, 6279–6286. [Google Scholar] [CrossRef]
- Cardoso-Martínez, F.; De La Rosa, J.M.; Díaz-Marrero, A.R.; Darias, J.; D’Croz, L.; Cerella, C.; Diederich, M.; Cueto, M. Oximoaspergillimide, a fungal derivative from a marine isolate of Aspergillus sp. Eur. J. Org. Chem. 2015, 2015, 2256–2261. [Google Scholar] [CrossRef]
- Karplus, M. Contact electron-spin coupling of nuclear magnetic moments. J. Chem. Phys. 1959, 30, 11–15. [Google Scholar] [CrossRef]
- Yu, L.; Liu, J.; Yu, L.; Chen, L.; Qiu, F. Chemical constituents of seed oil leavings of xanthoceras sorbifolia. Chem. Nat. Compd. 2018, 54, 769–771. [Google Scholar] [CrossRef]
- Netz, N.; Opatz, T. Marine indole alkaloids. Mar. Drugs 2015, 13, 4814–4914. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.W.; Cordell, G.A. Metabolism studies of indole derivatives using a staurosporine producer, Streptomyces staurosporeus. J. Nat. Prod. 1997, 60, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Lacret, R.; Oves-Costales, D.; Gómez, C.; Díaz, C.; De La Cruz, M.; Pérez-Victoria, I.; Vicente, F.; Genilloud, O.; Reyes, F. New ikarugamycin derivatives with antifungal and antibacterial properties from streptomyces zhaozhouensis. Mar. Drugs 2015, 13, 128–140. [Google Scholar] [CrossRef]
- Cartus, A.T.; Stegmüller, S.; Simson, N.; Wahl, A.; Neef, S.; Kelm, H.; Schrenk, D. Hepatic metabolism of carcinogenic β-asarone. Chem. Res. Toxicol. 2015, 28, 1760–1773. [Google Scholar] [CrossRef]
- Xie, C.L.; Niu, S.W.; Zhou, T.T.; Zhang, G.Y.; Yang, Q.; Yang, X.W. Chemical constituents and chemotaxonomic study on the marine actinomycete Williamsia sp. MCCC 1A11233. Biochem. Syst. Ecol. 2016, 67, 129–133. [Google Scholar] [CrossRef]
- Böttcher, C.; Chapman, A.; Fellermeier, F.; Choudhary, M.; Scheel, D.; Glawischnig, E. The biosynthetic pathway of indole-3-carbaldehyde and indole-3-carboxylic acid derivatives in arabidopsis. Plant Physiol. 2014, 165, 841–853. [Google Scholar] [CrossRef]
- Khan, A.R.; Park, G.S.; Asaf, S.; Hong, S.J.; Jung, B.K.; Shin, J.H. Complete genome analysis of Serratia marcescens RSC-14: A plant growth-promoting bacterium that alleviates cadmium stress in host plants. PLoS ONE 2017, 12, 1–17. [Google Scholar] [CrossRef]
- Zhang, L.S.; Davies, S.S. Microbial metabolism of dietary components to bioactive metabolites: Opportunities for new therapeutic interventions. Genome Med. 2016, 8, 46. [Google Scholar] [CrossRef]
- Hudzicki, J. Kirby-bauer disk diffusion susceptibility test protocol author information. Am. Soc. Microbiol. 2009, 1–23. [Google Scholar]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. [Google Scholar] [CrossRef] [PubMed]
- Nothias, L.F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H. Feature-based molecular networking in the GNPS analysis environment. BiorXiv 2019, 812404. [Google Scholar]
- Pluskal, T.; Castillo, S.; Villar-briones, A.; Orešič, M.; Ore, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef]
- Carpenter, D.E.; Karen Anderson Diane Citron, C.M.; JoAnn Dzink-Fox, B.L.; Meredith Hackel, M.; Jenkins, S.G.; Cindy Knapp, F.; Laura Koeth, M.; Audrey Schuetz, M.N.; Hannah Wexler, D. Methods for Antimicrobial Susceptibility Testing of Anaerobic Bacteria; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; pp. 11–19. [Google Scholar]
Sample Availability: Not available. |
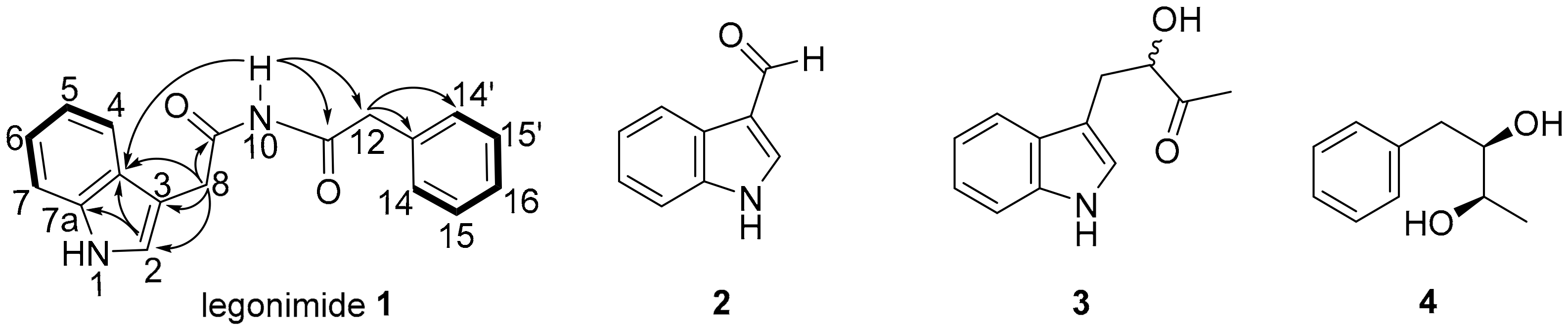
| Position | 1H (mult., J in Hz) | 13C |
|---|---|---|
| 1-NH | 10.96 (1H, s) | - |
| 2 | 7.18 (1H, s) | 123.6 |
| 3 | - | 109.2 |
| 4a | - | 127.3 |
| 4 | 7.54 (1H, d, J = 7.85) | 118.7 |
| 5 | 7.02 (1H, t, J = 7.39) | 118.5 |
| 6 | 7.06 (1H, t, J = 7.32) | 120.8 |
| 7 | 7.34 (1H, d, J = 7.80) | 111.3 |
| 7a | - | 136.5 |
| 8 | 3.46(2H, s) | 32.4 |
| 9 | - | 173.1 |
| 10-NH | 7.37 (1H, s) | - |
| 11 | - | 172.3 |
| 12 | 3.35(2H, s) | 42.0 |
| 13 | - | 136.6 |
| 14,14′ | 7.28 (2H, d, J = 7.67) | 128.1 |
| 15,15′ | 7.21 (2H, m) | 126.4 |
| 16 | 7.31 (1H, m) | 126.3 |
| C. albicans ATCC 10231 MIC (µg/mL) | |
| Legonimide 1 | 21.54 |
| Compound 2 | 11.47 |
| Compound 3 | >50 |
| Compound 4 | >50 |
| Ampicillin | 3.193 |
| Tetracycline | 0.3615 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, Q.; Maglangit, F.; Mugat, M.; Urwald, C.; Kyeremeh, K.; Deng, H. Targeted Isolation of Indole Alkaloids from Streptomyces sp. CT37. Molecules 2020, 25, 1108. https://doi.org/10.3390/molecules25051108
Fang Q, Maglangit F, Mugat M, Urwald C, Kyeremeh K, Deng H. Targeted Isolation of Indole Alkaloids from Streptomyces sp. CT37. Molecules. 2020; 25(5):1108. https://doi.org/10.3390/molecules25051108
Chicago/Turabian StyleFang, Qing, Fleurdeliz Maglangit, Morgane Mugat, Caroline Urwald, Kwaku Kyeremeh, and Hai Deng. 2020. "Targeted Isolation of Indole Alkaloids from Streptomyces sp. CT37" Molecules 25, no. 5: 1108. https://doi.org/10.3390/molecules25051108
APA StyleFang, Q., Maglangit, F., Mugat, M., Urwald, C., Kyeremeh, K., & Deng, H. (2020). Targeted Isolation of Indole Alkaloids from Streptomyces sp. CT37. Molecules, 25(5), 1108. https://doi.org/10.3390/molecules25051108







