Interaction of Coumarin Phytoestrogens with ERα and ERβ: A Molecular Dynamics Simulation Study
Abstract
1. Introduction
2. Results and Discussion
2.1. Docking Analysis
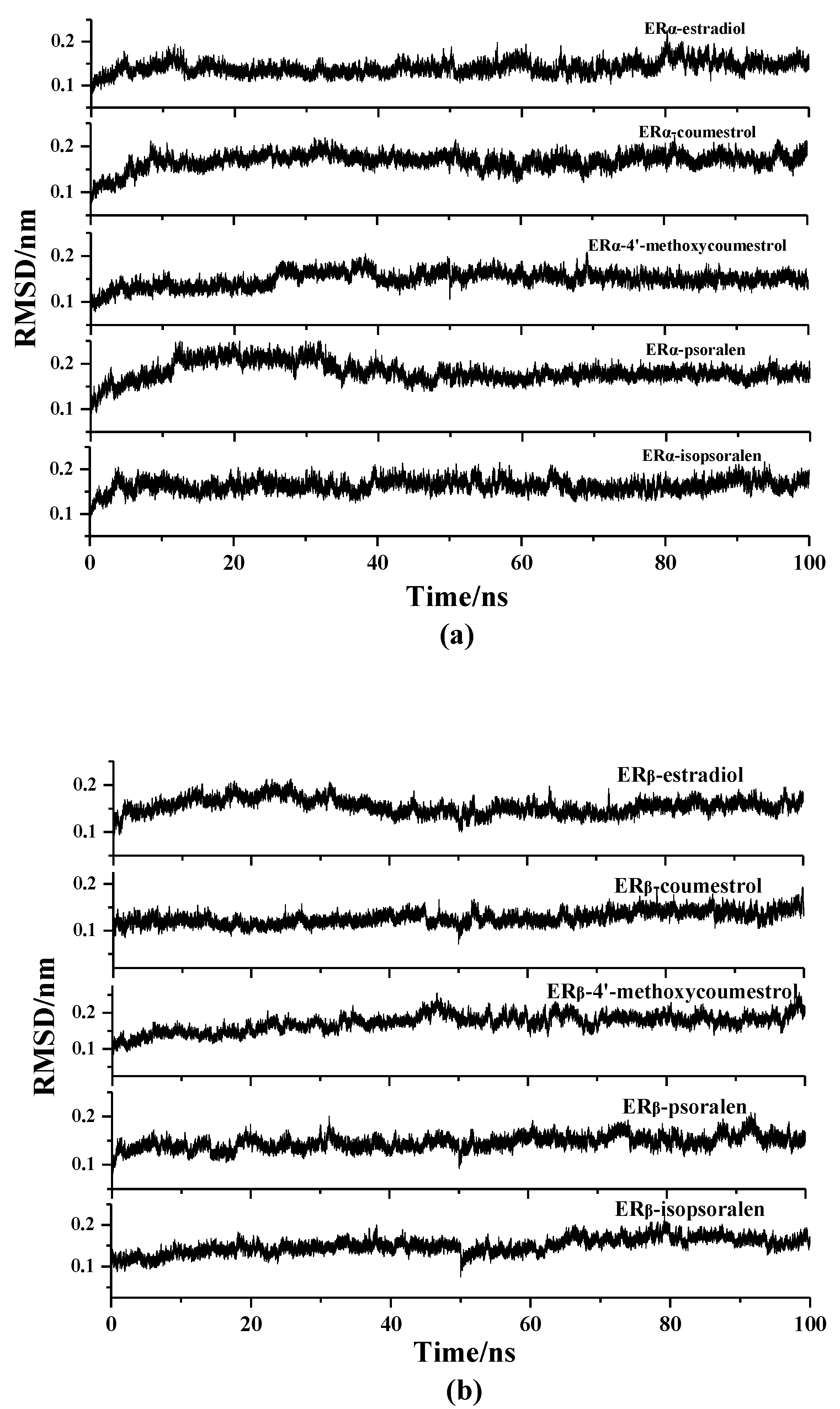
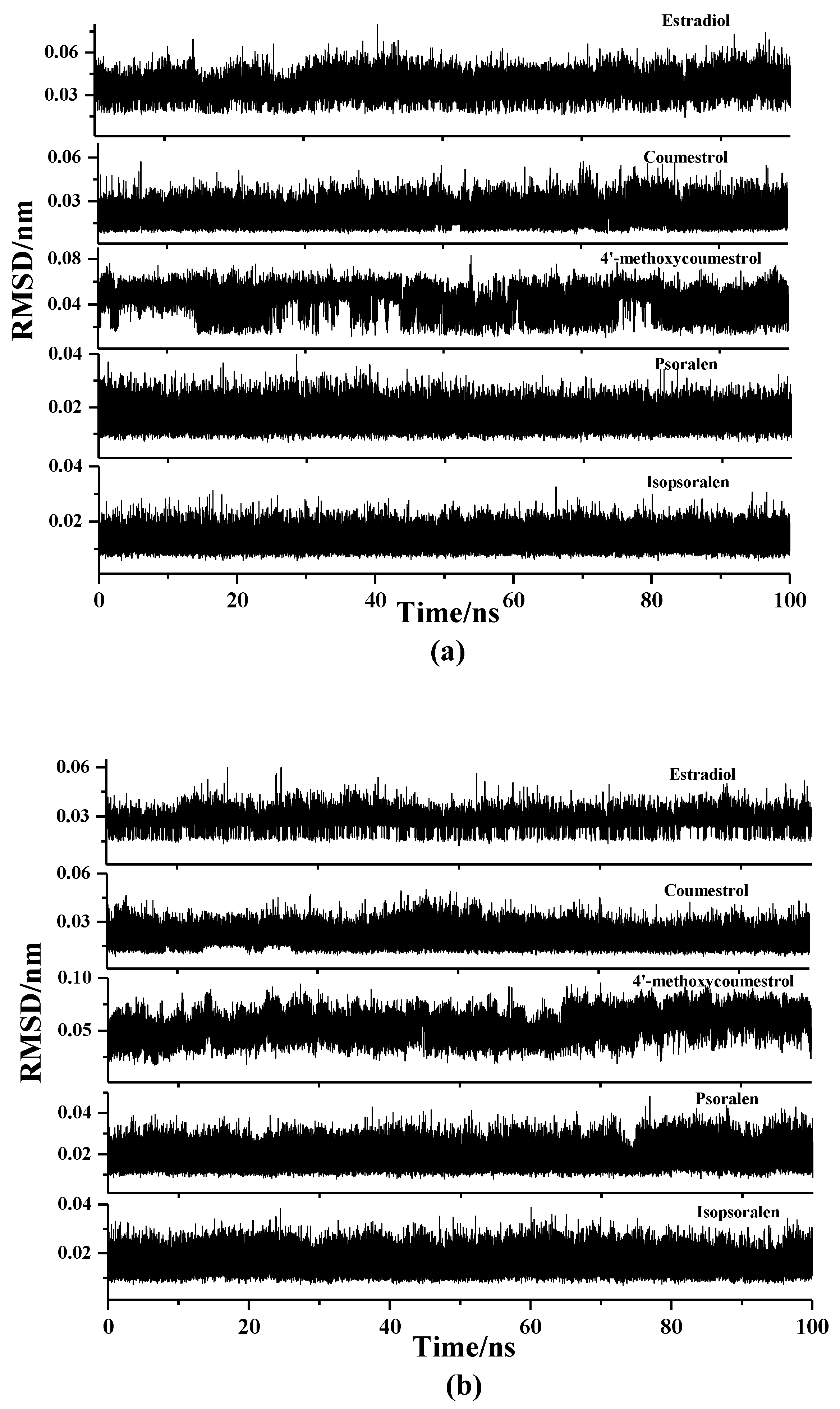
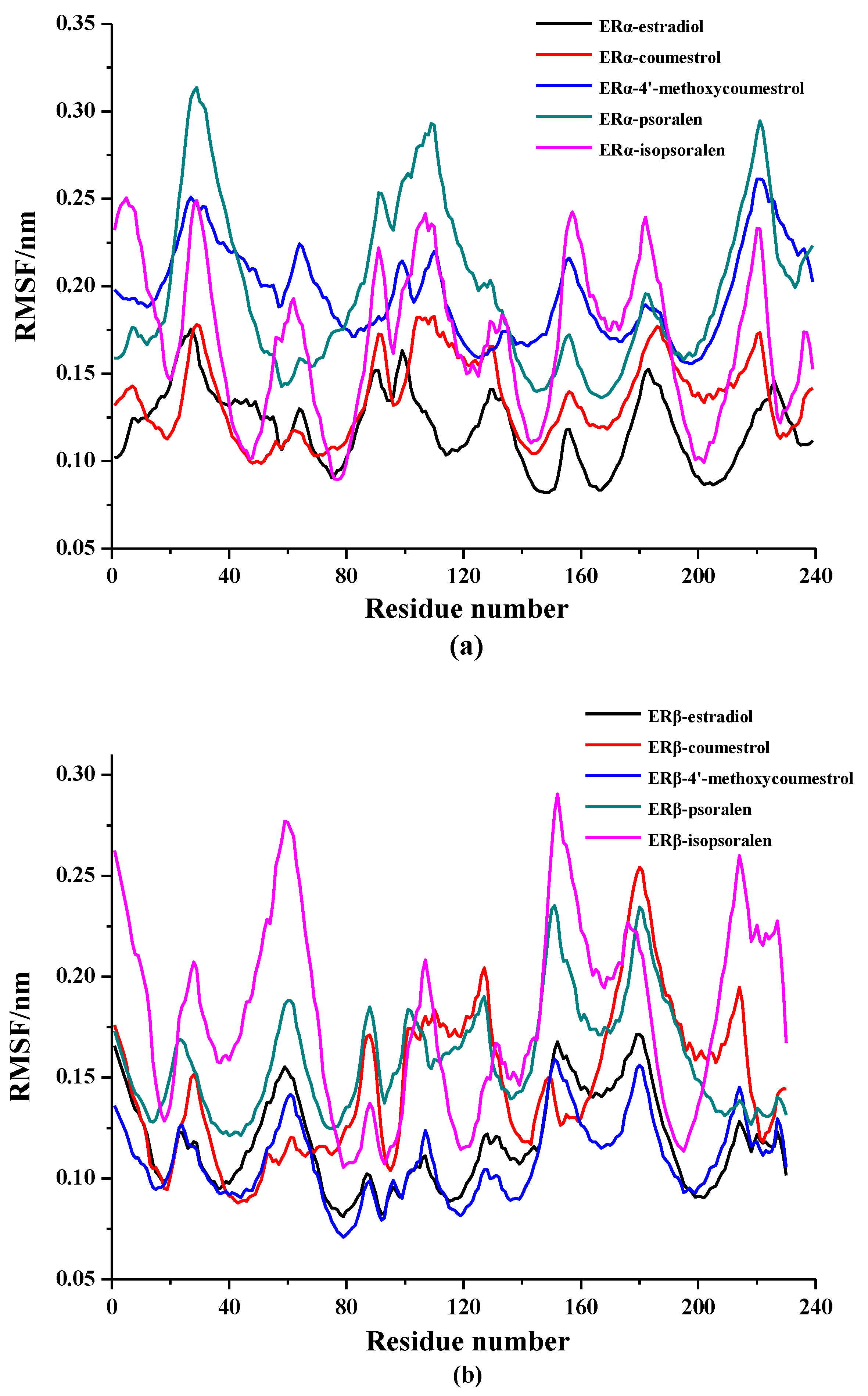
2.2. Molecular Dynamics Simulation Analysis
2.3. Binding Patterns Analysis
2.4. Binding Free Energy Analysis
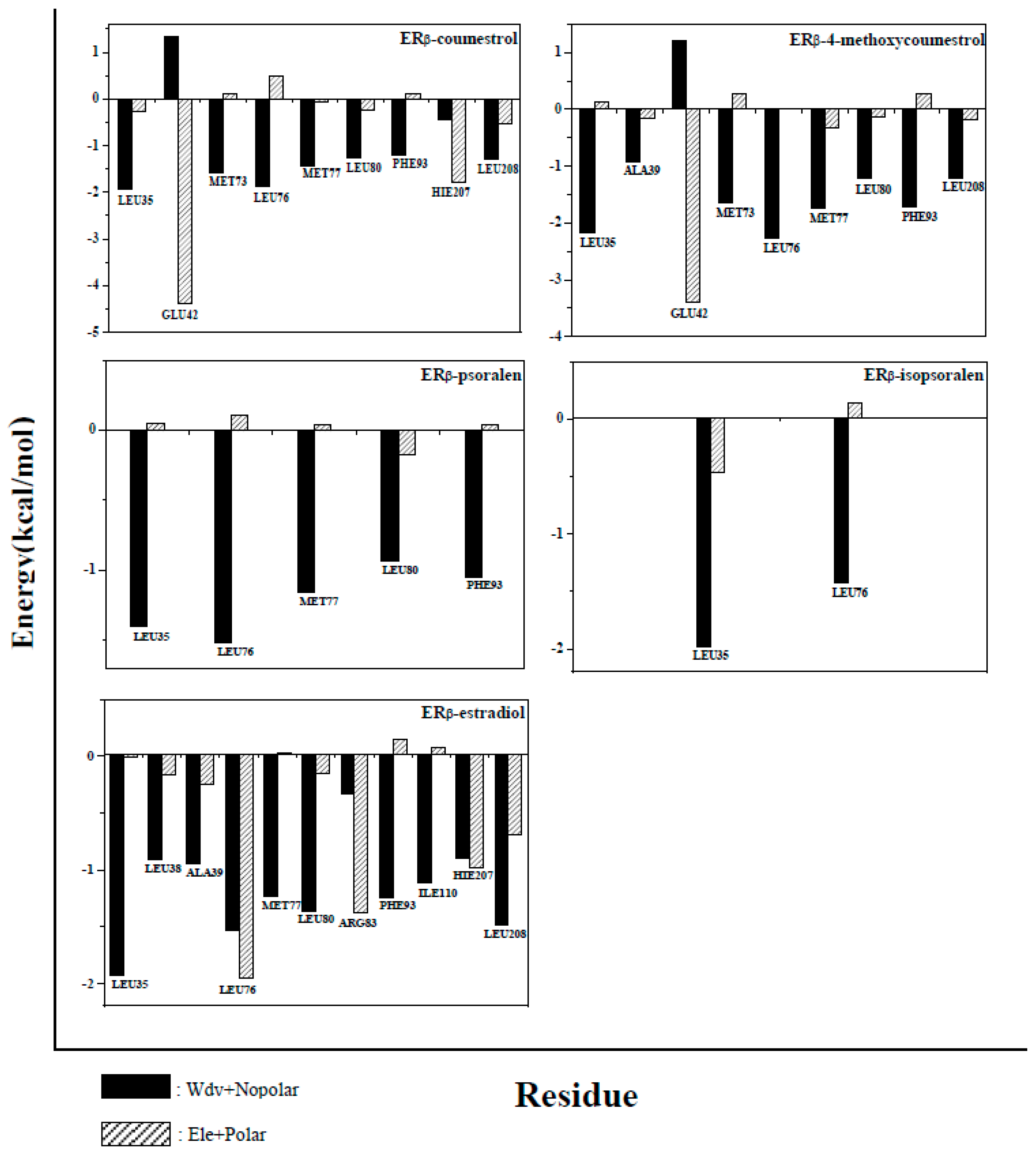
2.5. Binding Free Energy Decomposition
3. Materials and Methods
3.1. Protein and Ligand Preparation
3.2. Molecular Docking Studies
3.3. Molecular Dynamics Simulations
3.4. Binding Free Energy Calculations and Energy Decomposition
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ososki, A.L.; Kennelly, E.J. Phytoestrogens: A review of the present state of research. Phytother. Res. 2003, 17, 845–869. [Google Scholar] [CrossRef]
- Adlercreutz, H. Phyto-oestrogens and cancer. Lancet Oncol. 2002, 3, 364–373. [Google Scholar] [CrossRef]
- Gaynor, M.L. Isoflavones and the prevention and treatment of prostate disease: Is there a role? Cleve. Clin. J. Med. 2003, 70, 203–204. [Google Scholar] [CrossRef]
- Van Der Schouw, Y.T.; Raats, M.; Groot, L.D.; Staveren, W.V. Phytoestrogens and the health of older women. Food Ageing Popul. 2009, 21, 430–457. [Google Scholar]
- Lecomte, S.; Lelong, M.; Bourgine, G.; Efstathiou, T.; Saligaut, C.; Pakdel, F. Assessment of the potential activity of major dietary compounds as selective estrogen receptor modulators in two distinct cell models for proliferation and differentiation. Toxicol. Appl. Pharmacol. 2017, 325, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Murkies, A.L.; Wilcox, G.; Davis, S.R. Clinical review 92: Phytoestrogens. J. Clin. Endocrinol. Metab. 1998, 83, 297–303. [Google Scholar] [PubMed]
- Ibarreta, D.; Daxenberger, A.; Meyer, H.H. Possible health impact of phytoestrogens and xenoestrogens in food. APMIS 2001, 109, 161–184. [Google Scholar] [CrossRef] [PubMed]
- Ayubi, E.; Safiri, S. Dietary isoflavone intake and all-cause mortality in breast cancer survivors: The Breast Cancer Family Registry-methodological issues. Cancer 2017, 123, 3638–3639. [Google Scholar] [CrossRef]
- Tina, S.; Niemeyer, H.B.; Honig, D.M.; Manfred, M. Identification and stereochemical characterization of lignans in flaxseed and pumpkin seeds. J. Agric. Food Chem. 2003, 51, 1181–1188. [Google Scholar]
- Alekel, D.L.; Germain, A.S.; Peterson, C.T.; Hanson, K.B.; Stewart, J.W.; Toda, T. Isoflavone-rich soy protein isolate attenuates bone loss in the lumbar spine of perimenopausal women. Am. J. Clin. Nutr. 2000, 72, 844–852. [Google Scholar] [CrossRef]
- Goodman-Gruen, D.; Kritz-Silverstein, D. Usual dietary isoflavone intake is associated with cardiovascular disease risk factors in postmenopausal women. J. Nutr. 2001, 131, 1202–1206. [Google Scholar] [CrossRef] [PubMed]
- Tice, J.A.; Ettinger, B.; Ensrud, K.; Wallace, R.; Blackwell, T.; Cummings, S.R. Phytoestrogen supplements for the treatment of hot flashes: The Isoflavone Clover Extract (ICE) Study: A randomized controlled trial. J. Am. Med. Assoc. 2003, 290, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.Y.; Qin, L.Q. Soy isoflavones consumption and risk of breast cancer incidence or recurrence: A meta-analysis of prospective studies. Breast Cancer Res. Treat. 2011, 125, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Reger, M.K.; Zollinger, T.W.; Liu, Z.; Jones, J.; Zhang, J. Dietary intake of isoflavones and coumestrol and the risk of prostate cancer in the prostate, lung, colorectal and ovarian cancer screening trial. Int. J. Cancer 2018, 142, 719–728. [Google Scholar] [CrossRef] [PubMed]
- Prossnitz, E.R.; Arterburn, J.B. International union of basic and clinical pharmacology. XCVII. G-protein-coupled estrogen receptor and its pharmacologic modulators. Pharmacol. Rev. 2015, 67, 505–540. [Google Scholar] [CrossRef] [PubMed]
- Mazur, W. Phytoestrogen content in foods. Baillieres Clin. Endocrinol. Metab. 1998, 12, 729–742. [Google Scholar] [CrossRef]
- Zhang, S.Y.; Meng, L.; Gao, W.Y.; Song, N.N.; Jia, W.; Duan, H.Q. Advances on biological activities of coumarins. China J. Chin. Mater. Med. 2005, 30, 410–414. [Google Scholar]
- Wu, C.T.; Tzeng, J.N.; Lai, J.N.; Tsan, S.H.; Wang, J.D. Prescription profile of Chinese herbal products containing coumestrol, genestein, and/or daidzein among female users: An analysis of national health insurance data in Taiwan between 1997 and 2007. Chin. Med. 2012, 7, 22–28. [Google Scholar] [CrossRef]
- Zhao, P.; Niu, J.; Wang, J.; Wang, L. The role of phytoestrogens in psoralen and its mechanism. J. Tradit. Chin. Med. 2008, 33, 59–63. [Google Scholar]
- Patil, R.; Sawant, S. Molecular dynamics guided receptor independent 4D QSAR studies of substituted coumarins as anticancer agents. Curr. Comput. Aided Drug Des. 2015, 11, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Mcinnes, C.; Zhu, B.T. Structural characterization of the binding interactions of various endogenous estrogen metabolites with human estrogen receptor α and β subtypes: A molecular modeling study. PLoS ONE 2013, 8, 74615–74626. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Baker, M.E.; Lathe, R. The promiscuous estrogen receptor: Evolution of physiological estrogens and response to phytochemicals and endocrine disruptors. J. Steroid Biochem. Mol. Biol. 2018, 184, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Gutendorf, B.; Westendorf, J. Comparison of an array of in vitro assays for the assessment of the estrogenic potential of natural and synthetic estrogens, phytoestrogens and xenoestrogens. Toxicol 2001, 166, 79–89. [Google Scholar] [CrossRef]
- Chrzan, B.G.; Bradford, P.G. Phytoestrogens activate estrogen receptor beta1 and estrogenic responses in human breast and bone cancer cell lines. Mol. Nutr. Food Res. 2010, 51, 171–177. [Google Scholar] [CrossRef]
- Zafar, A.; Ahmad, S.; Naseem, I. Insight into the structural stability of coumestrol with human estrogen receptor α and β subtypes: A combined approach involving docking and molecular dynamics simulation studies. RSC Adv. 2015, 5, 81295–81312. [Google Scholar] [CrossRef]
- Acharya, R.; Chacko, S.; Bose, P.; Lapenna, A.; Pattanayak, S.P. Structure based multitargeted molecular docking analysis of selected furanocoumarins against breast cancer. Sci. Rep. 2019, 9, 15743. [Google Scholar] [CrossRef]
- Rohan, P.; Suranjana, D.; Ashley, S.; Lumbani, Y.; Akulapalli, S.; Varma, A.K. Optimized hydrophobic interactions and hydrogen bonding at the target-ligand interface leads the pathways of drug-designing. PLoS ONE 2010, 5, 12029–12039. [Google Scholar]
- Lu, Y.; Wang, Y.; Xu, Z.; Yan, X.; Luo, X.; Jiang, H.; Zhu, W. C−X···H contacts in biomolecular systems: How they contribute to protein−ligand binding affinity. J. Phys. Chem. B 2009, 113, 12615–12621. [Google Scholar] [CrossRef]
- Zhang, T.; Zhong, S.; Meng, Y.; Deng, W.; Hou, L.; Wang, Y.; Xing, X.; Guan, T.; Zhang, J.; Li, T. Quantitative structure-activity relationship for estrogenic flavonoids from Psoralea corylifolia. J. Pharm. Biomed. Anal. 2018, 161, 129–135. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, W.; Wang, Y.; Xing, X.; Zhong, S.; Guan, T.; Zhang, T.; Hou, L.; Li, T. Estrogen receptor-based fluorescence polarization assay for bisphenol analogues and molecular modeling study of their complexation mechanism. Anal. Chim. Acta 2018, 1032, 107–113. [Google Scholar] [CrossRef]
- Zhao, L.; Brinton, R.D. Structure-based virtual screening for plant-based ERβ-selective ligands as potential preventative therapy against age-related neurodegenerative diseases. J. Med. Chem. 2005, 48, 3463–3466. [Google Scholar] [CrossRef] [PubMed]
- Enyedy, I.J.; Wang, J.; Zaman, W.A.; Johnson, K.M.; Wang, S. Discovery of substituted 3,4-Diphenyl-thiazoles as a novel class of monoamine transporter inhibitors through 3-D pharmacophore search using a new pharmacophore model derived from mazindol. Biorg. Med. Chem. Lett. 2002, 12, 1775–1778. [Google Scholar] [CrossRef]
- Berman, H.; Battistuz, T.; Bhat, T.; Bluhm, W.; Bourne, P.; Burkhardt, K.; Feng, Z.; Gilliland, G.; Iype, L.; Fagan, P.; et al. The Protein data Bank. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002, 58, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Shivakumar, D.; Williams, J.; Wu, Y.; Damm, W.; Shelley, J.; Sherman, W. Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J. Chem. Theory Comput. 2010, 6, 1509–1519. [Google Scholar] [CrossRef]
- Sunghwan, K.; Thiessen, P.A.; Bolton, E.E.; Jie, C.; Gang, F.; Asta, G.; Han, L.; He, J.; He, S.; Shoemaker, B.A. Pubchem substance and compound databases. Nucleic Acids Res. 2016, 44, 1202–1213. [Google Scholar]
- Shelley, J.C.; Cholleti, A.; Frye, L.L.; Greenwood, J.R.; Timlin, M.R.; Uchimaya, M. Epik: A software program for pK a prediction and protonation state generation for drug-like molecules. J. Comput. Aided Mol. Des. 2007, 21, 681–691. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Mee, S.; Perry, J.K. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Halgren, T.A.; Murphy, R.B.; Friesner, R.A.; Beard, H.S.; Frye, L.L.; Pollard, W.T.; Banks, J.L. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004, 47, 1750–1759. [Google Scholar] [CrossRef]
- Repasky, M.P.; Shelley, M.; Friesner, R.A. Flexible ligand docking with Glide. Curr. Protoc. Hum. Genet. 2007, 18, 1–36. [Google Scholar]
- Case, D.A.; Babin, V. AMBER 14; University of California: San Francisco, CA, USA, 2014. [Google Scholar]
- Case, D.A.; Cheatham, T.E.; Tom, D.; Holger, G.; Ray, L.; Merz, K.M.; Alexey, O.; Carlos, S.; Bing, W.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2010, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Joung, I.S.; Cheatham, R.T. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B 2008, 112, 9020–9041. [Google Scholar] [CrossRef] [PubMed]
- Darden, T.; York, D.M.; Pedersen, L.G. Particle mesh Ewald: An Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Roe, D.R. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Miller, I.B.; McGee, J.T.; Swails, J.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
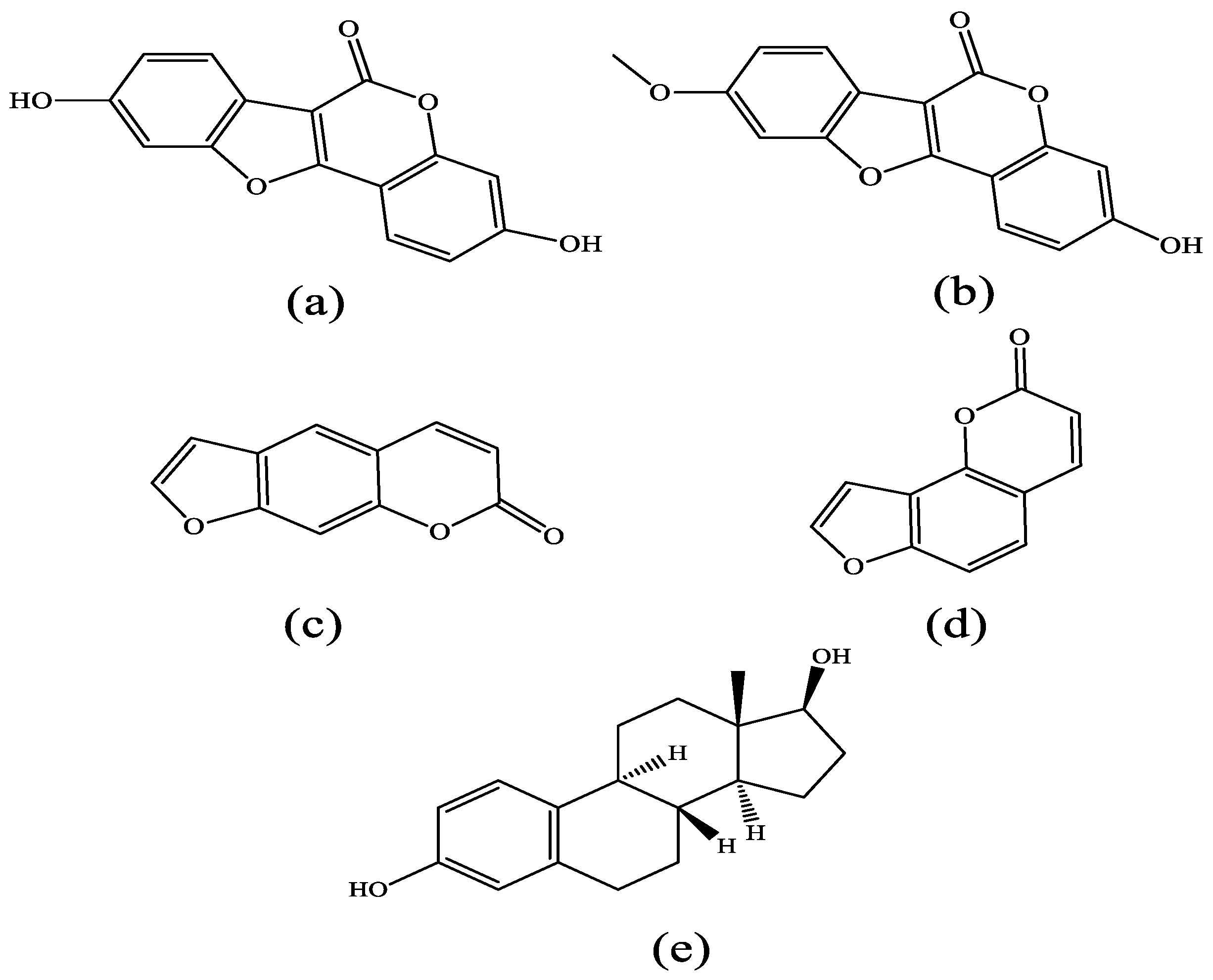
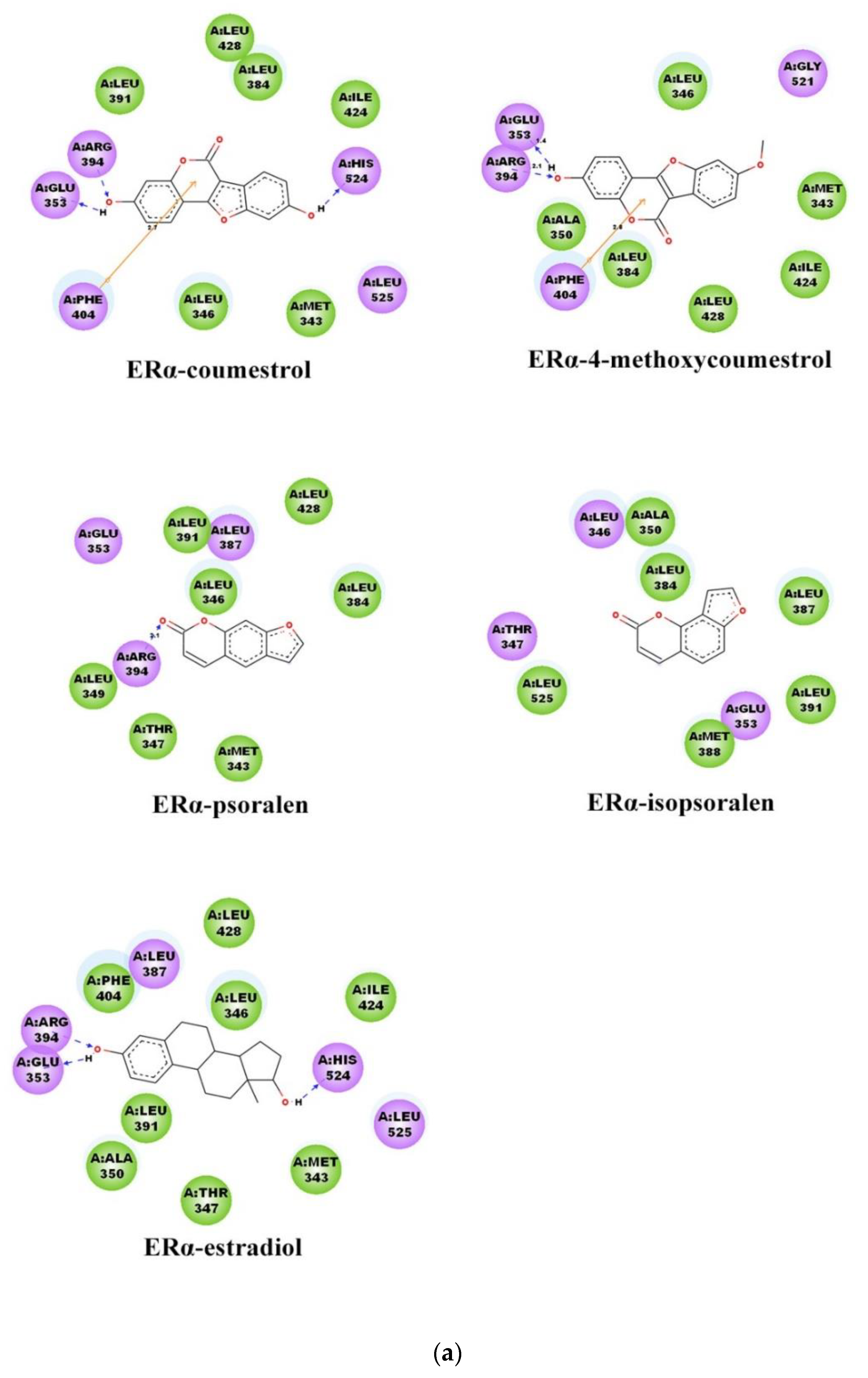

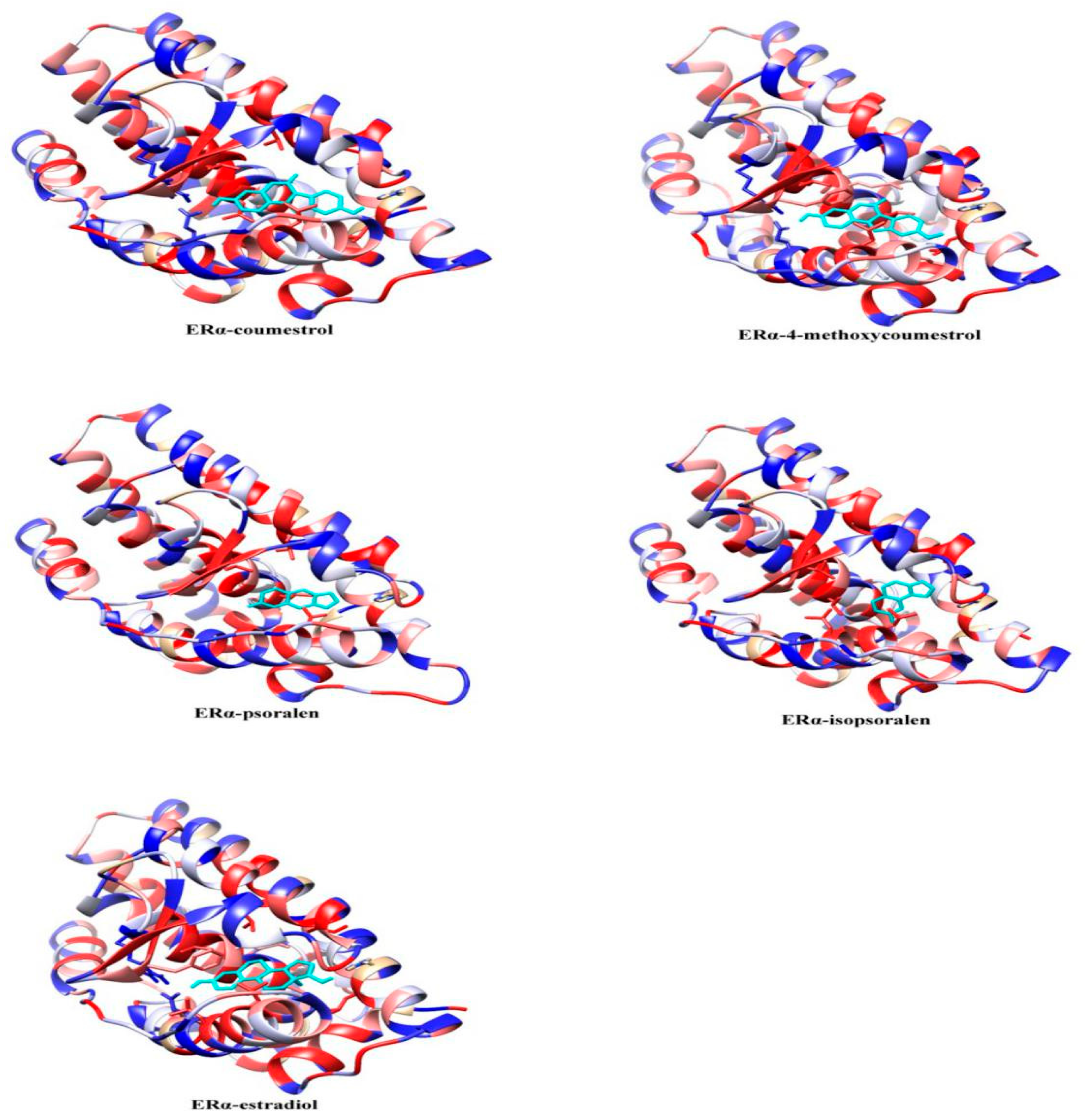
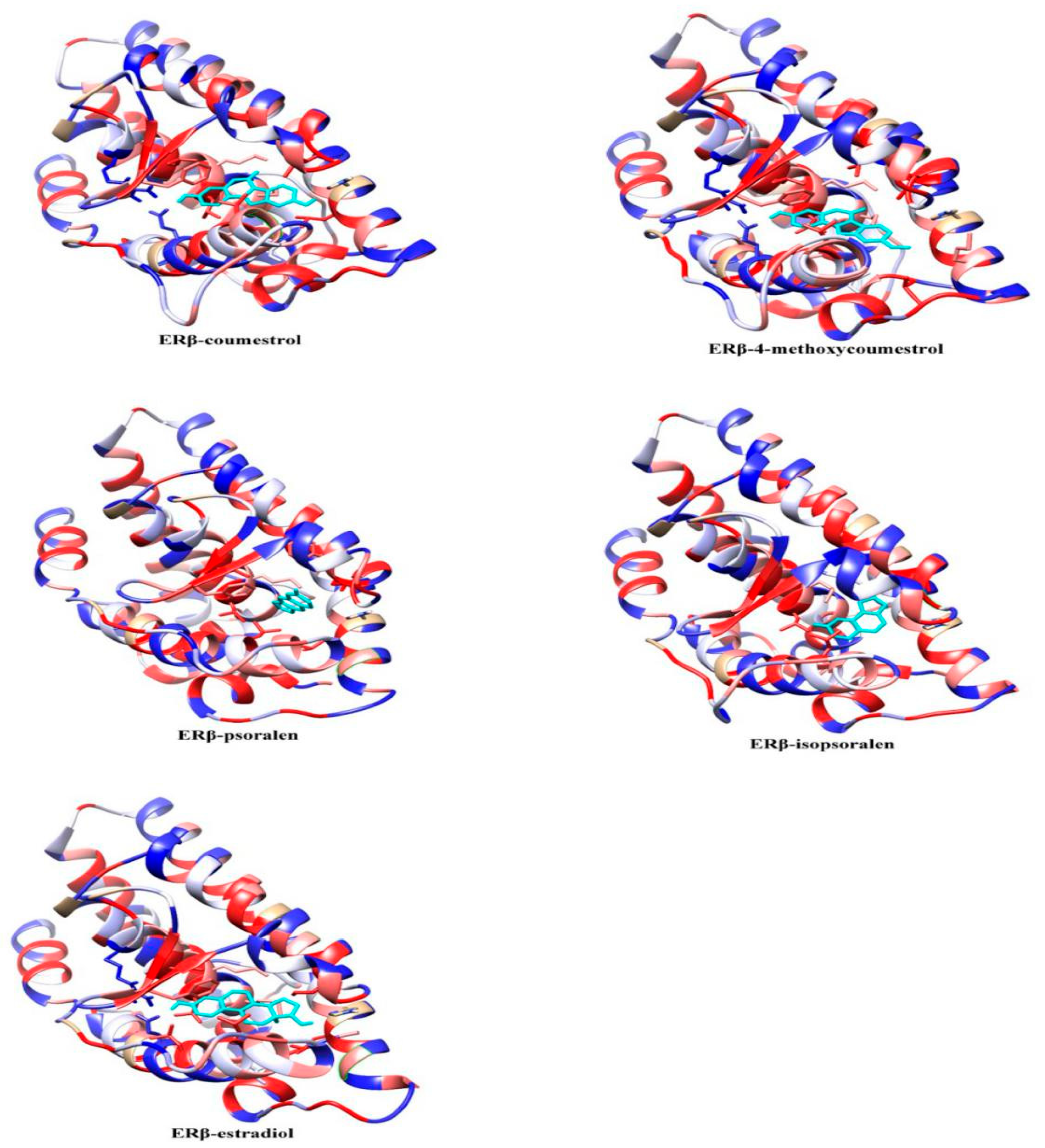
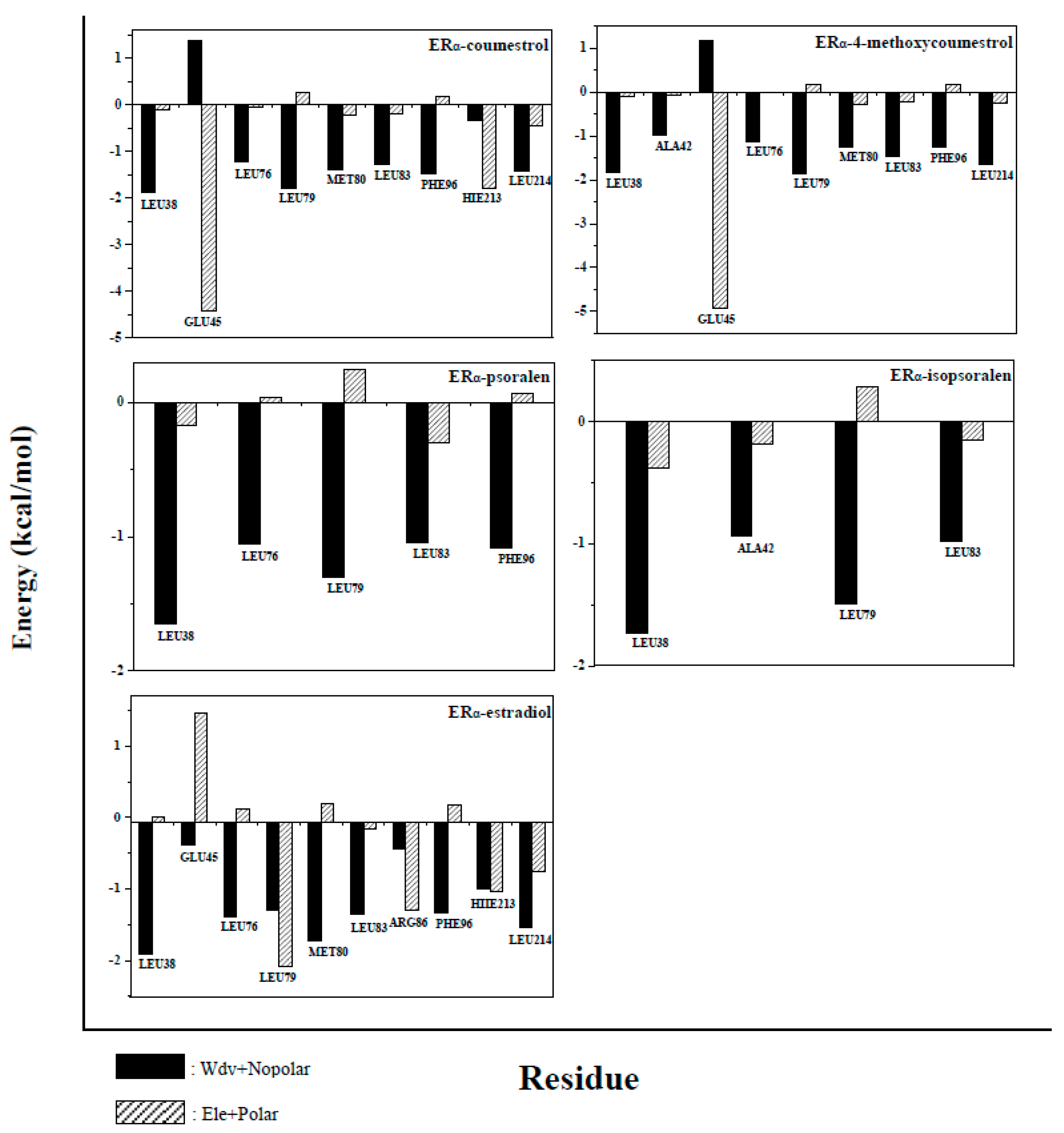
| ERα System Docking Score (kcal/mol) | ERβ System Docking Score (kcal/mol) | |
|---|---|---|
| Coumestrol | −9.805 | −10.489 |
| 4-methoxycoumestrol | −8.677 | −8.812 |
| Psoralen | −7.107 | −7.339 |
| Isopsoralen | −7.311 | −7.502 |
| 17β-estradiol | −10.784 | −10.943 |
| Energy (kcal/mol) | ERα-Estradiol | ERα-Coumestrol | ERα-4-Methoxycoumestrol | ERα-Psoralen | ERα-Isopsoralen |
|---|---|---|---|---|---|
| ΔEVDW | −41.3422 | −34.9511 | −34.9963 | −25.1273 | −25.3492 |
| ΔEELE | −8.8830 | −31.1511 | −26.4485 | −8.6063 | −4.4464 |
| ΔEPOL | 10.1237 | 37.4083 | 29.6470 | 16.1202 | 12.0756 |
| ΔENPOL | −5.2950 | −4.8390 | −5.2148 | −3.4632 | −3.4415 |
| ΔGGAS | −50.2298 | −66.1022 | −61.4449 | −33.7336 | −29.7956 |
| ΔGSOLV | 4.8287 | 32.5693 | 24.4321 | 12.6569 | 8.6341 |
| ΔGTOT | −45.4011 | −33.5329 | −37.0128 | −21.0766 | −21.1615 |
| Energy (kcal/mol) | ERβ-Estradiol | ERβ-Coumestrol | ERβ-4-Methoxycoumestrol | ERβ-Psoralen | ERβ-Isopsoralen |
|---|---|---|---|---|---|
| ΔEVDW | −41.6851 | −34.4483 | −36.8087 | −26.9760 | −27.5848 |
| ΔEELE | −12.8699 | −32.3476 | −33.5825 | −4.8503 | −3.2704 |
| ΔEPOL | 12.8858 | 35.7288 | 35.7121 | 11.8430 | 11.2447 |
| ΔENPOL | −5.1785 | −5.0680 | −5.2660 | −3.6656 | −3.6354 |
| ΔGGAS | −54.5584 | −66.7959 | −70.3912 | −31.8263 | −30.8552 |
| ΔGSOLV | 7.7074 | 30.6607 | 30.4461 | 8.1774 | 7.6093 |
| ΔGTOT | −46.8510 | −36.1352 | −39.9451 | −23.6490 | −23.2459 |
| Complexes | Standard Type | Standard Value | Document Report |
|---|---|---|---|
| ERα-coumestrol | IC50 | 75.7 nM | Ref [31] |
| ERα-4-methoxycoumestrol | NA | NA | NA |
| ERα-psoralen | NA | NA | NA |
| ERα-isopsoralen | NA | NA | NA |
| ERα-estradiol | IC50 | 2 nM | Ref [32] |
| ERβ-coumestrol | IC50 | 18.6 nM | Ref [31] |
| ERβ-4-methoxycoumestrol | NA | NA | NA |
| ERβ-psoralen | NA | NA | NA |
| ERβ-isopsoralen | NA | NA | NA |
| ERβ- estradiol | IC50 | 2 nM | Ref [32] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Wang, Y.; Zhuang, X.; Luan, F.; Zhao, C.; Cordeiro, M.N.D.S. Interaction of Coumarin Phytoestrogens with ERα and ERβ: A Molecular Dynamics Simulation Study. Molecules 2020, 25, 1165. https://doi.org/10.3390/molecules25051165
Wang T, Wang Y, Zhuang X, Luan F, Zhao C, Cordeiro MNDS. Interaction of Coumarin Phytoestrogens with ERα and ERβ: A Molecular Dynamics Simulation Study. Molecules. 2020; 25(5):1165. https://doi.org/10.3390/molecules25051165
Chicago/Turabian StyleWang, Ting, Yunfei Wang, Xuming Zhuang, Feng Luan, Chunyan Zhao, and M. Natália D. S. Cordeiro. 2020. "Interaction of Coumarin Phytoestrogens with ERα and ERβ: A Molecular Dynamics Simulation Study" Molecules 25, no. 5: 1165. https://doi.org/10.3390/molecules25051165
APA StyleWang, T., Wang, Y., Zhuang, X., Luan, F., Zhao, C., & Cordeiro, M. N. D. S. (2020). Interaction of Coumarin Phytoestrogens with ERα and ERβ: A Molecular Dynamics Simulation Study. Molecules, 25(5), 1165. https://doi.org/10.3390/molecules25051165







