Proteomic Analysis of the Early Development of the Phalaenopsis amabilis Flower Bud under Low Temperature Induction Using the iTRAQ/MRM Approach
Abstract
:1. Introduction
2. Results
2.1. Morphological Observation of Flower Bud Differentiation
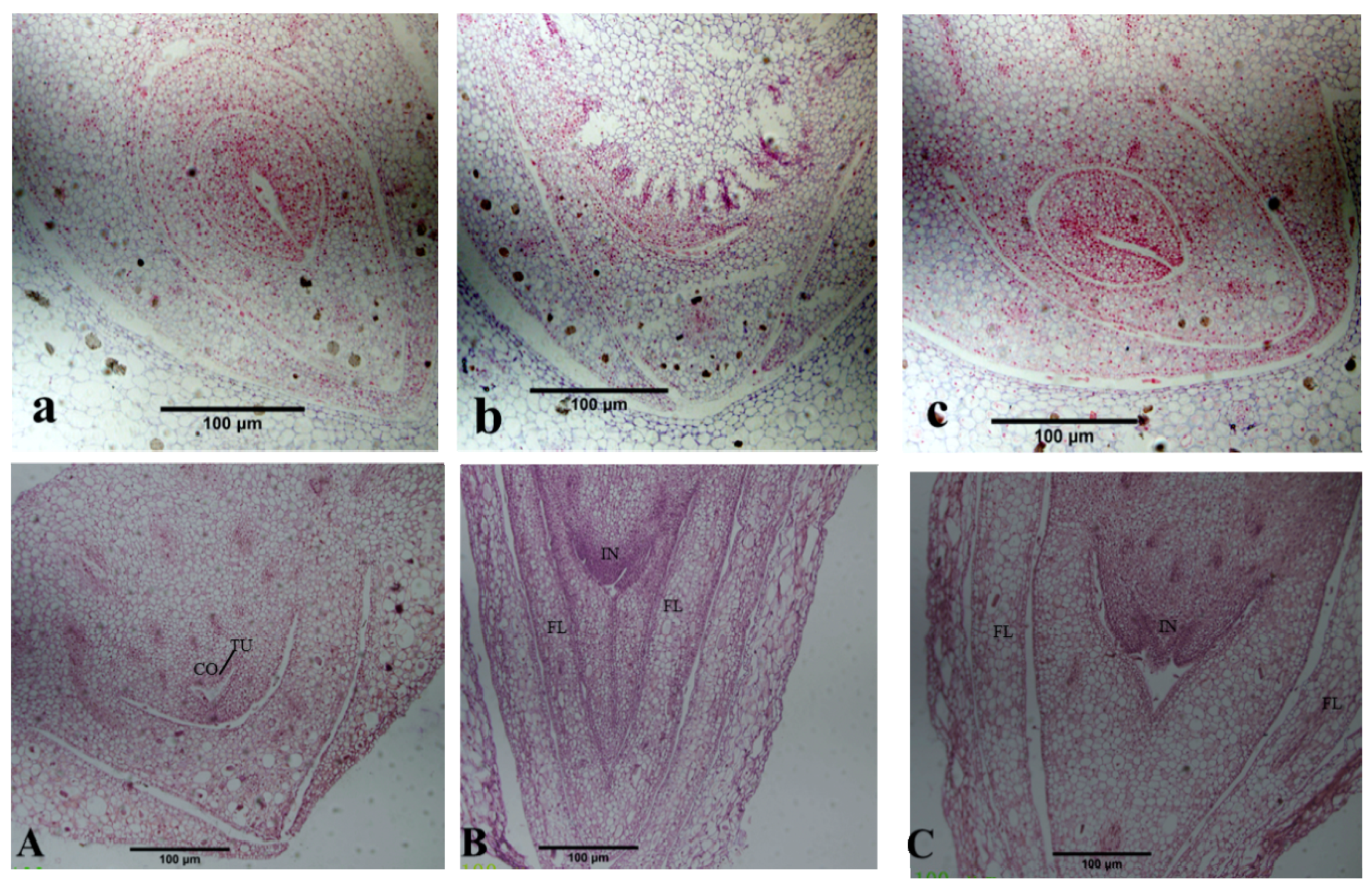
2.2. Histological Observation of Flower Bud Differentiation
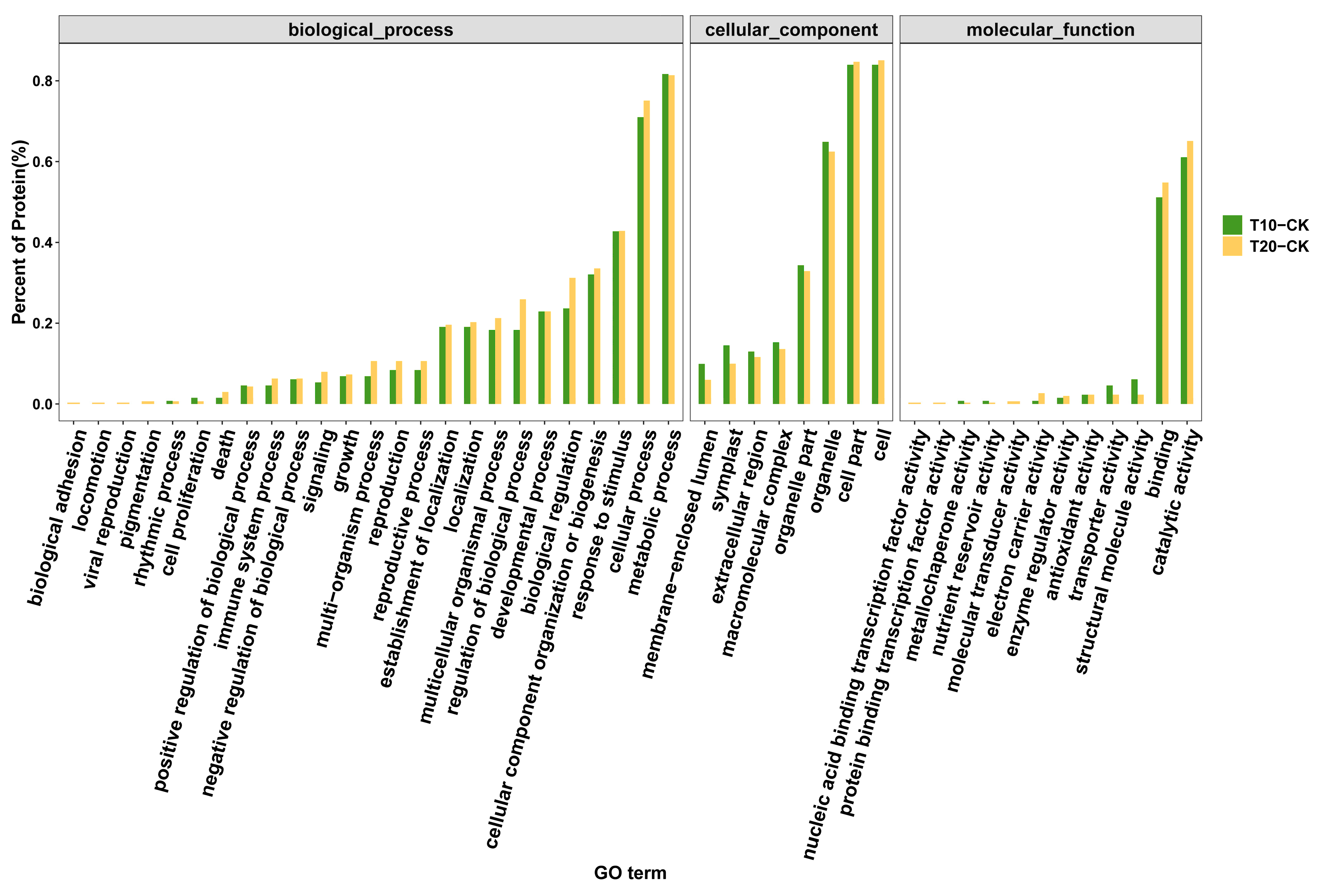
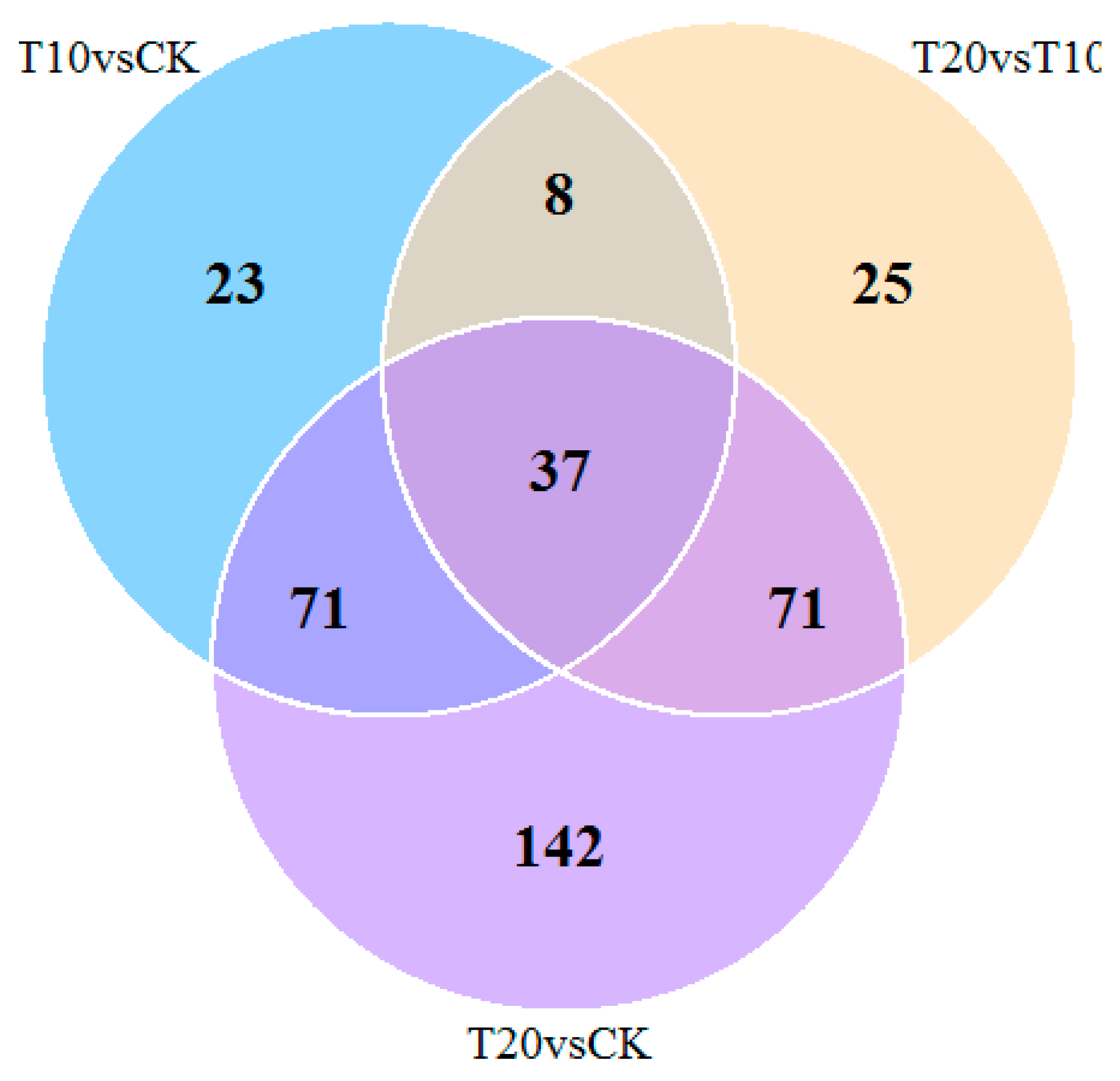
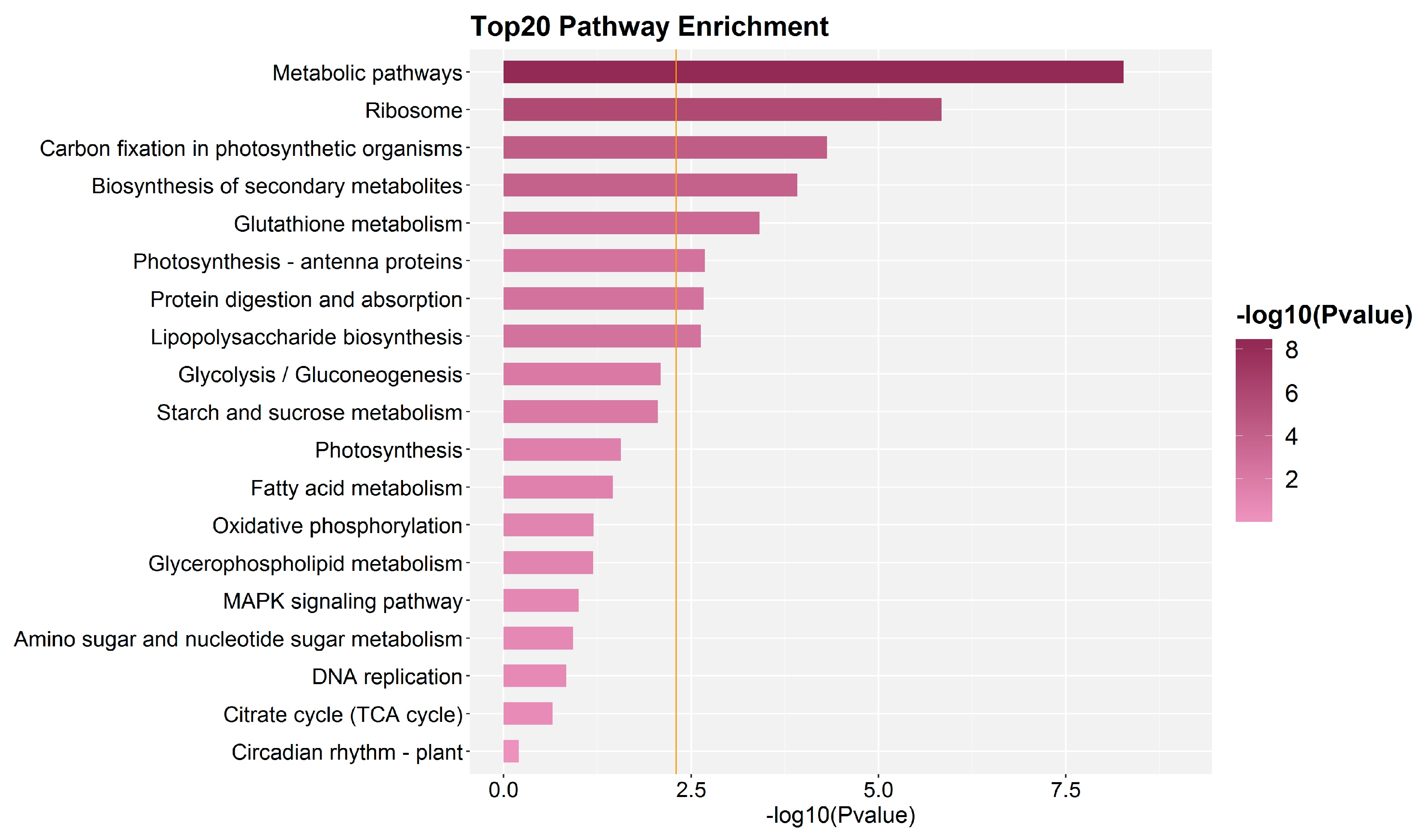
2.3. Quantitative Identification of Proteins by Isobaric Tags for Relative and Absolute Quantification (iTRAQ)
2.4. MRM Verification
3. Discussion
3.1. Photoperiod-Related Proteins
3.2. Vernalization-Related Proteins
3.3. Ubiquitin-Related Proteins
3.4. Other Proteins
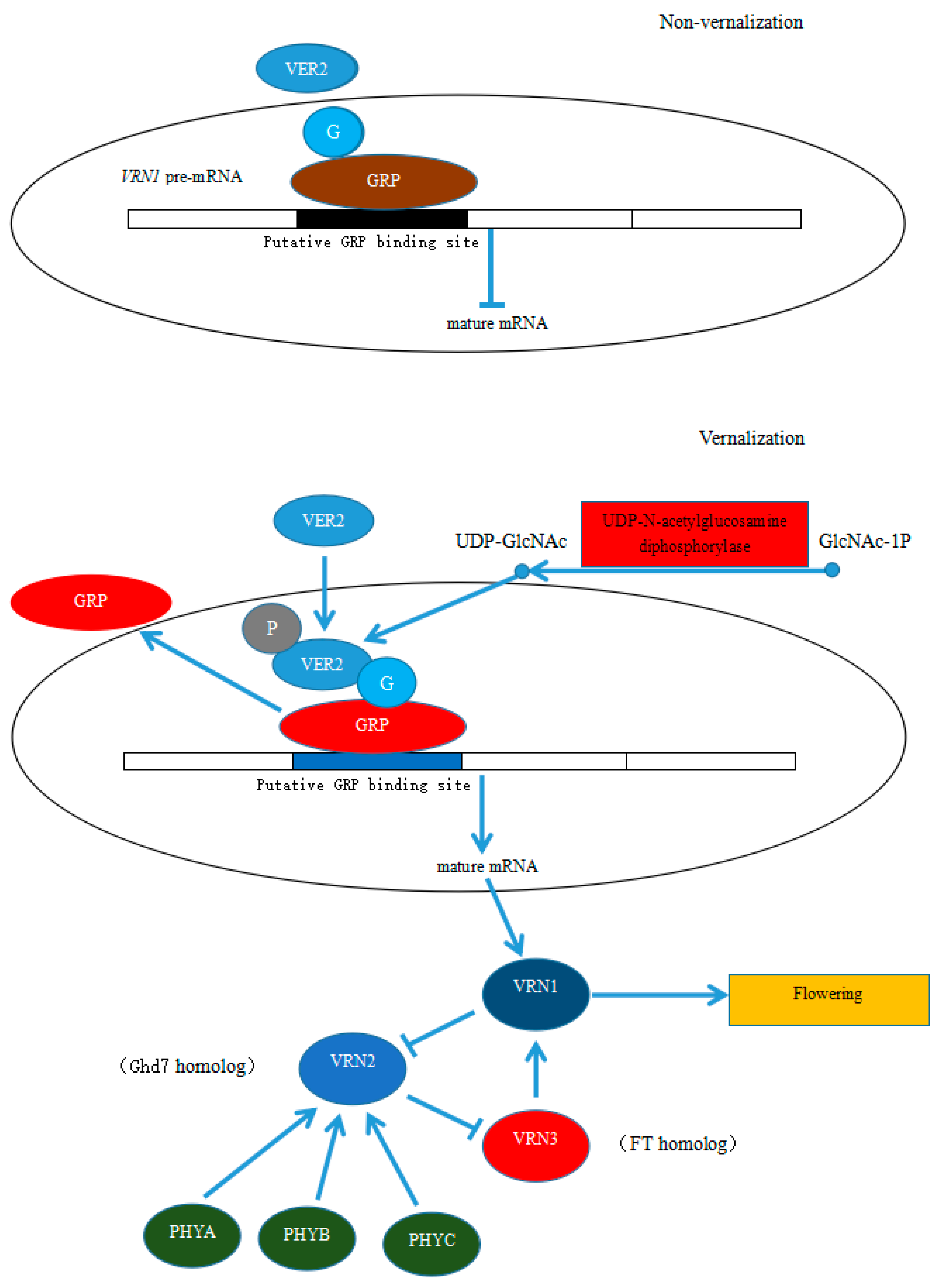
3.5. Possible Mechanism for the Regulation of the Early Development of Flower Bud in P. amabilis under Low Temperature
4. Materials and Methods
4.1. Plant Materials
4.2. Histological Observation of Flower Bud Development
4.3. Protein Extraction
4.4. iTRAQ Labeling and LC-ESI MS/MS Analysis
4.5. Protein Identification and Proteome Analysis
4.6. MRM Analyses
4.7. Data Analysis
4.8. Functional Analysis of Proteins
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ream, T.S.; Woods, D.P.; Amasino, R.M. The molecular basis of vernalization in different plant groups. Cold Spring Harb. Symp. 2012, 77, 105–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouché, F.; Woods, D.P.; Amasino, R.M. Winter memory throughout the plant kingdom: Different paths to flowering. Plant Physiol. 2017, 173, 27–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunner, A.M.; Evans, L.M.; Hsu, C.Y.; Sheng, X. Vernalization and the chilling requirement to exit bud dormancy: Shared or separate regulation? Front. Plant Sci. 2014, 5, 732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komeda, Y. Genetic regulation of time to flower in Arabidopsis Thaliana. Annu. Rev. Plant Biol. 2004, 55, 521–535. [Google Scholar] [CrossRef] [Green Version]
- Smyth, D.R.; Bowman, J.L.; Meyerowitz, E.M. Early flower development in Arabidopsis. Plant Cell 1990, 2, 755–767. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.; Xu, S.J.; Li, C.H.; Xu, Y.Y.; Xing, L.J.; Niu, Y.D.; Huan, Q.; Tang, Y.M.; Zhao, C.P.; Wagner, D.; et al. O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during vernalization in winter wheat. Nat. Commun. 2014, 5, 4572. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, J.P.; Huang, S.Q.; Pan, S.Q.; Ye, Q.S. Construction and identification of a cDNA expression library from Dendrobium nobile. Acta Hortic. Sin. 2006, 33, 895–897. [Google Scholar] [CrossRef]
- Huang, J.Z.; Lin, C.P.; Cheng, T.C.; Chang, C.H.; Cheng, S.Y.; Chen, Y.W. A de novo floral transcriptome reveals clues into phalaenopsis orchid flower development. PLoS ONE 2015, 10, e0123474. [Google Scholar] [CrossRef] [Green Version]
- Chong, K.; Yong, W.D.; TAN, K.H. Advances on research of vernalization in higher plants. Chin. Bull. Bot. 1999, 16, 481–487. [Google Scholar] [CrossRef]
- Su, W.R.; Chen, W.S.; Koshioka, M. Changes in gibberellin levels in the flowering shoot of Phalaenopsis hybrida under high temperature conditions when flower development is blocked. Plant Physiol. Biochem. 2001, 39, 45–50. [Google Scholar] [CrossRef]
- Chen, W.H.; Hsu, C.Y.; Cheng, H.Y. Downregulation of putative UDP-glucose: Flavonoid 3-O-glucosyltransferase gene alters flower coloring in Phalaenopsis. Plant Cell Rep. 2011, 30, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Lopez, P.; Wheatley, K.; Robson, F.; Onouchi, H.; Valverde, F.; Coupland, G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 2001, 410, 1116–1120. [Google Scholar] [CrossRef] [PubMed]
- Ben-Naim, O.; Eshed, R.; Parnis, A.; Teper-Bamnolker, P.; Shalit, A.; Coupland, G. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 2006, 46, 462–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valverde, F.; Mouradov, A.; Soppe, W. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 2004, 303, 1003–1006. [Google Scholar] [CrossRef] [Green Version]
- Fornara, F.; Panigrahi, K.C.S.; Gissot, L. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 2009, 17, 75–86. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.H.; Estrada, D.A.; Johnson, R.S. Distinct roles of FKF1, GIGANTEA, and ZEITLUPE proteins in the regulation of CONSTANS stability in Arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. USA 2014, 111, 17672–17677. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.J.; Zhang, Y.C.; Li, Q.H. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 2008, 20, 292–306. [Google Scholar] [CrossRef]
- Song, Y.H.; Smith, R.W.; To, B.J. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 2012, 336, 1045–1049. [Google Scholar] [CrossRef] [Green Version]
- Quesada, V.; Dean, C.; Simpson, G.G. Regulated RNA processing in the control of Arabidopsis flowering. Int. J. Dev. Biol. 2005, 49, 773–780. [Google Scholar] [CrossRef]
- Rodriguez-Cazorla, E.; Ripoll, J.J.; Andujar, A.; Bailey, L.J.; Martinez-Laborda, A.; Yanofsky, M.F. K-homology nuclear ribonucleoproteins regulate floral organ identity and determinacy in Arabidopsis. PLoS Genet. 2015, 11, e1004983. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, P.; Hart, G.W. O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine: Polypeptide-N-acetylglucosaminyl transferase) (OGT). In Handbook of Glycosyltransferases and Related Genes; Springer: Tokyo, Japan, 2014; Volume 1, pp. 393–408. [Google Scholar] [CrossRef]
- Murase, K.; Hirano, Y.; Sun, T.P.; Hakoshima, T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 2008, 456, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Daviere, J.M.; Achard, P. Gibberellin signaling in plants. Development 2013, 140, 1147–1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, S.; Galvao, V.C.; Zhang, Y.C.; Horrer, D.; Zhang, T.Q.; Hao, Y.H. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 2012, 24, 3320–3332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.; Chong, K. Ubiquitin-proteasome pathway and regulation of auxin. Plant Physiol. Commun. 2004, 40, 653–658. [Google Scholar] [CrossRef]
- Zhuang, W.; Gao, Z.; Wang, L.; Zhong, W.; Ni, Z.; Zhang, Z. Comparative proteomic and transcriptomic approaches to address the active role of GA4 in Japanese apricot flower bud dormancy release. J. Exp. Bot. 2013, 64, 4953–4966. [Google Scholar] [CrossRef]
- Chouard, P. Vernalization and its relation to dormancy. Annu. Rev. Plant Physiol. 1960, 11, 191–238. [Google Scholar] [CrossRef]
- Michaels, S.D.; Amasino, R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 1999, 11, 949–956. [Google Scholar] [CrossRef] [Green Version]
- Danyluk, J. TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol. 2003, 132, 1849–1860. [Google Scholar] [CrossRef] [Green Version]
- Trevaskis, B.; Bagnall, D.J.; Ellis, M.H.; Peacock, W.J.; Dennis, E.S. MADS box genes control vernalization-induced flowering in cereals. Proc. Natl Acad. Sci. USA 2003, 100, 13099–13104. [Google Scholar] [CrossRef] [Green Version]
- Yan, L. Positional cloning of the wheat vernalization gene VRN1. Proc. Natl Acad. Sci. USA 2003, 100, 6263–6268. [Google Scholar] [CrossRef] [Green Version]
- Distelfeld, A.; Li, C.; Dubcovsky, J. Regulation of flowering in temperate cereals. Curr. Opin. Plant Biol. 2009, 12, 178–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.; Angel, A.; Howard, M.; Dean, C. Vernalization—A cold-induced epigenetic switch. J. Cell Sci. 2012, 125, 3723–3731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenup, A.; Peacock, W.J.; Dennis, E.S.; Trevaskis, B. The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Ann. Bot. 2009, 103, 1165–1172. [Google Scholar] [CrossRef] [Green Version]
- Trevaskis, B. The central role of the VERNALIZATION1 gene in the vernalization response of cereals. Funct. Plant Biol. 2010, 37, 479–487. [Google Scholar] [CrossRef] [Green Version]
- Liang, S.; Ye, Q.S.; Li, R.H.; Leng, J.Y.; Li, M.R.; Wang, X.J.; Li, H.Q. Transcriptional regulations on the low-temperature-induced floral transition in an orchidaceae species, dendrobium nobile: An expressed sequence tags analysis. Comp. Funct. Genom. 2012, 3, 757–801. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.H.; Doyle, M.R.; Sung, S.; Amasino, R.M. Vernalization: Winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 2009, 25, 277–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangeon, A.; Junqueira, R.M.; Sachetto-Martins, G. Functional diversity of the plant glycine-rich proteins superfamily. Plant Signal. Behav. 2010, 5, 99–104. [Google Scholar] [CrossRef]
- Streitner, C.; Koster, T.; Simpson, C.G.; Shaw, P.; Danisman, S.; Brown, J.W. An hnRNP-like RNA-binding protein affects alternative splicing by in vivo interaction with transcripts in Arabidopsis thaliana. Nucleic Acids Res. 2012, 40, 11240–11255. [Google Scholar] [CrossRef]
- Zachara, N.E.; Hart, G.W. O-GlcNAc a sensor of cellular state: The role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim. Biophys. Acta 2004, 1673, 13–28. [Google Scholar] [CrossRef]
- Yan, L.L.; Loukoianov, A.; Blechl, A. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 2004, 303, 1640–1644. [Google Scholar] [CrossRef] [Green Version]
- Xue, W.Y.; Xing, Y.Z.; Weng, X.Y. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Osugi, A.; Itoh, H.; Ikeda-Kawakatsu, K.; Takano, M.; Izawa, T. Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice. Plant Physiol. 2011, 157, 1128–1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strasser, B.; Alvarez, M.J.; Califano, A.; Cerdan, P.D. A complementary role for ELF3 and TFL1 in the regulation of flowering time by ambient temperature. Plant J. 2009, 58, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Yasui, Y.; Mukougawa, K.; Uemoto, M.; Yokofuji, A.; Suzuri, R.; Nishitani, A. The phytochrome-interacting vascular plant one-zinc finger1 and VOZ2 redundantly regulate flowering in Arabidopsis. Plant Cell 2012, 24, 3248–3263. [Google Scholar] [CrossRef] [Green Version]
- Yasui, Y.; Kohchi, T. VASCULAR PLANT ONE-ZINC FINGER1 and VOZ2 repress the FLOWERING LOCUS C clade members to control flowering time in Arabidopsis. Biosci. Biotechnol. Biochem. 2014, 78, 1850–1855. [Google Scholar] [CrossRef] [Green Version]
- Yan, L.; Fu, D.; Li, C. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. USA 2006, 103, 19581–19586. [Google Scholar] [CrossRef] [Green Version]
- Shitsukawa, N.; Ikari, C.; Shimada, S. The einkorn wheat (Triticum monococcum) mutant, maintained vegetative phase, is caused by a deletion in the VRN1 gene. Genes Genet. Syst. 2007, 82, 167–170. [Google Scholar] [CrossRef] [Green Version]
- Mertins, P.; Udeshi, N.D.; Clauser, K.R. iTRAQ labeling is superior to mTRAQ for quantitative global proteomics and phosphoproteomics. Mol. Cell. Proteomics 2011, 11, M111.014423. [Google Scholar] [CrossRef] [Green Version]
- Ow, S.Y.; Salim, M.; Noirel, J. iTRAQ underestimation in simple and complex mixtures: “The good, the bad and the ugly”. J. Proteome Res. 2009, 8, 5347–5355. [Google Scholar] [CrossRef]
- Huang, S.Q.; Guo, J.J.; Ye, Q.S.; Lin, J.Y. Floral Induction and development in Phalaenopsis under different temperatures. Acta Scientiarum Naturalium Universitatis Sunyatseni 2003, 42, 132–134. [Google Scholar] [CrossRef]
- Yang, S.H.; Ji, J.; Wang, G. Effect of nutrient formulation on growth of water culture Phalaenopsis. J. Agric. Univ. Hebei 2008, 31, 30–33. (In Chinese) [Google Scholar] [CrossRef]
- Sun, C.Q.; Chen, F.D.; Fang, W.M.; Liu, Z.L.; Ma, J.; Teng, N.J. Cellular mechanism of reproductive barrier during cross breeding between Dendranthema grandiflorum cv. Aoyuntianshi and D. japonense. Sci. Agric. Sin. 2009, 42, 2085–2091. [Google Scholar] [CrossRef]
- Li, M.; Wang, K.; Li, S.Q. Exploration of rice pistil responses during early post-pollination through a combined proteomic and transcriptomic analysis. J. Proteomics 2016, 131, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Cai, J.; Liu, X.; Vanneste, K. The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 2015, 47, 65–72. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Aguilar, J.; Molloy, M.P. Label-free selected reaction monitoring enables multiplexed quantitation of S100 protein isoforms in cancer cells. J. Proteome Res. 2013, 12, 3679–3688. [Google Scholar] [CrossRef]
- Tang, H.Y.; Beer, L.A.; Barnhart, K.T.; Speicheret, D.W. Rapid verification of candidate serological biomarkers using gel-based, label-free multiple reaction monitoring. J. Proteome Res. 2011, 10, 4005–4017. [Google Scholar] [CrossRef] [Green Version]
- Martins-de-Souza, D.; Alsaif1, M.; Ernst, A. The application of selective reaction monitoring confirms dysregulation of glycolysis in a preclinical model of schizophrenia. BMC Res. Notes 2012, 5, 146. [Google Scholar] [CrossRef] [Green Version]
- MacLean, B.; Tomazela, D.M.; Shulman, N. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef] [Green Version]
- Escher, C.; Reiter, L.; MacLean, B. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics 2012, 12, 1111–1121. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: Samples of the compounds are not available from the authors. |

| Accession Code | GI Name | % Cov (95) | PI | MW (kDa) | # of Peptides | Fold Change | ||
|---|---|---|---|---|---|---|---|---|
| T10/CK | T20/CK | T20/T10 | ||||||
| Photoperiod-Related Proteins | ||||||||
| PEQU_11519 | LON peptidase N-terminal domain and RING finger protein 1 | 21.560 | 6.9 | 39.96 | 2 | 1.146 ± 0.127 | 1.141 ± 0.119 | 0.986 ± 0.006 |
| PEQU_27404 | Protein BTR1 | 23.560 | 5.72 | 26.84 | 12 | 1.295 ± 0.067 | 1.654 ± 0.258 | 1.275 ± 0.149 |
| PEQU_18278 | Chromatin-remodeling protein EBS-like isoform X1 | 25.560 | 7.9 | 24.72 | 4 | 0.428 ± 0.053 | 0.316 ± 0.016 | 0.696 ± 0.059 |
| PEQU_06861 | PHD finger protein ALFIN-LIKE 9-like | 26.560 | 5.64 | 30.18 | 3 | 0.936 ± 0.083 | 0.875 ± 0.003 | 1.034 ± 0.061 |
| PEQU_02392 | Zinc finger CCCH domain-containing protein 4-like | 34.560 | 6.87 | 113.85 | 2 | 0.838 ± 0.093 | 0.742 ± 0.106 | 0.875 ± 0.028 |
| PEQU_16431 | PHD finger protein ALFIN-LIKE 6-like isoform X1 | 35.560 | 4.75 | 28.43 | 3 | 0.963 ± 0.419 | 0.723 ± 0.587 | 0.662 ± 0.319 |
| PEQU_03363 | Probable ADP-ribosylation factor GTPase-activating protein AGD9 | 36.560 | 8.03 | 44.08 | 9 | 1.035 ± 0.087 | 1.000 ± 0.013 | 0.958 ± 0.100 |
| PEQU_09548 | Cryptochrome-1-like | 12.560 | 5.1 | 71.84 | 6 | 0.836 ± 0.016 | 0.825 ± 0.043 | 0.977 ± 0.032 |
| PEQU_22307 | Cryptochrome-1 isoform X1 | 13.560 | 5.47 | 73.66 | 14 | 0.823 ± 0.505 | 0.473 ± 0.101 | 0.644 ± 0.265 |
| PEQU_41401 | Phytochrome C isoform X2 | 14.560 | 5.73 | 125.83 | 11 | 0.777 ± 0.277 | 0.529 ± 0.065 | 0.688 ± 0.151 |
| PEQU_13449 | Phytochrome A | 15.560 | 6.26 | 125.84 | 7 | 0.694 ± 0.121 | 0.904 ± 0.024 | 1.305 ± 0.237 |
| PEQU_09249 | Phytochrome B isoform X1 | 16.560 | 5.4 | 98.54 | 5 | 1.068 ± 0.221 | 0.802 ± 0.042 | 0.752 ± 0.112 |
| PEQU_06485 | Bibenzyl synthase | 27.560 | 6.43 | 42.59 | 4 | 1.017 ± 0.742 | 1.927 ± 0.175 | 2.399 ± 1.511 |
| PEQU_07192 | Cryptochrome-1 | 32.560 | 5.06 | 77.8 | 7 | 1.067 ± 0.000 | 1.008 ± 0.124 | 0.937 ± 0.116 |
| PEQU_35378 | Scarecrow-like transcription factor PAT1 | 33.560 | 6.59 | 60.22 | 6 | 0.774 ± 0.359 | 0.568 ± 0.018 | 0.813 ± 0.387 |
| PEQU_12445 | Cytochrome P450 71A1-like | 31.560 | 8.1 | 57.79 | 2 | 1.018 ± 0.126 | 0.933 ± 0.006 | 0.921 ± 0.126 |
| PEQU_41539 | Allene oxide synthase 2-like | 41.560 | 7.84 | 47.64 | 18 | 2.376 ± 0.991 | 3.853 ± 1.445 | 1.587 ± 0.103 |
| PEQU_19521 | FHA domain-containing protein DDL | 20.560 | 10.86 | 44.49 | 1 | 1.194 ± 0.124 | 1.273 ± 0.263 | 1.050 ± 0.109 |
| PEQU_02676 | Oxygen-evolving enhancer protein 2, chloroplastic-like | 54.560 | 8.95 | 28.5 | 9 | 1.446 ± 0.075 | 4.743 ± 0.124 | 3.368 ± 0.372 |
| PEQU_02776 | Oxygen-evolving enhancer protein 1, chloroplastic-like | 55.560 | 7.89 | 35.73 | 17 | 1.217 ± 0.213 | 3.265 ± 0.424 | 2.783 ± 0.181 |
| Vernalization-related proteins | ||||||||
| PEQU_11434 | Glycine-rich RNA-binding protein GRP1A-like | 24.560 | 6.86 | 13.65 | 5 | 1.741 ± 2.093 | 2.479 ± 1.676 | 2.969 ± 2.633 |
| PEQU_11045 | UDP-N-acetylglucosamine diphosphorylase 1-like | 33.270 | 8.39 | 56.17 | 13 | 1.092 ± 0.156 | 1.71 ± 0.430 | 1.569 ± 0.194 |
| PEQU_19304 | Flowering locus T | 22.560 | 7.51 | 19.75 | 7 | 2.240 ± 1.602 | 2.030 ± 0.649 | 1.062 ± 0.487 |
| Ubiquitin-related proteins | ||||||||
| PEQU_03892 | Auxin response factor 17-like | 17.560 | 6.44 | 99.46 | 7 | 0.977 ± 0.366 | 1.683 ± 0.033 | 1.788 ± 0.594 |
| PEQU_28956 | Auxin response factor 17-like | 28.560 | 8.37 | 22.66 | 7 | 0.895 ± 0.000 | 0.974 ± 0.063 | 1.078 ± 0.070 |
| PEQU_26378 | Ubiquitin-conjugating enzyme E2 36-like | 29.560 | 7.51 | 17.18 | 8 | 0.836 ± 0.368 | 1.079 ± 0.538 | 1.283 ± 0.050 |
| PEQU_21711 | Ubiquitin-conjugating enzyme E2 7-like isoform X1 | 37.560 | 4.16 | 16.48 | 3 | 1.124 ± 0.380 | 1.113 ± 0.065 | 1.030 ± 0.291 |
| PEQU_12056 | Probable ubiquitin-conjugating enzyme E2 18 | 38.560 | 7.11 | 17.47 | 1 | 1.040 ± 0.095 | 1.082 ± 0.021 | 1.034 ± 0.074 |
| PEQU_13455 | NEDD8-conjugating enzyme Ubc12-like | 39.560 | 8.31 | 20.85 | 2 | 0.997 ± 0.155 | 0.951 ± 0.031 | 0.958 ± 0.112 |
| PEQU_22283 | Cytochrome P450 90A1-like | 2.679 | 6.94 | 51.26 | 1 | 0.943 ± 0.055 | 0.884 ± 0.052 | 0.825 ± 0.006 |
| Carbon- and energy metabolism-related proteins | ||||||||
| PEQU_03049 | Probable flavin-containing monooxygenase 1 | 19.560 | 7.59 | 61.7 | 2 | 0.829 ± 0.070 | 0.637 ± 0.025 | 0.763 ± 0.045 |
| PEQU_21336 | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic | 42.560 | 8.91 | 42.57 | 10 | 2.533 ± 0.459 | 11.914 ± 0.310 | 5.380 ± 1.821 |
| PEQU_18900 | LOW-QUALITY PROTEIN: ribulose bisphosphate carboxylase small chain clone 512-like | 43.560 | 9.3 | 19.85 | 8 | 2.620 ± 1.336 | 12.538 ± 0.571 | 6.228 ± 2.635 |
| PEQU_34221 | Ferredoxin--NADP reductase, leaf-type isozyme, chloroplastic-like | 44.560 | 8.72 | 40.83 | 10 | 0.963 ± 0.338 | 2.823 ± 0.439 | 3.110 ± 1.389 |
| PEQU_02056 | ATP synthase CF1 beta subunit (chloroplast) | 45.560 | 5.25 | 53.5 | 27 | 1.225 ± 0.048 | 1.845 ± 0.012 | 1.507 ± 0.049 |
| PEQU_01887 | Ribulose bisphosphate carboxylase/oxygenase activase, chloroplastic-like isoform X2 | 46.560 | 7.36 | 51.78 | 15 | 2.071 ± 1.462 | 8.066 ± 1.256 | 5.560 ± 4.683 |
| Stress-related proteins | ||||||||
| PEQU_18289 | Beta-glucosidase 1-like | 47.560 | 6.72 | 52.91 | 27 | 0.817 ± 0.608 | 0.368 ± 0.048 | 0.583 ± 0.373 |
| PEQU_08910 | GDP-mannose 3,5-epimerase 2 | 48.560 | 6.44 | 42.78 | 25 | 3.665 ± 0.095 | 3.158 ± 1.011 | 0.844 ± 0.260 |
| PEQU_14594 | Catalase isozyme A | 49.560 | 7.06 | 56.35 | 28 | 3.370 ± 0.762 | 5.550 ± 1.325 | 1.652 ± 0.032 |
| PEQU_05138 | S-Adenosylmethionine synthase | 50.560 | 6.74 | 37.76 | 12 | 1.905 ± 2.536 | 3.589 ± 1.389 | 6.971 ± 8.110 |
| PEQU_15901 | Alcohol dehydrogenase class-3 | 51.560 | 6.71 | 40.77 | 20 | 0.475 ± 0.316 | 0.199 ± 0.058 | 0.618 ± 0.567 |
| PEQU_10731 | Protein IN2-1 homolog B-like | 52.560 | 7.58 | 47.15 | 18 | 2.813 ± 1.972 | 4.755 ± 1.955 | 1.876 ± 0.647 |
| Protein | GI Name | Fold Change of T10/CK | Adj. pvalue | Fold Change of T20/CK | Adj. pvalue | Fold Change of T20/T10 | Adj. pvalue |
|---|---|---|---|---|---|---|---|
| PEQU_01887 | Ribulose bisphosphate carboxylase/oxygenase activase, chloroplastic-like isoform X2 | 2.938 | 0.0001 | 23.137 | <0.0001 | 7.876 | <0.0001 |
| PEQU_02056 | ATP synthase CF1 beta subunit (chloroplast) | 0.709 | 0.0023 | 1.344 | 0.0027 | 1.896 | <0.0001 |
| PEQU_02676 | Oxygen-evolving enhancer protein 2, chloroplastic-like | 0.966 | 0.4428 | 1.018 | 0.6831 | 1.054 | 0.2243 |
| PEQU_02776 | Oxygen-evolving enhancer protein 1, chloroplastic-like | 0.801 | 0.0092 | 3.040 | <0.0001 | 3.795 | <0.0001 |
| PEQU_03102 | DEAD-box ATP-dependent RNA helicase 3, chloroplastic | 2.445 | 0.0015 | 3.096 | 0.0002 | 1.266 | 0.1502 |
| PEQU_07151 | Probable methyltransferase PMT2 | 2.587 | <0.0001 | 4.321 | <0.0001 | 1.670 | <0.0001 |
| PEQU_08910 | GDP-mannose 3,5-epimerase 2 | 2.247 | <0.0001 | 3.929 | <0.0001 | 1.749 | 0.0003 |
| PEQU_10731 | Protein IN2-1 homolog B-like | 1.181 | 0.0218 | 3.182 | <0.0001 | 2.693 | <0.0001 |
| PEQU_11434 | Glycine-rich RNA-binding protein GRP1A-like | 4.438 | <0.0001 | 6.590 | <0.0001 | 1.485 | 0.0010 |
| PEQU_14594 | Catalase isozyme A | 1.508 | 0.0017 | 3.903 | <0.0001 | 2.589 | <0.0001 |
| PEQU_15901 | Alcohol dehydrogenase class-3 | 0.733 | 0.0079 | 0.479 | <0.0001 | 0.654 | 0.0011 |
| PEQU_16988 | Homospermidine synthase | 2.376 | <0.0001 | 4.517 | <0.0001 | 1.901 | <0.0001 |
| PEQU_18289 | Beta-glucosidase 1-like | 0.872 | 0.0260 | 0.350 | <0.0001 | 0.402 | <0.0001 |
| PEQU_18900 | LOW-QUALITY PROTEIN: ribulose bisphosphate carboxylase small chain clone 512-like | 1.222 | 0.0346 | 3.444 | <0.0001 | 2.818 | <0.0001 |
| PEQU_19304 | Flowering locus T | 2.438 | <0.0001 | 2.586 | <0.0001 | 1.061 | 0.1502 |
| PEQU_21336 | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic | 1.385 | 0.0150 | 10.707 | <0.0001 | 7.731 | <0.0001 |
| PEQU_22307 | Cryptochrome-1 isoform X1 | 0.775 | 0.0196 | 0.432 | <0.0001 | 0.557 | 0.0003 |
| PEQU_34221 | Ferredoxin--NADP reductase, leaf-type isozyme, chloroplastic-like | 1.057 | 0.4850 | 3.078 | <0.0001 | 2.911 | <0.0001 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.; Zeng, L.; Zhao, H.; Ye, Q. Proteomic Analysis of the Early Development of the Phalaenopsis amabilis Flower Bud under Low Temperature Induction Using the iTRAQ/MRM Approach. Molecules 2020, 25, 1244. https://doi.org/10.3390/molecules25051244
Chen C, Zeng L, Zhao H, Ye Q. Proteomic Analysis of the Early Development of the Phalaenopsis amabilis Flower Bud under Low Temperature Induction Using the iTRAQ/MRM Approach. Molecules. 2020; 25(5):1244. https://doi.org/10.3390/molecules25051244
Chicago/Turabian StyleChen, Cong, Lanting Zeng, Haiyi Zhao, and Qingsheng Ye. 2020. "Proteomic Analysis of the Early Development of the Phalaenopsis amabilis Flower Bud under Low Temperature Induction Using the iTRAQ/MRM Approach" Molecules 25, no. 5: 1244. https://doi.org/10.3390/molecules25051244
APA StyleChen, C., Zeng, L., Zhao, H., & Ye, Q. (2020). Proteomic Analysis of the Early Development of the Phalaenopsis amabilis Flower Bud under Low Temperature Induction Using the iTRAQ/MRM Approach. Molecules, 25(5), 1244. https://doi.org/10.3390/molecules25051244






