The Secondary Structure of a Major Wine Protein is Modified upon Interaction with Polyphenols
Abstract
:1. Introduction
2. Results and Discussion
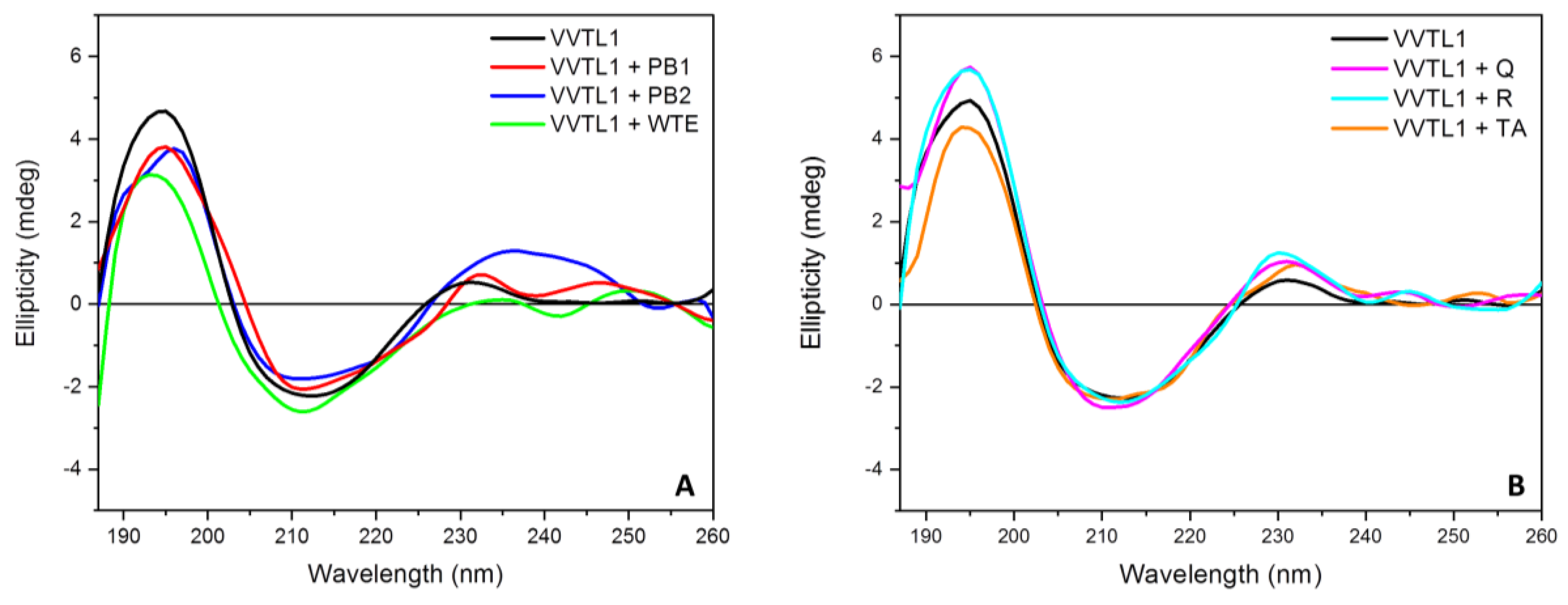
2.1. Polyphenols-VVTL1 Interactions
2.2. Thermal and UV-Denaturation Assays
3. Materials and Methods
3.1. Polyphenols and Protein Preparation
3.2. Synchrotron Radiation Circular Dichroism
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Han, X.; Shen, T.; Lou, H.-X. Dietary Polyphenols and Their Biological Significance. Int. J. Mol. Sci. 2007, 8, 950–988. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.L.; Sacks, G.; Jeffery, D.W. Maceration and Extraction of Grape Components. In Understanding Wine Chemistry; John Wiley & Sons: Chichester, West Sussex, UK, 2016; pp. 179–193. [Google Scholar]
- Smith, P.; McRae, J.; Bindon, K. Impact of winemaking practices on the concentration and composition of tannins in red wine. Aust. J. Grape Wine Res. 2015, 21, 601–614. [Google Scholar] [CrossRef]
- Maury, C.; Sarni-Manchado, P.; Lefebvre, S.; Cheynier, V.; Moutounet, M. Influence of fining with different molecular weight gelatins on proanthocyanidin composition and perception of wines. Am. J. Enol. Vitic. 2001, 52, 140–145. [Google Scholar]
- Gazzola, D.; Vincenzi, S.; Marangon, M.; Pasini, G.; Curioni, A. Grape seed extract: The first protein-based fining agent endogenous to grapes. Aust. J. Grape Wine Res. 2017, 23, 215–225. [Google Scholar] [CrossRef]
- Cosme, F.; Ricardo-Da-Silva, J.M.; Laureano, O. Interactions between protein fining agents and proanthocyanidins in white wine. Food Chem. 2008, 106, 536–544. [Google Scholar] [CrossRef]
- Marangon, M.; Vincenzi, S.; Curioni, A. Wine Fining with Plant Proteins. Molecules 2019, 24, 2186. [Google Scholar] [CrossRef] [Green Version]
- Siebert, K.J. Effects of protein-polyphenol interactions on beverage haze, stabilization, and analysis. J. Agric. Food Chem. 1999, 47, 353–362. [Google Scholar] [CrossRef]
- Springer, L.F.; Sherwood, R.W.; Sacks, G. Pathogenesis-Related Proteins Limit the Retention of Condensed Tannin Additions to Red Wines. J. Agric. Food Chem. 2016, 64, 1309–1317. [Google Scholar] [CrossRef]
- Springer, L.F.; Sacks, G. Protein-Precipitable Tannin in Wines fromVitis viniferaand Interspecific Hybrid Grapes (Vitisssp.): Differences in Concentration, Extractability, and Cell Wall Binding. J. Agric. Food Chem. 2014, 62, 7515–7523. [Google Scholar] [CrossRef]
- Li, S.M.A.; Thome, J.R. Boiling of Ethanol-Water and Ethanol-Benzene Mixtures on an Enhanced Boiling Surface. Heat Transf. Eng. 1984, 5, 70–81. [Google Scholar] [CrossRef]
- Van Sluyter, S.C.; McRae, J.; Falconer, R.J.; Smith, P.; Bacic, A.; Waters, E.J.; Marangon, M. Wine Protein Haze: Mechanisms of Formation and Advances in Prevention. J. Agric. Food Chem. 2015, 63, 4020–4030. [Google Scholar] [CrossRef]
- Esteruelas, M.; Kontoudakis, N.; Gil Cortiella, M.H.; Fort, F.; Canals, J.M.; Zamora, F. Phenolic compounds present in natural haze protein of Sauvignon white wine. Food Res. Int. 2011, 44, 77–83. [Google Scholar] [CrossRef]
- Marangon, M.; Vincenzi, S.; Lucchetta, M.; Curioni, A. Heating and reduction affect the reaction with tannins of wine protein fractions differing in hydrophobicity. Anal. Chim. Acta 2010, 660, 110–118. [Google Scholar] [CrossRef]
- Marangon, M.; Sauvage, F.-X.; Waters, E.J.; Vernhet, A. Effects of Ionic Strength and Sulfate upon Thermal Aggregation of Grape Chitinases and Thaumatin-like Proteins in a Model System. J. Agric. Food Chem. 2011, 59, 2652–2662. [Google Scholar] [CrossRef]
- Smith, M.R.; Penner, M.H.; Bennett, S.E.; Bakalinsky, A. Quantitative Colorimetric Assay for Total Protein Applied to the Red Wine Pinot Noir. J. Agric. Food Chem. 2011, 59, 6871–6876. [Google Scholar] [CrossRef]
- Casassa, F. Proteins and Bentonite in Winemaking: Chemical and Practical Aspects and Sensory Consequences. In Post-Fermentation and -Distillation Technology. Stabilization, Aging, and Spoilage; CRC Press: Boca Raton, FL, USA, 2017; pp. 81–112. [Google Scholar]
- Batista, L.; Monteiro, S.; Loureiro, V.B.; Teixeira, A.R.; Ferreira, R.B. Protein haze formation in wines revisited. The stabilising effect of organic acids. Food Chem. 2010, 122, 1067–1075. [Google Scholar] [CrossRef] [Green Version]
- Gazzola, D.; Van Sluyter, S.C.; Curioni, A.; Waters, E.J.; Marangon, M. Roles of Proteins, Polysaccharides, and Phenolics in Haze Formation in White Wine via Reconstitution Experiments. J. Agric. Food Chem. 2012, 60, 10666–10673. [Google Scholar] [CrossRef]
- Waters, E.J.; Alexander, G.; Muhlack, R.; Pocock, K.; Colby, C.; O’Neill, B.; Høj, P.; Jones, P. Preventing protein haze in bottled white wine. Aust. J. Grape Wine Res. 2005, 11, 215–225. [Google Scholar] [CrossRef]
- Cosme, F.; Fernandes, C.; Ribeiro, T.; Filipe-Ribeiro, L.; Nunes, F. White Wine Protein Instability: Mechanism, Quality Control and Technological Alternatives for Wine Stabilisation—An Overview. Beverages 2020, 6, 19. [Google Scholar] [CrossRef] [Green Version]
- Marangon, M.; Van Sluyter, S.C.; Neilson, K.A.; Chan, C.; Haynes, P.A.; Waters, E.J.; Falconer, R.J. Roles of Grape Thaumatin-like Protein and Chitinase in White Wine Haze Formation. J. Agric. Food Chem. 2011, 59, 733–740. [Google Scholar] [CrossRef]
- Wigand, P.; Tenzer, S.; Schild, H.; Decker, H. Analysis of Protein Composition of Red Wine in Comparison with Rosé and White Wines by Electrophoresis and High-Pressure Liquid Chromatography−Mass Spectrometry (HPLC-MS). J. Agric. Food Chem. 2009, 57, 4328–4333. [Google Scholar] [CrossRef] [PubMed]
- Marangon, M.; Van Sluyter, S.C.; Waters, E.J.; Menz, R.I. Structure of Haze Forming Proteins in White Wines: Vitis vinifera Thaumatin-Like Proteins. PLoS ONE 2014, 9, e113757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falconer, R.J.; Marangon, M.; Van Sluyter, S.C.; Neilson, K.A.; Chan, C.; Waters, E.J. Thermal Stability of Thaumatin-Like Protein, Chitinase, and Invertase Isolated from Sauvignon blanc and Semillon Juice and Their Role in Haze Formation in Wine. J. Agric. Food Chem. 2010, 58, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Toledo, L.; Aquino, A.; Salazar, F. A Theoretical Approach for Understanding the Haze Phenomenon in Bottled White Wines at Molecular Level. South Afr. J. Enol. Vitic. 2017, 38, 64–71. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Le Bourvellec, C.; Renard, C.M. Interactions between Polyphenols and Macromolecules: Quantification Methods and Mechanisms. Crit. Rev. Food Sci. Nutr. 2012, 52, 213–248. [Google Scholar] [CrossRef]
- Murray, N.J.; Williamson, M.P.; Lilley, T.H.; Haslam, E. Study of the interaction between salivary proline-rich proteins and a polyphenol by 1H-NMR spectroscopy. JBIC J. Boil. Inorg. Chem. 1994, 219, 923–935. [Google Scholar] [CrossRef]
- McRae, J.; Falconer, R.J.; Kennedy, J.A. Thermodynamics of Grape and Wine Tannin Interaction with Polyproline: Implications for Red Wine Astringency. J. Agric. Food Chem. 2010, 58, 12510–12518. [Google Scholar] [CrossRef]
- Ozdal, T.; Capanoglu, E.; Altay, F. A review on protein–phenolic interactions and associated changes. Food Res. Int. 2013, 51, 954–970. [Google Scholar] [CrossRef]
- Richard, T.; Lefeuvre, D.; Descendit, A.; Quideau, S.; Monti, J.-P. Recognition characters in peptide–polyphenol complex formation. Biochim. et Biophys. Acta (BBA) Gen. Subj. 2006, 1760, 951–958. [Google Scholar] [CrossRef]
- Silva, M.A.; Ky, I.; Jourdes, M.; Teissedre, P.-L. Rapid and simple method for the quantification of flavan-3-ols in wine. Eur. Food Res. Technol. 2011, 234, 361–365. [Google Scholar] [CrossRef]
- Cheynier, V. Polyphenols in foods are more complex than often thought. Am. J. Clin. Nutr. 2005, 81, 223S–229S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cala, O.; Pinaud, N.; Simon, C.; Fouquet, E.; Laguerre, M.; Dufourc, E.J.; Pianet, I. NMR and molecular modeling of wine tannins binding to saliva proteins: Revisiting astringency from molecular and colloidal prospects. FASEB J. 2010, 24, 4281–4290. [Google Scholar] [CrossRef] [PubMed]
- Tarascou, I.; Barathieu, K.; Simon, C.; Ducasse, M.-A.; André, Y.; Fouquet, E.; Varma, G.; De Freitas, V.; Laguerre, M.; Pianet, I. A 3D structural and conformational study of procyanidin dimers in water and hydro-alcoholic media as viewed by NMR and molecular modeling. Magn. Reson. Chem. 2006, 44, 868–880. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Muñoz, N.; Gómez-Alonso, S.; García-Romero, E.; Hermosín-Gutiérrez, I. Flavonol profiles of Vitis vinifera white grape cultivars. J. Food Compos. Anal. 2010, 23, 699–705. [Google Scholar] [CrossRef]
- Rogers, D.; Jasim, S.B.; Dyer, N.T.; Auvray, F.; Réfrégiers, M.; Hirst, J. Electronic Circular Dichroism Spectroscopy of Proteins. Chem 2019, 5, 2751–2774. [Google Scholar] [CrossRef]
- Kumagai, P.; Araujo, A.P.U.; Lopes, J. Going deep into protein secondary structure with synchrotron radiation circular dichroism spectroscopy. Biophys. Rev. 2017, 9, 517–527. [Google Scholar] [CrossRef] [Green Version]
- Van Sluyter, S.C.; Marangon, M.; Stranks, S.D.; Neilson, K.; Hayasaka, Y.; Haynes, P.A.; Menz, R.I.; Waters, E.J. Two-Step Purification of Pathogenesis-Related Proteins from Grape Juice and Crystallization of Thaumatin-like Proteins. J. Agric. Food Chem. 2009, 57, 11376–11382. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: New York, NY, USA, 2005; pp. 571–607. [Google Scholar]
- Ruzza, P.; Honisch, C.; Marangon, M.; Curioni, A.; Bakalinsky, A.; Vincenzi, S. Influence of the reducing environment in the misfolding of wine proteins. In Protein Misfolding; Elsevier Academic Press: London, UK, 2019; Volume 118, pp. 413–436. [Google Scholar]
- Di Gaspero, M.; Ruzza, P.; Hussain, R.; Vincenzi, S.; Biondi, B.; Gazzola, D.; Siligardi, G.; Curioni, A. Spectroscopy reveals that ethyl esters interact with proteins in wine. Food Chem. 2017, 217, 373–378. [Google Scholar] [CrossRef]
- Platt, J.R. Classification of Spectra of Cata-Condensed Hydrocarbons. J. Chem. Phys. 1949, 17, 484. [Google Scholar] [CrossRef]
- Hussain, R.; Benning, K.; Jávorfi, T.; Longo, E.; Rudd, T.; Pulford, B.; Siligardi, G. CDApps: Integrated software for experimental planning and data processing at beamline B23, Diamond Light Source. J. Synchrotron Radiat. 2015, 22, 465–468. [Google Scholar] [CrossRef] [Green Version]
- Provencher, S.W.; Gloeckner, J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 1981, 20, 33–37. [Google Scholar] [CrossRef]
- Waters, E.J.; Hayasaka, Y.; Tattersall, D.B.; Adams, K.S.; Williams, P.J. Sequence Analysis of Grape (Vitis vinifera) Berry Chitinases That Cause Haze Formation in Wines. J. Agric. Food Chem. 1998, 46, 4950–4957. [Google Scholar] [CrossRef]
- Butzke, C.E.; Vogt, E.E.; Chacón-Rodríguez, L. Effects of heat exposure on wine quality during transport and storage. J. Wine Res. 2012, 23, 15–25. [Google Scholar] [CrossRef]
- Patel, A.R.; Heussen, P.C.; Hazekamp, J.; Drost, E.; Imhof, A. Quercetin loaded biopolymeric colloidal particles prepared by simultaneous precipitation of quercetin with hydrophobic protein in aqueous medium. Food Chem. 2012, 133, 423–429. [Google Scholar] [CrossRef]
- Gambuti, A.; Picariello, L.; Rinaldi, A.; Forino, M.; Blaiotta, G.; Moine, V.; Moio, L. New insights into the formation of precipitates of quercetin in Sangiovese wines. J. Food Sci. Technol. 2020, 1–10. [Google Scholar] [CrossRef]
- Hussain, R.; Jávorfi, T.; Siligardi, G. 8.23 Spectroscopic Analysis: Synchrotron Radiation Circular Dichroism. Comprehensive Chirality 2012, 8, 438–448. [Google Scholar]
- Hussain, R.; Longo, E.; Siligardi, G. UV-Denaturation Assay to Assess Protein Photostability and Ligand-Binding Interactions Using the High Photon Flux of Diamond B23 Beamline for SRCD. Molecules 2018, 23, 1906. [Google Scholar] [CrossRef] [Green Version]
- Xu, G.; Chance, M.R. Hydroxyl Radical-Mediated Modification of Proteins as Probes for Structural Proteomics. Chem. Rev. 2007, 107, 3514–3543. [Google Scholar] [CrossRef]
- Grosvenor, A.J.; Morton, J.D.; Dyer, J. Profiling of residue-level photo-oxidative damage in peptides. Amino Acids 2009, 39, 285–296. [Google Scholar] [CrossRef]
- Ruzza, P.; Hussain, R.; Biondi, B.; Calderan, A.; Tessari, I.; Bubacco, L.; Siligardi, G. Effects of Trehalose on Thermodynamic Properties of Alpha-synuclein Revealed through Synchrotron Radiation Circular Dichroism. Biomolecules 2015, 5, 724–734. [Google Scholar] [CrossRef] [Green Version]
- Ruzza, P.; Vitale, R.M.; Hussain, R.; Biondi, B.; Amodeo, P.; Sechi, G.; Siligardi, G. Interactions of GFAP with ceftriaxone and phenytoin: SRCD and molecular docking and dynamic simulation. Biochim. et Biophys. Acta (BBA) Gen. Subj. 2016, 1860, 2239–2248. [Google Scholar] [CrossRef] [Green Version]
- Morales, J.; Günther, G.; Zanocco, A.; Lemp, E. Singlet Oxygen Reactions with Flavonoids. A Theoretical—Experimental Study. PLoS ONE 2012, 7, e40548. [Google Scholar] [CrossRef]
- Honisch, C.; Donadello, V.; Hussain, R.; Peterle, D.; De Filippis, V.; Arrigoni, G.; Gatto, C.; Giurgola, L.; Siligardi, G.; Ruzza, P. Application of Circular Dichroism and Fluorescence Spectroscopies to Assess Photostability of Water-Soluble Porcine Lens Proteins. ACS Omega 2020, 5, 4293–4301. [Google Scholar] [CrossRef] [PubMed]
- Mayr, C.M.; De Rosso, M.; Vedova, A.D.; Flamini, R. High-Resolution Mass Spectrometry Identification of Secondary Metabolites in Four Red Grape Varieties Potentially Useful as Traceability Markers of Wines. Beverages 2018, 4, 74. [Google Scholar] [CrossRef] [Green Version]
- Rodger, A.; Marrington, R.; Roper, D.I.; Windsor, S.; Nienhaus, G.U. Circular Dichroism Spectroscopy for the Study of Protein–Ligand Interactions. In Protein-Ligand Interactions; Humana Press: Totowa, NJ, USA, 2005; Volume 305, pp. 343–364. [Google Scholar]
- Siebert, K.J.; Troukhanova, N.V.; Lynn, P.Y. Nature of Polyphenol−Protein Interactions. J. Agric. Food Chem. 1996, 44, 80–85. [Google Scholar] [CrossRef]
- Mateus, N.; Carvalho, E.; Luís, C.; De Freitas, V. Influence of the tannin structure on the disruption effect of carbohydrates on protein–tannin aggregates. Anal. Chim. Acta 2004, 513, 135–140. [Google Scholar] [CrossRef]
- Kemp, B.; Condé, B.; Jégou, S.; Howell, K.; Vasserot, Y.; Marchal, R. Chemical compounds and mechanisms involved in the formation and stabilization of foam in sparkling wines. Crit. Rev. Food Sci. Nutr. 2018, 59, 2072–2094. [Google Scholar] [CrossRef]
- Esteruelas, M.; Poinsaut, P.; Sieczkowski, N.; Manteau, S.; Fort, M.F.; Canals, J.M.; Zamora, F. Comparison of methods for estimating protein stability in white wines. Am. J. Enol. Vitic. 2009, 60, 302–311. [Google Scholar]
Sample Availability: Samples of the compounds are available from the authors. |




| VVTL1 | % Secondary Structure Content | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 °C | 20 °C | 70 °C | ||||||||||
| α-helix | β-sheet | turns | Unord. | α-helix | β-sheet | turns | Unord. | α-helix | β-sheet | turns | Unord. | |
| Alone | 2 | 44 | 22 | 32 | 3 | 45 | 22 | 29 | 6 | 30 | 19 | 49 |
| + PB1 | 0 | 44 | 21 | 30 | 0 | 37 | 22 | 38 | 0 | 18 | 12 | 64 |
| + PB2 | 4 | 41 | 22 | 32 | 4 | 41 | 22 | 33 | 2 | 15 | 14 | 53 |
| + Q | 5 | 44 | 22 | 29 | 5 | 43 | 22 | 30 | 5 | 33 | 16 | 46 |
| + R | 5 | 43 | 22 | 30 | 4 | 44 | 22 | 30 | 4 | 34 | 16 | 46 |
| + WTE | 4 | 41 | 22 | 33 | 4 | 43 | 22 | 31 | 7 | 20 | 12 | 60 |
| + TA | 4 | 46 | 23 | 27 | 5 | 44 | 22 | 29 | 7 | 24 | 14 | 55 |
| Sample | TM 1 | TM 2 |
|---|---|---|
| VVTL1 alone | 32.2 | 53.9 |
| VVTL1 + PB1 | 42.8 | 58.5 |
| VVTL1 + PB2 | 19.5 | 55.4 |
| VVTL1 + Q | 18.8 | 47.0 |
| VVTL1 + R | 12.4 | 48.0 |
| VVTL1 + WTE | 11.2 | 58.3 |
| VVTL1 + TA | 29.3 | 53.3 |
| VVTL1 | % Secondary Structure | |||||||
|---|---|---|---|---|---|---|---|---|
| After 1 Scan | After 20 Scans | |||||||
| α-Helix | β-Sheet | Turns | Unordered | α-Helix | β-Sheet | Turns | Unordered | |
| alone | 6 | 41 | 22 | 31 | 6 | 32 | 17 | 45 |
| + PB1 | 5 | 43 | 24 | 28 | 7 | 25 | 24 | 44 |
| + PB2 | 4 | 43 | 22 | 31 | 9 | 38 | 23 | 30 |
| + Q | 5 | 43 | 22 | 30 | 6 | 39 | 20 | 35 |
| + R | 5 | 43 | 22 | 30 | 6 | 41 | 20 | 33 |
| + WTE | 4 | 43 | 23 | 31 | 17 | 21 | 19 | 44 |
| + TA | 4 | 46 | 23 | 27 | 4 | 38 | 19 | 38 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Gaspero, M.; Ruzza, P.; Hussain, R.; Honisch, C.; Biondi, B.; Siligardi, G.; Marangon, M.; Curioni, A.; Vincenzi, S. The Secondary Structure of a Major Wine Protein is Modified upon Interaction with Polyphenols. Molecules 2020, 25, 1646. https://doi.org/10.3390/molecules25071646
Di Gaspero M, Ruzza P, Hussain R, Honisch C, Biondi B, Siligardi G, Marangon M, Curioni A, Vincenzi S. The Secondary Structure of a Major Wine Protein is Modified upon Interaction with Polyphenols. Molecules. 2020; 25(7):1646. https://doi.org/10.3390/molecules25071646
Chicago/Turabian StyleDi Gaspero, Mattia, Paolo Ruzza, Rohanah Hussain, Claudia Honisch, Barbara Biondi, Giuliano Siligardi, Matteo Marangon, Andrea Curioni, and Simone Vincenzi. 2020. "The Secondary Structure of a Major Wine Protein is Modified upon Interaction with Polyphenols" Molecules 25, no. 7: 1646. https://doi.org/10.3390/molecules25071646







