Design of Calcium-Binding Proteins to Sense Calcium
Abstract
:1. Introduction
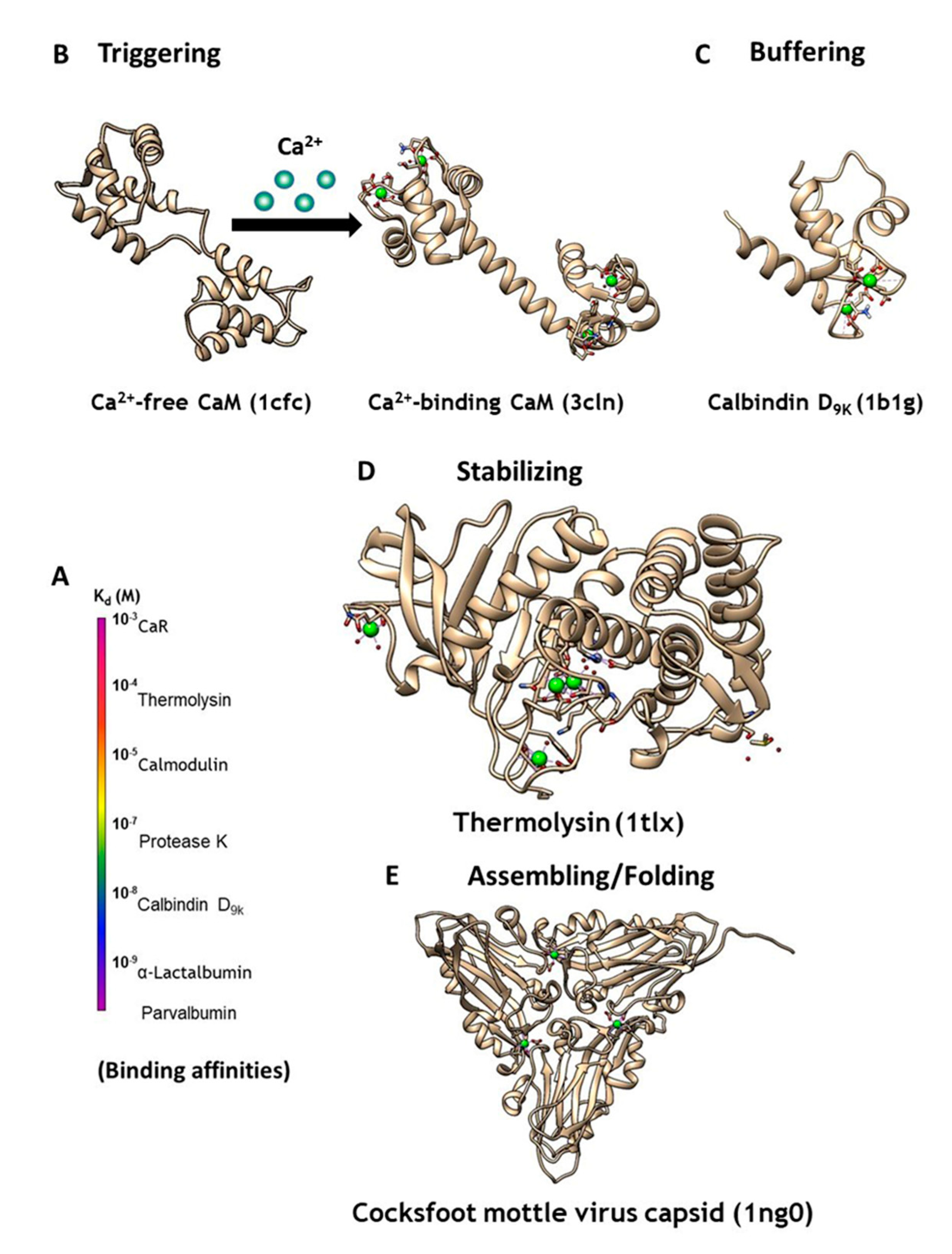
2. Classification and Prediction of Calcium Binding Sites in Proteins
3. Development of Grafting and Design Approaches to Discover Key Factors that Contribute to Ca2+ Binding Affinities and Selectivities, and Ca2+-Induced Conformational Change
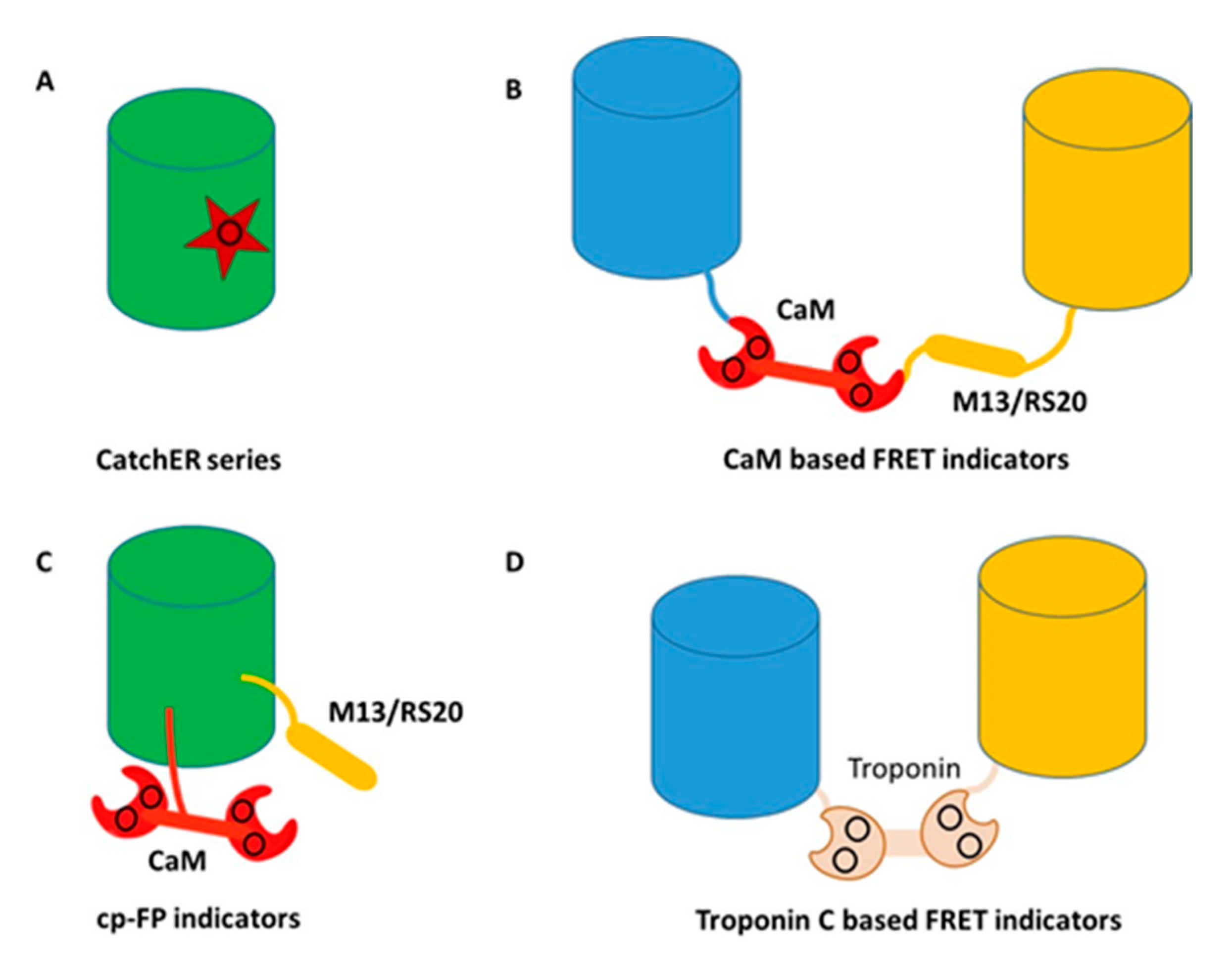
4. Development of Genetically Encoded Calcium Indicators (GECIs) Using Protein Engineering
5. Development of Calcium Sensors Using Rational Design
6. Perspectives, Future Research, and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Miyawaki, A.; Llopis, J.; Heim, R.; Mccaffery, J.M.; Adams, J.A.; Ikura, M.; Tsien, R.Y. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 1997, 388, 882–887. [Google Scholar] [CrossRef] [PubMed]
- Rudolf, R.; Magalhaes, P.J.; Pozzan, T. Direct in vivo monitoring of sarcoplasmic reticulum Ca2+ and cytosolic camp dynamics in mouse skeletal muscle. J. Cell Biol. 2006, 173, 187–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bate, N.; Caves, R.E.; Skinner, S.P.; Goult, B.T.; Basran, J.; Mitcheson, J.S.; Vuister, G.W. A novel mechanism for calmodulin-dependent inactivation of transient receptor potential vanilloid 6. Biochemistry 2018, 57, 2611–2622. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.X. Multiple roles of calmodulin and other Ca2+-binding proteins in the functional regulation of TRP channels. Pflug. Arch Eur. J. Physiol. 2005, 451, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Hofer, A.M.; Brown, E.M. Extracellular calcium sensing and signaling. Nat. Rev. Mol. Cell Biol. 2003, 4, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Miller, C.L.; Brown, E.M.; Yang, J.J. The calcium sensing receptor: From calcium sensing to signaling. Sci. China Life Sci. 2015, 58, 14–27. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Zhang, T.; Zou, J.; Miller, C.L.; Gorkhali, R.; Yang, J.Y.; Schilmiller, A.; Wang, S.; Huang, K.; Brown, E.M.; et al. Structural basis for regulation of human calcium-sensing receptor by magnesium ions and an unexpected tryptophan derivative co-agonist. Sci. Adv. 2016, 2, E1600241. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Huang, Y.; Wong, H.C.; Zhou, Y.; Wang, X.; Yang, J.; Hall, R.A.; Brown, E.M.; Yang, J.J. Elucidation of a novel extracellular calcium-binding site on metabotropic glutamate receptor 1{alpha} (mglur1{alpha}) that controls receptor activation. J. Biol. Chem. 2010, 285, 33463–33474. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Frey, T.K.; Yang, J.J. Viral calciomics: Interplays between Ca2+ and virus. Cell Calcium 2009, 46, 1–17. [Google Scholar] [CrossRef]
- Xue, S.; Qiao, J.; Jiang, J.; Hubbard, K.; White, N.; Wei, L.; Li, S.; Liu, Z.R.; Yang, J.J. Design of procas (protein-based Gd(3+) mri contrast agents) with high dose efficiency and capability for molecular imaging of cancer biomarkers. Med. Res. Rev. 2014, 34, 1070–1099. [Google Scholar] [CrossRef]
- Xue, S.; Qiao, J.; Pu, F.; Cameron, M.; Yang, J.J. Design of a novel class of protein-based magnetic resonance imaging contrast agents for the molecular imaging of cancer biomarkers. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2013, 5, 163–179. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Li, S.; Wei, L.; Jiang, J.; Long, R.; Mao, H.; Wei, L.; Wang, L.; Yang, H.; Grossniklaus, H.E.; et al. HER2 targeted molecular MR imaging using a de novo designed protein contrast agent. PLoS ONE 2011, 6, E18103. [Google Scholar] [CrossRef] [PubMed]
- Pu, F.; Salarian, M.; Xue, S.; Qiao, J.; Feng, J.; Tan, S.; Patel, A.; Li, X.; Mamouni, K.; Hekmatyar, K.; et al. Prostate-specific membrane antigen targeted protein contrast agents for molecular imaging of prostate cancer by MRI. Nanoscale 2016, 8, 12668–12682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salarian, M.; Yang, H.; Turaga, R.C.; Tan, S.; Qiao, J.; Xue, S.; Gui, Z.; Peng, G.; Han, H.; Mittal, P.; et al. Precision detection of liver metastasis by collagen-targeted protein MRI contrast agent. Biomaterials 2019, 224, 119478. [Google Scholar] [CrossRef]
- Salarian, M.; Ibhagui, O.Y.; Yang, J.J. Molecular imaging of extracellular matrix proteins with targeted probes using magnetic resonance imaging. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020, E1622. [Google Scholar] [CrossRef]
- Kirberger, M.; Wang, X.; Deng, H.; Yang, W.; Chen, G.; Yang, J.J. Statistical analysis of structural characteristics of protein Ca2+-binding sites. J. Biol. Inorg. Chem. 2008, 13, 1169–1181. [Google Scholar] [CrossRef]
- Kirberger, M.; Yang, J.J. Structural differences between Pb2+- and Ca2+-binding sites in proteins: Implications with respect to toxicity. J. Inorg. Biochem. 2008. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Yang, W.; Kirberger, M.; Lee, H.W.; Ayalasomayajula, G.; Yang, J.J. Prediction of EF-hand calcium-binding proteins and analysis of bacterial EF-hand proteins. Proteins 2006, 65, 643–655. [Google Scholar] [CrossRef]
- Deng, H.; Chen, G.; Yang, W.; Yang, J.J. Predicting Calcium-binding sites in proteins—A graph theory and geometry approach. Proteins 2006, 64, 34–42. [Google Scholar] [CrossRef]
- Wang, X.; Kirberger, M.; Qiu, F.; Chen, G.; Yang, J.J. Towards predicting Ca2+-binding sites with different coordination numbers in proteins with atomic resolution. Proteins 2009, 75, 787–798. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhao, K.; Kirberger, M.; Wong, H.; Chen, G.; Yang, J. Analysis and prediction of calcium-binding pockets from apo protein structures exhibiting calcium-induced localized conformational changes. Protein Sci. 2010, 19, 1180–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, K.; Wang, X.; Wong, H.C.; Wohlhueter, R.; Kirberger, M.P.; Chen, G.; Yang, J.J. Predicting Ca2+-binding sites using refined carbon clusters. Proteins 2012, 80, 2666–2679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.J.; Yang, J.; Wei, L.; Zurkiya, O.; Yang, W.; Li, S.; Zou, J.; Zhou, Y.; Maniccia, A.L.; Mao, H.; et al. Rational design of protein-based MRI contrast agents. J. Am. Chem. Soc. 2008, 130, 9260–9267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Tzeng, W.P.; Yang, W.; Zhou, Y.; Ye, Y.; Lee, H.W.; Frey, T.K.; Yang, J. Identification of a Ca2+-binding domain in the rubella virus nonstructural protease. J. Virol. 2007, 81, 7517–7528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Zhou, Y.; Castiblanco, A.; Yang, W.; Brown, E.M.; Yang, J.J. Multiple Ca2+-binding sites in the extracellular domain of the Ca2+-sensing receptor corresponding to cooperative Ca2+ response. Biochemistry 2009, 48, 388–398. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Zhou, Y.; Yang, W.; Butters, R.; Lee, H.W.; Li, S.; Castiblanco, A.; Brown, E.M.; Yang, J.J. Identification and dissection of Ca2+-binding sites in the extracellular domain of Ca2+-sensing receptor. J. Biol. Chem. 2007, 282, 19000–19010. [Google Scholar] [CrossRef] [Green Version]
- Ye, Y.; Lee, H.W.; Yang, W.; Shealy, S.; Yang, J.J. Probing site-specific calmodulin calcium and lanthanide affinity by grafting. J. Am. Chem. Soc. 2005, 127, 3743–3750. [Google Scholar] [CrossRef]
- Ye, Y.; Shealy, S.; Lee, H.W.; Torshin, I.; Harrison, R.; Yang, J.J. A grafting approach to obtain site-specific metal-binding properties of EF-hand proteins. Protein Eng. 2003, 16, 429–434. [Google Scholar] [CrossRef]
- Wilkins, A.L.; Ye, Y.; Yang, W.; Lee, H.W.; Liu, Z.R.; Yang, J.J. Metal-binding studies for a de novo designed calcium-binding protein. Protein Eng. 2002, 15, 571–574. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.W.; Yang, W.; Ye, Y.; Liu, Z.R.; Glushka, J.; Yang, J.J. Isolated Ef-loop III of calmodulin in a scaffold protein remains unpaired in solution using pulsed-field-gradient NMR spectroscopy. Biochim. Biophys. Acta 2002, 1598, 80–87. [Google Scholar] [CrossRef]
- Franz, K.J.; Nitz, M.; Imperiali, B. Lanthanide-binding tags as versatile protein coexpression probes. Chembiochem 2003, 4, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Jones, L.M.; Isley, L.; Ye, Y.; Lee, H.W.; Wilkins, A.; Liu, Z.R.; Hellinga, H.W.; Malchow, R.; Ghazi, M.; et al. Rational design of a calcium-binding protein. J. Am. Chem. Soc. 2003, 125, 6165–6171. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Wilkins, A.L.; Li, S.; Ye, Y.; Yang, J.J. The effects of Ca2+ binding on the dynamic properties of a designed Ca2+-binding protein. Biochemistry 2005, 44, 8267–8273. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Wilkins, A.L.; Ye, Y.; Liu, Z.R.; Li, S.Y.; Urbauer, J.L.; Hellinga, H.W.; Kearney, A.; Van Der Merwe, P.A.; Yang, J.J. Design of a calcium-binding protein with desired structure in a cell adhesion molecule. J. Am. Chem. Soc. 2005, 127, 2085–2093. [Google Scholar] [CrossRef]
- Jones, L.M.; Yang, W.; Maniccia, A.W.; Harrison, A.; Van Der Merwe, P.A.; Yang, J.J. Rational design of a novel calcium-binding site adjacent to the ligand-binding site on CD2 increases its CD48 affinity. Protein Sci. 2008, 17, 439–449. [Google Scholar] [CrossRef] [Green Version]
- Maniccia, A.W.; Yang, W.; Li, S.Y.; Johnson, J.A.; Yang, J.J. Using protein design to dissect the effect of charged residues on metal binding and protein stability. Biochemistry 2006, 45, 5848–5856. [Google Scholar] [CrossRef]
- Ye, Y.; Lee, H.W.; Yang, W.; Yang, J.J. Calcium and lanthanide affinity of the Ef-loops from the C-terminal domain of calmodulin. J. Inorg. Biochem. 2005, 99, 1376–1383. [Google Scholar] [CrossRef]
- Li, S.; Yang, W.; Maniccia, A.W.; Barrow, D., Jr.; Tjong, H.; Zhou, H.X.; Yang, J.J. Rational design of a conformation-switchable Ca2+- and Tb3+-binding protein without the use of multiple coupled metal-binding sites. FEBS J. 2008, 275, 5048–5061. [Google Scholar] [CrossRef] [Green Version]
- Giepmans, B.N.; Adams, S.R.; Ellisman, M.H.; Tsien, R.Y. The fluorescent toolbox for assessing protein location and function. Science 2006, 312, 217–224. [Google Scholar] [CrossRef] [Green Version]
- Lippincott-Schwartz, J.; Patterson, G.H. Development and use of fluorescent protein markers in living cells. Science 2003, 300, 87–91. [Google Scholar] [CrossRef] [Green Version]
- Rizzuto, R.; Simpson, A.W.; Brini, M.; Pozzan, T. Rapid changes of mitochondrial Ca2+ revealed by specifically targeted recombinant aequorin. Nature 1992, 358, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Hofer, A.M.; Landolfi, B.; Debellis, L.; Pozzan, T.; Curci, S. Free [Ca2+] dynamics measured in agonist-sensitive stores of single living intact cells: A new look at the refilling process. EMBO J. 1998, 17, 1986–1995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Persechini, A.; Lynch, J.A.; Romoser, V.A. Novel fluorescent indicator proteins for monitoring free intracellular Ca2+. Cell Calcium 1997, 22, 209–216. [Google Scholar] [CrossRef]
- Heim, R.; Cubitt, A.B.; Tsien, R.Y. Improved green fluorescence. Nature 1995, 373, 663–664. [Google Scholar] [CrossRef] [PubMed]
- Palmer, A.E.; Tsien, R.Y. Measuring calcium signaling using genetically targetable fluorescent indicators. Nat. Protoc. 2006, 1, 1057–1065. [Google Scholar] [CrossRef]
- Heim, N.; Griesbeck, O. Genetically encoded indicators of cellular calcium dynamics based on troponin C and green fluorescent protein. J. Biol. Chem. 2004, 279, 14280–14286. [Google Scholar] [CrossRef] [Green Version]
- Barykina, N.V.; Subach, O.M.; Piatkevich, K.D.; Jung, E.E.; Malyshev, A.Y.; Smirnov, I.V.; Bogorodskiy, A.O.; Borshchevskiy, V.I.; Varizhuk, A.M.; Pozmogova, G.E.; et al. Green fluorescent genetically encoded calcium indicator based on calmodulin/M13-peptide from fungi. PLoS ONE 2017, 12, E0183757. [Google Scholar] [CrossRef] [Green Version]
- Nakai, J.; Ohkura, M.; Imoto, K. A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein. Nat. Biotechnol. 2001, 19, 137–141. [Google Scholar] [CrossRef]
- Tallini, Y.N.; Ohkura, M.; Choi, B.R.; Ji, G.; Imoto, K.; Doran, R.; Lee, J.; Plan, P.; Wilson, J.; Xin, H.B.; et al. Imaging cellular signals in the heart in vivo: Cardiac expression of the high-signal Ca2+ indicator GCaMP2. Proc. Natl. Acad. Sci. USA 2006, 103, 4753–4758. [Google Scholar] [CrossRef] [Green Version]
- Tian, L.; Hires, S.A.; Mao, T.; Huber, D.; Chiappe, M.E.; Chalasani, S.H.; Petreanu, L.; Akerboom, J.; Mckinney, S.A.; Schreiter, E.R.; et al. Imaging neural activity in worms, flies and mice with improved gcamp calcium indicators. Nat. Methods 2009, 6, 875–881. [Google Scholar] [CrossRef] [Green Version]
- Akerboom, J.; Chen, T.W.; Wardill, T.J.; Tian, L.; Marvin, J.S.; Mutlu, S.; Calderon, N.C.; Esposti, F.; Borghuis, B.G.; Sun, X.R.; et al. Optimization of a gcamp calcium indicator for neural activity imaging. J. Neurosci. 2012, 32, 13819–13840. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.W.; Wardill, T.J.; Sun, Y.; Pulver, S.R.; Renninger, S.L.; Baohan, A.; Schreiter, E.R.; Kerr, R.A.; Orger, M.B.; Jayaraman, V.; et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 2013, 499, 295–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, J.H.; Swanson, C.J.; Chen, J.; Li, A.; Lippert, L.G.; Boye, S.E.; Rose, K.; Sivaramakrishnan, S.; Chuong, C.M.; Chow, R.H. The GCaMP-R family of genetically encoded ratiometric calcium indicators. ACS Chem. Biol. 2017, 12, 1066–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Juan-Sanz, J.; Holt, G.T.; Schreiter, E.R.; De Juan, F.; Kim, D.S.; Ryan, T.A. Axonal endoplasmic reticulum Ca2+ content controls release probability in cns nerve terminals. Neuron 2017, 93, 867–881 E866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Araki, S.; Wu, J.; Teramoto, T.; Chang, Y.F.; Nakano, M.; Abdelfattah, A.S.; Fujiwara, M.; Ishihara, T.; Nagai, T.; et al. An Expanded palette of genetically encoded Ca2+ indicators. Science 2011, 333, 1888–1891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, M.J.; Baldwin, H.A.; Werley, C.A.; Boccardo, S.; Whitaker, L.R.; Yan, X.; Holt, G.T.; Schreiter, E.R.; Looger, L.L.; Cohen, A.E.; et al. A low affinity gcamp3 variant (GCaMPer) for imaging the endoplasmic reticulum calcium store. PLoS ONE 2015, 10, E0139273. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, J.; Kanemaru, K.; Ishii, K.; Ohkura, M.; Okubo, Y.; Iino, M. Imaging intraorganellar Ca2+ at subcellular resolution using CEPIA. Nat. Commun. 2014, 5, 4153. [Google Scholar] [CrossRef]
- Helassa, N.; Zhang, X.H.; Conte, I.; Scaringi, J.; Esposito, E.; Bradley, J.; Carter, T.; Ogden, D.; Morad, M.; Torok, K. Fast-response calmodulin-based fluorescent indicators reveal rapid intracellular calcium dynamics. Sci. Rep. 2015, 5, 15978. [Google Scholar] [CrossRef] [Green Version]
- Ohkura, M.; Sasaki, T.; Sadakari, J.; Gengyo-Ando, K.; Kagawa-Nagamura, Y.; Kobayashi, C.; Ikegaya, Y.; Nakai, J. Genetically encoded green fluorescent Ca2+ indicators with improved detectability for neuronal Ca2+ signals. PLoS ONE 2012, 7, E51286. [Google Scholar] [CrossRef] [Green Version]
- Muto, A.; Ohkura, M.; Kotani, T.; Higashijima, S.; Nakai, J.; Kawakami, K. Genetic visualization with an improved GCaMP calcium indicator reveals spatiotemporal activation of the spinal motor neurons in zebrafish. Proc. Natl. Acad. Sci. USA 2011, 108, 5425–5430. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Liu, L.; Matsuda, T.; Zhao, Y.; Rebane, A.; Drobizhev, M.; Chang, Y.F.; Araki, S.; Arai, Y.; March, K.; et al. Improved orange and red Ca²± indicators and photophysical considerations for optogenetic applications. ACS Chem. Neurosci. 2013, 4, 963–972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dana, H.; Sun, Y.; Mohar, B.; Hulse, B.K.; Kerlin, A.M.; Hasseman, J.P.; Tsegaye, G.; Tsang, A.; Wong, A.; Patel, R.; et al. High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat. Methods 2019, 16, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Hendel, T.; Mank, M.; Schnell, B.; Griesbeck, O.; Borst, A.; Reiff, D.F. Fluorescence changes of genetic calcium indicators and OGB-1 correlated with neural activity and calcium in vivo and in vitro. J. Neurosci. 2008, 28, 7399–7411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badura, A.; Sun, X.R.; Giovannucci, A.; Lynch, L.A.; Wang, S.S. Fast calcium sensor proteins for monitoring neural activity. Neurophotonics 2014, 1, 025008. [Google Scholar] [CrossRef]
- Kerruth, S.; Coates, C.; Durst, C.D.; Oertner, T.G.; Torok, K. The kinetic mechanisms of fast-decay red-fluorescent genetically encoded calcium indicators. J. Biol. Chem. 2019, 294, 3934–3946. [Google Scholar] [CrossRef] [Green Version]
- Barykina, N.V.; Doronin, D.A.; Subach, O.M.; Sotskov, V.P.; Plusnin, V.V.; Ivleva, O.A.; Gruzdeva, A.M.; Kunitsyna, T.A.; Ivashkina, O.I.; Lazutkin, A.A.; et al. Ntnc-like genetically encoded calcium indicator with a positive and enhanced response and fast kinetics. Sci. Rep. 2018, 8, 15233. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Dana, H.; Abdelfattah, A.S.; Patel, R.; Shea, J.; Molina, R.S.; Rawal, B.; Rancic, V.; Chang, Y.F.; Wu, L.; et al. A genetically encoded Ca2+ indicator based on circularly permutated sea anemone red fluorescent protein eqFP578. BMC Biol. 2018, 16, 9. [Google Scholar] [CrossRef] [Green Version]
- Qian, Y.; Piatkevich, K.D.; Mc Larney, B.; Abdelfattah, A.S.; Mehta, S.; Murdock, M.H.; Gottschalk, S.; Molina, R.S.; Zhang, W.; Chen, Y.; et al. A genetically encoded near-infrared fluorescent calcium ion indicator. Nat. Methods 2019, 16, 171–174. [Google Scholar] [CrossRef]
- Subach, O.M.; Barykina, N.V.; Anokhin, K.V.; Piatkevich, K.D.; Subach, F.V. Near-infrared genetically encoded positive calcium indicator based on GAF-FP bacterial phytochrome. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef] [Green Version]
- Witte, M.E.; Schumacher, A.M.; Mahler, C.F.; Bewersdorf, J.P.; Lehmitz, J.; Scheiter, A.; Sanchez, P.; Williams, P.R.; Griesbeck, O.; Naumann, R.; et al. Calcium influx through plasma-membrane nanoruptures drives axon degeneration in a model of multiple sclerosis. Neuron 2019, 101, 615–624. [Google Scholar] [CrossRef] [Green Version]
- Mank, M.; Reiff, D.F.; Heim, N.; Friedrich, M.W.; Borst, A.; Griesbeck, O. A fret-based calcium biosensor with fast signal kinetics and high fluorescence change. Biophys. J. 2006, 90, 1790–1796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thestrup, T.; Litzlbauer, J.; Bartholomaus, I.; Mues, M.; Russo, L.; Dana, H.; Kovalchuk, Y.; Liang, Y.; Kalamakis, G.; Laukat, Y.; et al. Optimized ratiometric calcium sensors for functional in vivo imaging of neurons and T lymphocytes. Nat. Methods 2014, 11, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Horikawa, K.; Yamada, Y.; Matsuda, T.; Kobayashi, K.; Hashimoto, M.; Matsu-Ura, T.; Miyawaki, A.; Michikawa, T.; Mikoshiba, K.; Nagai, T. Spontaneous network activity visualized by ultrasensitive Ca2+ indicators, yellow cameleon-nano. Nat. Methods 2010, 7, 729–732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fosque, B.F.; Sun, Y.; Dana, H.; Yang, C.T.; Ohyama, T.; Tadross, M.R.; Patel, R.; Zlatic, M.; Kim, D.S.; Ahrens, M.B.; et al. Neural circuits. labeling of active neural circuits in vivo with designed calcium integrators. Science 2015, 347, 755–760. [Google Scholar] [CrossRef] [PubMed]
- Zolnik, T.A.; Sha, F.; Johenning, F.W.; Schreiter, E.R.; Looger, L.L.; Larkum, M.E.; Sachdev, R.N. All-optical functional synaptic connectivity mapping in acute brain slices using the calcium integrator campari. J. Physiol. 2017, 595, 1465–1477. [Google Scholar] [CrossRef] [Green Version]
- Tang, S.; Wong, H.C.; Wang, Z.M.; Huang, Y.; Zou, J.; Zhuo, Y.; Pennati, A.; Gadda, G.; Delbono, O.; Yang, J.J. Design and application of a class of sensors to monitor Ca2+ dynamics in high Ca2+ concentration cellular compartments. Proc. Natl. Acad. Sci. USA 2011, 108, 16265–16270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddish, F.N.; Miller, C.L.; Gorkhali, R.; Yang, J.J. Monitoring Er/Sr Calcium release with the targeted Ca2+ sensor catcher. J. Vis. Exp. 2017. [Google Scholar] [CrossRef]
- Zhang, Y.; Reddish, F.; Tang, S.; Zhuo, Y.; Wang, Y.F.; Yang, J.J.; Weber, I.T. Structural basis for a hand-like site in the calcium sensor catcher with fast kinetics. Acta Crystallogr. D 2013, 69, 2309–2319. [Google Scholar] [CrossRef] [Green Version]
- Zhuo, Y.; Solntsev, K.M.; Reddish, F.; Tang, S.; Yang, J.J. Effect of Ca2+ on the steady-state and time-resolved emission properties of the genetically encoded fluorescent sensor Catcher. J. Phys. Chem. B 2015, 119, 2103–2111. [Google Scholar] [CrossRef] [Green Version]
- Zou, J.; Hofer, A.M.; Lurtz, M.M.; Gadda, G.; Ellis, A.L.; Chen, N.; Huang, Y.; Holder, A.; Ye, Y.; Louis, C.F.; et al. Developing sensors for real-time measurement of high Ca2+ concentrations. Biochemistry 2007, 46, 12275–12288. [Google Scholar] [CrossRef]
- Zhou, Y.; Xue, S.; Yang, J.J. Calciomics: Integrative studies of Ca2+-binding proteins and their interactomes in biological systems. Metallomics 2013, 5, 29–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Yang, W.; Lurtz, M.M.; Ye, Y.; Huang, Y.; Lee, H.W.; Chen, Y.; Louis, C.F.; Yang, J.J. Identification of the calmodulin binding domain of connexin 43. J. Biol. Chem. 2007, 282, 35005–35017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, J.; Salarian, M.; Chen, Y.; Zhuo, Y.; Brown, N.E.; Hepler, J.R.; Yang, J.J. Direct visualization of interaction between calmodulin and connexin45. Biochem. J. 2017, 474, 4035–4051. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, Y.; Lin, X.; Wong, H.C.; Xu, Q.; Jiang, J.; Wang, S.; Lurtz, M.M.; Louis, C.F.; Veenstra, R.D.; et al. Molecular interaction and functional regulation of connexin50 gap junctions by calmodulin. Biochem. J. 2011, 435, 711–722. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yang, W.; Lurtz, M.M.; Chen, Y.; Jiang, J.; Huang, Y.; Louis, C.F.; Yang, J.J. Calmodulin mediates the Ca2+-dependent regulation of Cx44 gap junctions. Biophys. J. 2009, 96, 2832–2848. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Huang, Y.; Jiang, Y.; Mulpuri, N.; Wei, L.; Hamelberg, D.; Brown, E.M.; Yang, J.J. Identification of an l-phenylalanine binding site enhancing the cooperative responses of the calcium-sensing receptor to calcium. J. Biol. Chem. 2014, 289, 5296–5309. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Mulpuri, N.; Hannan, F.M.; Nesbit, M.A.; Thakker, R.V.; Hamelberg, D.; Brown, E.M.; Yang, J.J. Role of Ca2+ and l-Phe in regulating functional cooperativity of disease-associated “toggle” calcium-sensing receptor mutations. PLoS ONE 2014, 9, E113622. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Zhuo, Y.; Moniz, H.A.; Wang, S.; Moremen, K.W.; Prestegard, J.H.; Brown, E.M.; Yang, J.J. Direct determination of multiple ligand interactions with the extracellular domain of the calcium-sensing receptor. J. Biol. Chem. 2014, 289, 33529–33542. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Xu, X.; Li, B.; Brown, E.; Farris, A.B.; Sun, S.Y.; Yang, J.J. Prostate cancer metastatic to bone has higher expression of the calcium-sensing receptor (CaSR) than primary prostate cancer. Recept. Clin. Investig. 2014, 1. [Google Scholar] [CrossRef]
- Huang, Y.; Zhou, Y.; Wong, H.C.; Castiblanco, A.; Chen, Y.; Brown, E.M.; Yang, J.J. Calmodulin regulates Ca2+-sensing receptor-mediated Ca2+ signaling and its cell surface expression. J. Biol. Chem. 2010, 285, 35919–35931. [Google Scholar] [CrossRef] [Green Version]
- Crotti, L.; Johnson, C.N.; Graf, E.; De Ferrari, G.M.; Cuneo, B.F.; Ovadia, M.; Papagiannis, J.; Feldkamp, M.D.; Rathi, S.G.; Kunic, J.D.; et al. Calmodulin mutations associated with recurrent cardiac arrest in infants. Circulation 2013, 127, 1009–1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makita, N.; Yagihara, N.; Crotti, L.; Johnson, C.N.; Beckmann, B.M.; Roh, M.S.; Shigemizu, D.; Lichtner, P.; Ishikawa, T.; Aiba, T.; et al. Novel calmodulin mutations associated with congenital arrhythmia susceptibility. Circ. Cardiovasc. Genet. 2014, 7, 466–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boczek, N.J.; Gomez-Hurtado, N.; Ye, D.; Calvert, M.L.; Tester, D.J.; Kryshtal, D.; Hwang, H.S.; Johnson, C.N.; Chazin, W.J.; Loporcaro, C.G.; et al. Spectrum and prevalence of CALM1-, CALM2-, and CALM3-encoded calmodulin variants in long QT syndrome and functional characterization of a novel long QT syndrome-associated calmodulin missense variant, E141G. Circ. Cardiovasc. Genet. 2016, 9, 136–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez-Hurtado, N.; Boczek, N.J.; Kryshtal, D.O.; Johnson, C.N.; Sun, J.; Nitu, F.R.; Cornea, R.L.; Chazin, W.J.; Calvert, M.L.; Tester, D.J.; et al. Novel CPVT-associated calmodulin mutation in CALM3 (CALM3-A103V) activates arrhythmogenic Ca waves and sparks. Circ. Arrhythm. Electrophysiol. 2016, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Xue, S.; Zou, J.; Lopez, J.R.; Yang, J.J.; Perez, C.F. Myoplasmic resting Ca2+ regulation by ryanodine receptors is under the control of a novel Ca2+-binding region of the receptor. Biochem. J. 2014, 460, 261–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, J.; Zhou, Y.; Zou, J.; Chen, Y.; Patel, P.; Yang, J.J.; Balog, E.M. Site-specific modification of calmodulin Ca2+ affinity tunes the skeletal muscle ryanodine receptor activation profile. Biochem. J. 2010, 432, 89–99. [Google Scholar] [CrossRef]
- Korendovych, I.V.; Degrado, W.F. De novo protein design, a retrospective. Q. Rev. Biophys 2020, 53, E3. [Google Scholar] [CrossRef]
- Korendovych, I.V. Rational and semirational protein design. Methods Mol. Biol. 2018, 1685, 15–23. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, S.; Deng, X.; Jiang, J.; Kirberger, M.; Yang, J.J. Design of Calcium-Binding Proteins to Sense Calcium. Molecules 2020, 25, 2148. https://doi.org/10.3390/molecules25092148
Tang S, Deng X, Jiang J, Kirberger M, Yang JJ. Design of Calcium-Binding Proteins to Sense Calcium. Molecules. 2020; 25(9):2148. https://doi.org/10.3390/molecules25092148
Chicago/Turabian StyleTang, Shen, Xiaonan Deng, Jie Jiang, Michael Kirberger, and Jenny J. Yang. 2020. "Design of Calcium-Binding Proteins to Sense Calcium" Molecules 25, no. 9: 2148. https://doi.org/10.3390/molecules25092148
APA StyleTang, S., Deng, X., Jiang, J., Kirberger, M., & Yang, J. J. (2020). Design of Calcium-Binding Proteins to Sense Calcium. Molecules, 25(9), 2148. https://doi.org/10.3390/molecules25092148




