Larvicidal Activity of Cinnamic Acid Derivatives: Investigating Alternative Products for Aedes aegypti L. Control
Abstract
1. Introduction
2. Results and Discussion
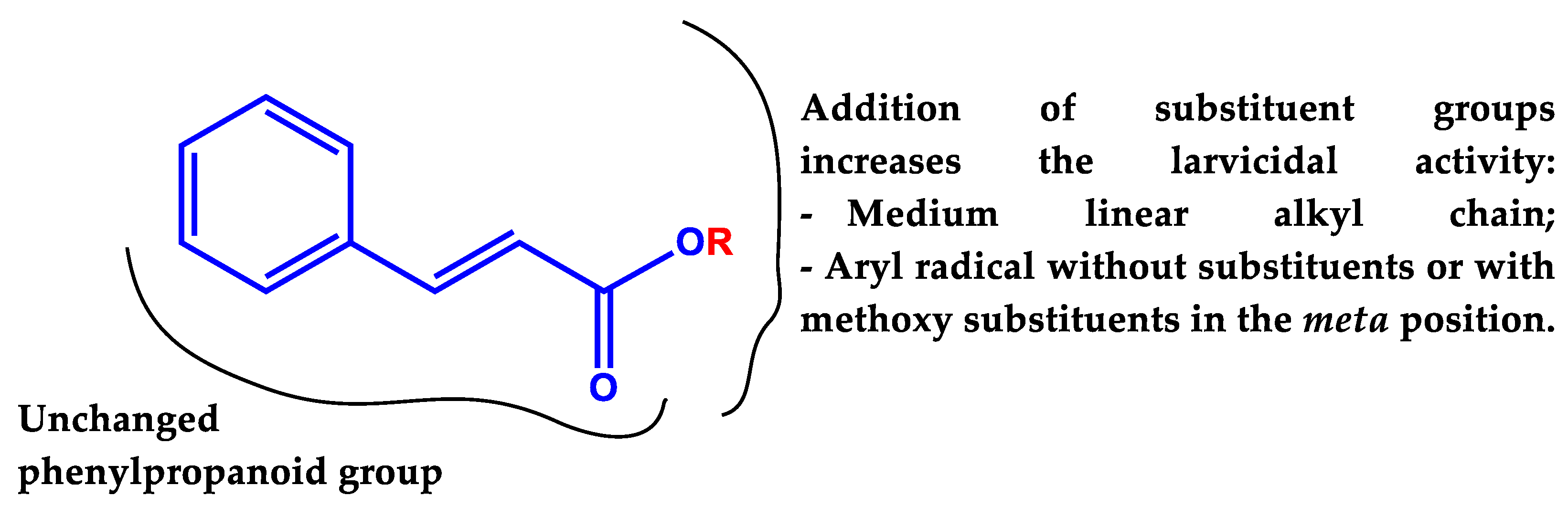
2.1. Effect of Cinnamic Acid Derivatives on Ae. aegypti Larvae
2.2. Computational Methods
3. Materials and Methods
3.1. Chemical Characterization and Reagents
3.2. Synthesis of Compounds 2–9; Fischer’s Esterification
3.3. Synthesis of Compound 10; Reaction with Halide
3.4. Synthesis of Compounds 11–18; the Mitsunobu Reaction
3.5. Chemical Characterization Compounds 2–18
3.6. Effect of Cinnamic Acid Derivatives on Ae. aegypti Larvae
3.7. Larvae Mortality Rate
3.8. Statistical Analysis
3.9. Computational Methods
3.9.1. Molecular Modeling
3.9.2. Targets Selection
3.9.3. Molecular Docking
3.9.4. Molecular Dynamics Simulations and Free Energies of Binding Estimation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Coelho, A.A.M.; De Paula, J.E.; Espíndola, L.S. Atividade larvicida de extratos vegetais sobre Aedes aegypti (L.) (Diptera: Culicidae), em condições de laboratório. BioAssay 2009, 4, 1–6. [Google Scholar] [CrossRef]
- Gibbons, R.V. Dengue conundrums. Int. J. Antimicrob. Agents 2010, 36, S36–S39. [Google Scholar] [CrossRef] [PubMed]
- Nunes, F.C.; Leite, J.A.; Oliveira, L.H.G.; Sousa, P.A.P.S.; Menezes, M.C.; Moraes, J.P.S.; Mascarenhas, S.R.; Braga, V.A. The larvicidal activity of Agave sisalana against L4 larvae of Aedes aegypti is mediated by internal necrosis and inhibition of nitric oxide production. Parasitol. Res. 2015, 114, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Simas, N.K.; Lima, E.C.; Conceição, S.R.; Kuster, R.M.; de Oliveira Filho, A.M. Produtos naturais para o controle da transmissão da dengue – atividade larvicida de Myroxylon balsamum (óleo vermelho) e de terpenóides e fenilpropanóides. Quím. Nova. 2004, 27, 46–49. [Google Scholar] [CrossRef]
- WHO. Dengue and Severe Dengue. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue (accessed on 17 September 2020).
- Yousaf, A.; Zuharah, W.F. Lethal response of the dengue vectors to the plant extracts from family Anacardiaceae. Asian Pac. J. Trop. Biomed. 2015, 5, 812–818. [Google Scholar] [CrossRef]
- Procópio, T.F.; Fernandes, K.M.; Pontual, E.V.; Ximenes, R.M.; de Oliveira, A.R.C.; Souza, C.S.; Melo, A.M.M.A.; Navarro, D.M.A.F.; Paiva, P.M.G.; Martins, G.F.; et al. Schinus terebinthifolius Leaf Extract Causes Midgut Damage, Interfering with Survival and Development of Aedes aegypti Larvae. PLoS ONE 2015, 14, 1–19. [Google Scholar] [CrossRef] [PubMed]
- da Silva-Alves, D.C.B.; dos Anjos, J.V.; Cavalcante, N.N.M.; Santos, G.K.N.; Navarro, D.M.A.F.; Srivastava, R.M. Larvicidal isoxazoles: Synthesis and their effective susceptibility towards Aedes aegypti larvae. Bioorg. Med. Chem. 2013, 21, 940–947. [Google Scholar] [CrossRef] [PubMed]
- Clifford, N.M. Chlorogenic acids and other cinnamates–nature, occurrence, dietary burden, absorption and metabolism. J. Sci. Food Agric. 2000, 80, 1033–1043. [Google Scholar] [CrossRef]
- Sova, M. Antioxidant and Antimicrobial Activities of Cinnamic Acid Derivatives. Mini. Rev. Med. Chem. 2012, 12, 749–767. [Google Scholar] [CrossRef]
- Kim, N.-J.; Byun, S.-G.; Cho, J.-E.; Chung, K.; Ahn, Y.-J. Larvicidal activity of Kaempferia galangal rhizome phenylpropanoids towards three mosquito species. Pest Manag. Sci. 2008, 64, 857–862. [Google Scholar]
- Panyakaew, J.; Sookkhee, S.; Rotarayanont, S.; Kittiwachana, S.; Wangkarn, S.; Mungkornasawakul, P. Chemical Variation and Potential of Kaempferia Oils as Larvicide against Aedes aegypti. J. Essent. Oil Bear. Plants 2017, 20, 1044–1056. [Google Scholar] [CrossRef]
- Othman, R.; Ibrahim, H.; Mohd, M.A.; Mustafa, M.R.; Awang, K. Bioassay-guided isolation of a vasorelaxant active compound from Kaempferia galanga L. Phytomedicine 2006, 13, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Baser, K.H.C.; Kurkcuoglu, M.; Demirci, B.; Erdogmus, H. Composition of the essential oil and the headspace sample of Mandragora autumnalis Bertol. fruits. J. Essent. Oil Res. 1998, 10, 632–634. [Google Scholar] [CrossRef]
- Hanus, L.O.; Rezanka, T.; Spízek, J.; Dembitsky, V.M. Substances isolated from Mandragora species. Phytochemistry 2005, 66, 2408–2417. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, P.A.; Van Genderen, M.H.P.; Thanh, L.; Khiên, P.V.; Dũng, N.X. Composition of the Essential Oil from the Aerial Parts of Piper pierrei C. DC. from Vietnam. J. Essent. Oil Res. 1997, 9, 721–724. [Google Scholar] [CrossRef]
- Seo, S.-M.; Park, H.-M.; Park, I.-K. Larvicidal Activity of Ajowan (Trachyspermum ammi) and Peru Balsam (Myroxylon pereira) Oils and Blends of Their Constituents against Mosquito, Aedes aegypti, Acute Toxicity on Water Flea, Daphnia magna, and Aqueous Residue. J. Agric. Food. Chem. 2012, 60, 5909–5914. [Google Scholar] [CrossRef] [PubMed]
- Cavalcanti, E.S.B.; Morais, S.M.; Lima, M.A.; Santana, E.W.P. Larvicidal activity of essential oils from Brazilian plants against Aedes aegypti L. Memórias Inst. Oswaldo Cruz. 2004, 99, 541–544. [Google Scholar] [CrossRef]
- Jung, E.H.; Kim, S.R.; Hwang, I.K.; Ha, T.Y. Hypoglycemic Effects of a Phenolic Acid Fraction of Rice Bran and Ferulic Acid in C57BL/KsJ-db/dbMice. J. Agric. Food. Chem. 2007, 55, 9800–9804. [Google Scholar] [CrossRef]
- Sharma, P. Cinnamic acid derivatives: A new chapter of various pharmacological activities. J. Chem. Pharm. Res. 2011, 3, 403–423. [Google Scholar]
- Auger, C.; Laurent, N.; Laurent, C.; Besançon, P.; Caporiccio, B.; Teissédre, P.L.; Rouanet, J.-M. Hydroxycinnamic acids do not prevent aortic atherosclerosis in hypercholesterolemic golden Syrian hamsters. Life Sci. 2004, 74, 2365–2377. [Google Scholar] [CrossRef]
- Korošec, B.; Sova, M.; Turk, S.; Kraševec, N.; Novak, M.; Lah, L.; Stojan, J.; Podobnik, B.; Berne, S.; Zupanec, N. Antifungal activity of cinnamic acid derivatives involves inhibition of benzoate 4-hydroxylase (CYP53). J. Appl. Microbiol. 2014, 116, 955–966. [Google Scholar] [CrossRef] [PubMed]
- Adisakwattana, S.; Moonsan, P.; Yibchok-Anun, S. Insulin-Releasing Properties of a Series of Cinnamic Acid Derivatives in Vitro and in Vivo. J. Agric. Food. Chem. 2008, 56, 7838–7844. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, J.; Mitsch, A.; Wiûner, P.; Jomaa, H.; Schlitzer, M. Structure-Activity Relationships of Novel Anti-Malarial Agents. Part 2: Cinnamic Acid Derivatives. Bioorg. Med. Chem. Lett. 2001, 11, 423–424. [Google Scholar] [CrossRef]
- Cheng, S.S.; Chua, M.T.; Chang, E.H.; Huang, C.G.; Chen, W.J.; Chang, S.T. Variations in insecticidal activity and chemical compositions of leaf essential oils from Cryptomeria japonica at different ages. Bioresour. Technol. 2009, 100, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, G.M.; Annies, V.; de Oliveira, C.F.; Lara, R.A.; Gabriel, M.M.; Betim, F.C.M.; Nadal, J.M.; Farago, P.V.; Dias, J.F.G.; Miguel, O.G.; et al. Evaluation of Larvicidal Activity and Ecotoxicity of Linalool, Methyl Cinnamate and Methyl cinnamate/linalool in Combination Against Aedes aegypti. Ecotoxicol. Environ. Saf. 2017, 139, 238–244. [Google Scholar] [CrossRef]
- Dias, C.N.; Moraes, D.F.C. Essential oils and their compounds as Aedes aegypti L. (Diptera: Culicidae) larvicides: Review. Parasitol. Res. 2014, 113, 565–592. [Google Scholar] [CrossRef]
- Cheng, S.-S.; Liu, J.-Y.; Tsai, K.-H.; Chen, W.-J.; Chang, S.-T. Chemical Composition and Mosquito Larvicidal Activity of Essential Oils from Leaves of Different Cinnamomum osmophloeum Provenances. J. Agric. Food. Chem. 2004, 52, 4395–4400. [Google Scholar] [CrossRef]
- Bateman, A. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar]
- Giraldo-Calderon, G.I.; Emrich, S.J.; MacCallum, R.M.; Maslen, G.; Dialynas, E.; Topalis, P.; Ho, N.; Gesing, S.; Madey, G.; Collins, F.H.; et al. VectorBase: An updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015, 43, D707–D713. [Google Scholar] [CrossRef]
- Nobsathian, S.; Bullangpoti, V.; Kumrungsee, N.; Wongsa, N.; Ruttanakum, D. Larvicidal efect of compounds isolated from Maerua siamensis (Capparidaceae) against Aedes aegypti (Diptera: Culicidae) larvae. Chem. Biol. Technol. Agric. 2018, 5, 1–7. [Google Scholar] [CrossRef]
- Engdahl, C.; Knutsson, S.; Ekström, F.; Linusson, A. Discovery of Selective Inhibitors Targeting Acetylcholinesterase 1 from Disease-Transmitting Mosquitoes. J. Med. Chem. 2016, 59, 9409–9421. [Google Scholar] [CrossRef]
- Knutsson, S.; Kindahl, T.; Engdahl, C.; Nikjoo, D.; Forsgren, N.; Kitur, S.; Ekström, F.; Kamau, L.; Linusson, A. N-Aryl-N’-ethyleneaminothioureas effectively inhibit acetylcholinesterase 1 from disease-transmitting mosquitoes. Eur. J. Med. Chem. 2017, 134, 415–442. [Google Scholar] [CrossRef]
- Otero, A.L.C.; Méndez, L.Y.V.; Duque, L.J.E.; Kouznetsov, V.V. Design, synthesis, acetylcholinesterase inhibition and larvicidal activity of girgensohnine analogs on Aedes aegypti, vector of dengue fever. Eur. J. Med. Chem. 2014, 78, 415–442. [Google Scholar]
- Lopes, S.P.; Castillo, Y.P.; Monteiro, M.L.; de Menezes, R.R.P.P.B.; Almeida, R.N.; Martins, A.M.C.; de Sousa, D.P. Trypanocidal Mechanism of Action and in silico Studies of p-Coumaric Acid Derivatives. Int. J. Mol. Sci. 2019, 20, 5916. [Google Scholar] [CrossRef]
- Turkez, H.; da Nóbrega, F.R.; Ozdemir, O.; Bezerra Filho, C.S.M.; de Almeida, R.N.; Tejera, E.; Castillo, Y.P.; de Sousa, D.P. NFBTA: A Potent Cytotoxic Agent against Glioblastoma. Molecules 2019, 24, 2411. [Google Scholar] [CrossRef]
- Francis, S.A.M.; Taylor-Wells, J.; Gross, A.D.; Bloomquist, J.R. Toxicity and Physiological Actions of Carbonic Anhydrase Inhibitors to Aedes aegypti and Drosophila melanogaster. Insects 2017, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.P.; Ekeris, L.V.; Linser, P.J. Characterization of Carbonic Anhydrase 9 in the Alimentary Canal of Aedes aegypti and Its Relationship to Homologous Mosquito Carbonic Anhydrases. Int. J. Environ. Res. Public Health 2017, 14, 213. [Google Scholar] [CrossRef] [PubMed]
- Kalsi, M.; Gillen, C.; Piermarini, P.M. Heterologous Expression of Aedes aegypti Cation Chloride Cotransporter 2 (aeCCC2) in Xenopus laevis Oocytes Induces an Enigmatic Na+/Li+ Conductance. Insects 2019, 10, 71. [Google Scholar] [CrossRef] [PubMed]
- Piermarini, P.M.; Akuma, D.C.; Crow, J.C.; Jamil, T.L.; Kerkhoff, W.G.; Viel, K.C.M.F.; Gillen, C.M. Differential expression of putative sodium-dependent cation-chloride cotransporters in Aedes aegypti. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2017, 214, 40–49. [Google Scholar] [CrossRef]
- de Farias, M.O.; Lima, T.C.; Pérez, A.L.A.L.; Silva, R.H.N.; Oliveira, A.J.M.S.; Lima, E.O.; de Sousa, D.P. Antifungal activity of ester derivatives from caffeic acid against Candida species. Int. J. Pharm. Pharm. Res. 2016, 7, 151–159. [Google Scholar]
- Araújo, M.O.; Freire Pessoa, H.L.; Lira, A.B.; Castillo, Y.P.; de Sousa, D.P. Synthesis, Antibacterial Evaluation, and QSAR of Caffeic Acid Derivatives. J. Chem. 2019, 2019, 1–9. [Google Scholar] [CrossRef]
- Handique, J.G.; Mahanta, D.; Devi, A.; Boruah, M.P. Synthesis and electrochemical behavior of some dendritic polyphenols as antioxidants. Lett. Org. Chem. 2013, 10, 53–59. [Google Scholar]
- Lutjen, A.B.; Quirk, M.A.; Barbera, A.M.; Kolonko, E.M. Synthesis of (E)-cinnamyl ester derivatives via a greener Steglich esterification. Bioorg. Med. Chem. 2018, 26, 5291–5298. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, B.; Belsare, D.; Pharande, D.; Mourya, V.; Dhake, A. Esters, amides and substituted derivatives of cinnamic acid: Synthesis, antimicrobial activity and QSAR investigations. Eur. J. Med. Chem. 2004, 39, 827–834. [Google Scholar] [CrossRef]
- Iranpoor, N.; Firouzabadi, H.; Riazi, A.; Pedrood, K. Regioselective hydrocarbonylation of phenylacetylene to α,β-unsaturatedesters and thioesters with Fe(CO)5 and Mo(CO)6. J. Organomet. Chem. 2016, 822, 67–73. [Google Scholar] [CrossRef]
- Choi, I.-H.; Park, J.-Y.; Shin, S.-C.; Park, I.-K. Nematicidal activity of medicinal plant extracts and two cinnamates isolated from Kaempferia galanga L. (Proh Hom) against the pine wood nematode, Bursaphelenchus xylophilus. J. Nematol. 2006, 8, 359–365. [Google Scholar] [CrossRef]
- Jakovetić, S.M.; Jugović, B.Z.; Gvozdenović, M.M.; Bezbradica, D.I.; Antov, M.G.; Mijin, D.Z.; Knežević-Jugović, Z.D. Synthesis of Aliphatic Esters of Cinnamic Acid as Potential Lipophilic Antioxidants Catalyzed by Lipase B from Candida Antarctica. Appl. Biochem. Biotechnol. 2013, 170, 1560–1573. [Google Scholar] [CrossRef] [PubMed]
- Fujita, H.; Kakuyama, S.; Fukuyoshi, S.; Hayakawa, N.; Oda, A.; Kunishima, M. Triazine-Based Cationic Leaving Group: Synergistic Driving Forces for Rapid Formation of Carbocation Species. J. Org. Chem. 2018, 83, 4568–4580. [Google Scholar] [CrossRef] [PubMed]
- Sova, M.; Perdih, A.; Kotnik, M.; Kristan, K.; Rizner, T.L.; Solmajer, T.; Gobec, S. Flavonoids and cinnamic acid esters as inhibitors of fungal 17β-hydroxysteroid dehydrogenase: A synthesis, QSAR and modelling study. Bioorg. Med. Chem. 2006, 14, 7404–7418. [Google Scholar] [CrossRef]
- Guntreddi, T.; Vanjari, R.; Singh, K.N. Direct conversion of methylarenes into dithiocarbamates, thioamides and benzyl esters. Tetrahedron 2014, 70, 3887–3892. [Google Scholar] [CrossRef]
- WHO. Guidelines for Laboratory and Field Testing of Mosquito Larvicides; WHO Technical Reports Series; World Health Organization: Geneva, Switzerland, 2005. [Google Scholar]
- WHO. Who-Guidelines for Efficacy Testing of Spatial Repellents, WHO Pesticide Evaluation Scheme; World Health Organization: Geneva, Switzerland, 2013; Volume 58. [Google Scholar]
- Rodriguez, S.D.; Drake, L.L.; Price, D.P.; Hammond, J.I.; Hansen, I.A. The Efficacy of Some Commercially Available Insect Repellents for Aedes aegypti (Diptera: Culicidae) and Aedes albopictus (Diptera: Culicidae). J. Insect Sci. 2015, 15, 1–5. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Keiser, M.J.; Roth, B.L.; Armbruster, B.N.; Ernsberger, P.; Irwin, J.J.; Shoichet, B.K. Relating protein pharmacology by ligand chemistry. Nat. Biotechnol. 2007, 25, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Hawkins, P.C.D.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. OMEGA. Santa Fe, NM: OpenEye Scientific Software. 2020. Available online: http://www.eyesopen.com. (accessed on 10 October 2020).
- OpenEye Scientific Software. QUACPAC. Santa Fe, NM: OpenEye Scientific Software. 2020. Available online: http://www.eyesopen.com (accessed on 23 October 2020).
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef]
- Case, D.A.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E.; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Ghoreishi, D.; Gilson, M.K.; et al. AMBER 2018; University of California: San Francisco, CA, USA, 2018. [Google Scholar]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory. Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef] [PubMed]


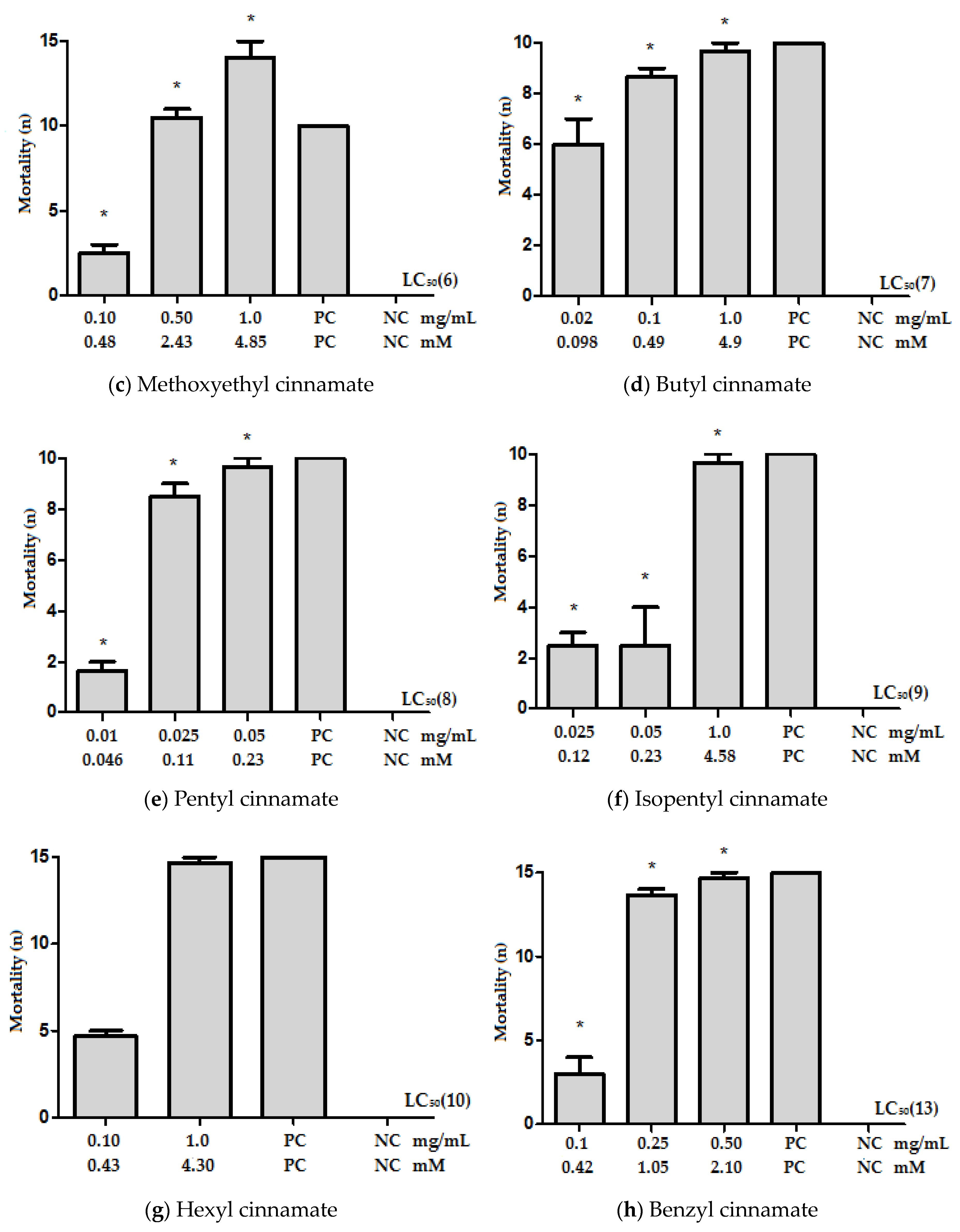
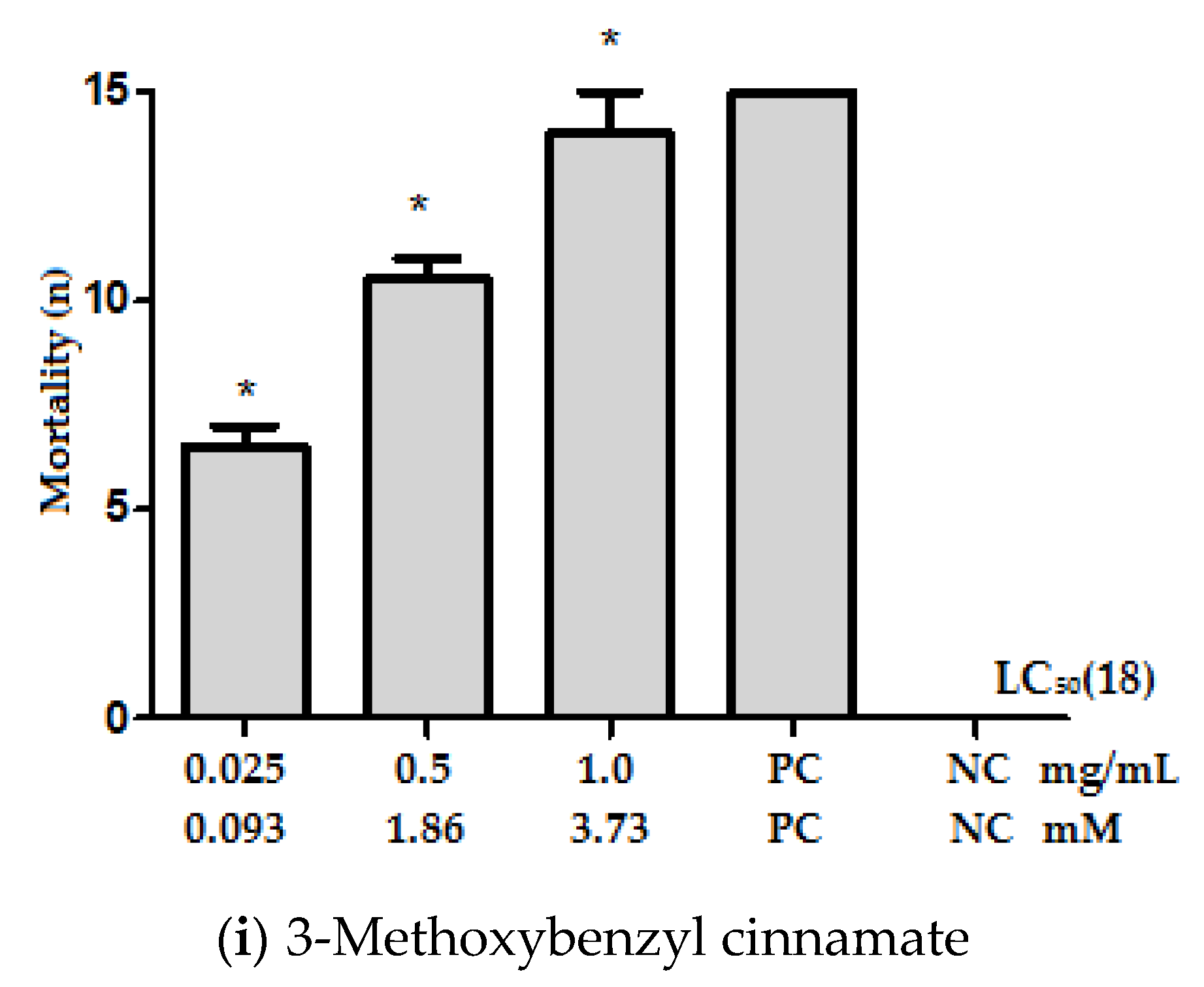


| Compounds | LC50 | |
|---|---|---|
| mg/mL | mM | |
| Cinnamic acid (1) | 0.69 (0.47–0.91) | 4.66 |
| Methyl cinnamate (2) | 0.26 (0.13–0.40) | 1.60 |
| Ethyl cinnamate (3) | 0.13 (0.0–0.32) | 0.74 |
| Propyl cinnamate (4) | 0.53 (0.50–0.56) | 2.79 |
| Isopropyl cinnamate (5) | 0.098 (0.0–0.21) | 0.52 |
| Methoxyethyl cinnamate (6) | 0.12 (0.0–0.33) | 0.58 |
| Butyl cinnamate (7) | 0.042 (0.019–0.091) | 0.21 |
| Pentyl cinnamate (8) | 0.036 (0.0–0.078) | 0.17 |
| Isopentyl cinnamate (9) | 0.18 (0.002–0.36) | 0.83 |
| Hexyl cinnamate (10) | 0.23 (0.0–0.47) | 0.99 |
| Dodecyl cinnamate (11) | 1.12 (1.07–1.17) | 3.54 |
| 4-Chlorobenzyl cinnamate (12) | 0.82 (0.62–1.02) | 3.01 |
| Benzyl cinnamate (13) | 0.13 (0.09–0.17) | 0.55 |
| 4-Methylbenzyl cinnamate (14) | 3.04 (0.0–8.7) | 12.06 |
| 4-Isopropylbenzyl cinnamate (15) | 3.14 (0.76–5.52) | 11.21 |
| 4-Nitrobenzyl cinnamate (16) | 3.44 (2.93–3.95) | 12.15 |
| 4-Methoxybenzyl cinnamate (17) | 11.42 (0.0–24.43) | 42.60 |
| 3-Methoxybenzyl cinnamate (18) | 0.22 (0.0–0.52) | 0.82 |
| Ae. aegypti Target a | Description | VetorBase Gene b | ID |
|---|---|---|---|
| Q174R0 | GTPase Rho | AAEL006786 | RHO |
| Q16YG0 | Cdc42 homolog | AAEL008543 | Cdc42 |
| Q1DH30 | Rac GTPase | AAEL015271 | RAC |
| Q16V02 | Ras-related protein Rac1 | AAEL009733 | RAC1 |
| A0A1S4G1K4 | Lactoylglutathione lyase | AAEL014393 | LGUL |
| Q16K18 | GTPase Rho | AAEL013139 | RHO1 |
| Q17BL7 | Carbonic anhydrase | AAEL004930 | CA |
| Q17DM5 | Aldo-keto reductase | AAEL004088 | AKR1 |
| Q17DM8 | Aldo-keto reductase | AAEL004102 | AKR2 |
| Q17DM9 | Aldo-keto reductase | AAEL004118 | AKR3 |
| Q17DN2 | Aldo-keto reductase | AAEL004086 | AKR4 |
| Q17DN0 | Aldo-keto reductase | AAEL004095 | AKR5 |
| A0A1S4F6Y1 | Aldo-keto reductase | AAEL004096 | AKR6 |
| Q16QZ8 | Histone deacetylase | AAEL011117 | HDAC1 |
| Q17CF0 | Histone deacetylase | AAEL004586 | HDAC2 |
| Q17GT4 | Thioredoxin reductase | AAEL002886 | TRXR |
| A0A1S4F1W5 | Glutamate receptor, ionotropic kainate 1, 2, 3 (glur5, glur6, glur7) | AAEL002506 | GLUR |
| A0A1S4FNU9 | Sodium-coupled cation-chloride cotransporter | AAEL009886 | CCC3 |
| A0A1S4FNN9 | Sodium-coupled cation-chloride cotransporter | AAEL009888 | CCC2 |
| H2L215 | SLC12-like K,Cl cotransporter | AAEL019507 | KCC1A |
| Q5PY77 | Glutathione transferase | AAEL007951 | GSTE2 |
| Q5PY78 | Glutathione transferase | AAEL007962 | GSTE4 |
| Q17MB0 | Glutathione transferase | AAEL026530 | GSTD2 |
| Q5PY76 | Glutathione transferase | AAEL009017 | GSTT1 |
| Q95W09 | Glutathione transferase | AAEL010500 | GSTX2 |
| Q5PY75 | Glutathione transferase | AAEL009016 | GSTT2 |
| Q6A2E2 | Acetylcholinesterase | AAEL000511 | ACE |
| Target | Conformer | CHEMPLP | GoldScore | ChemScore | ASP | Consensus Z-Score | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Score | Z-Score | Score | Z-Score | Score | Z-Score | Score | Z-Score | |||
| RHO | 1 | 58.90 | 1.77 | 33.80 | 1.49 | 20.58 | 1.86 | 21.71 | 2.19 | 1.83 |
| 2 | 60.45 | 2.22 | 29.36 | 0.35 | 20.12 | 1.60 | 19.45 | 1.27 | 1.36 | |
| 3 | 58.63 | 1.69 | 33.92 | 1.52 | 17.82 | 0.32 | 18.23 | 0.78 | 1.08 | |
| Cdc42 | 1 | 55.95 | 1.12 | 30.55 | 0.53 | 20.55 | 2.09 | 25.03 | 1.82 | 1.39 |
| 2 | 57.58 | 1.66 | 30.47 | 0.51 | 18.40 | 1.00 | 23.27 | 1.26 | 1.11 | |
| RAC | 1 | 64.39 | 1.82 | 39.30 | 1.85 | 20.91 | 1.78 | 22.54 | 1.55 | 1.75 |
| 2 | 60.10 | 0.78 | 35.55 | 0.74 | 20.07 | 1.42 | 22.60 | 1.57 | 1.13 | |
| RAC1 | 1 | 64.31 | 1.35 | 38.03 | 1.20 | 24.44 | 2.63 | 27.91 | 2.80 | 1.99 |
| 2 | 64.50 | 1.40 | 39.60 | 1.55 | 21.82 | 1.31 | 23.21 | 0.99 | 1.31 | |
| 3 | 62.32 | 0.83 | 40.51 | 1.76 | 21.89 | 1.34 | 23.61 | 1.14 | 1.27 | |
| LGUL | 1 | 69.59 | 3.01 | 28.25 | 0.49 | 33.02 | 2.77 | 30.47 | 1.75 | 2.01 |
| RHO1 | 1 | 60.44 | 1.90 | 38.37 | 2.35 | 20.55 | 1.36 | 19.10 | 1.17 | 1.70 |
| 2 | 56.37 | 1.23 | 35.55 | 1.63 | 18.01 | 0.76 | 20.78 | 1.49 | 1.28 | |
| 3 | 57.11 | 1.35 | 31.61 | 0.61 | 22.01 | 1.71 | 19.50 | 1.25 | 1.23 | |
| CA | 1 | 57.40 | 1.95 | 43.35 | 3.48 | 20.03 | 1.30 | 18.68 | −0.73 | 1.50 |
| 2 | 53.26 | 0.90 | 29.63 | 0.28 | 21.65 | 1.93 | 29.44 | 2.33 | 1.36 | |
| 3 | 58.64 | 2.27 | 31.04 | 0.61 | 21.26 | 1.77 | 21.29 | 0.02 | 1.17 | |
| AKR1 | 1 | 59.43 | 1.59 | 35.51 | 0.92 | 21.82 | 1.45 | 33.55 | 2.17 | 1.53 |
| 2 | 58.53 | 1.32 | 37.08 | 1.32 | 20.79 | 0.89 | 29.64 | 1.03 | 1.14 | |
| AKR2 | 1 | 64.38 | 1.71 | 36.58 | 0.49 | 21.94 | 1.41 | 29.62 | 1.27 | 1.22 |
| 2 | 60.40 | 0.68 | 39.87 | 1.27 | 21.92 | 1.41 | 28.09 | 0.75 | 1.03 | |
| AKR3 | 1 | 55.03 | 1.49 | 10.39 | 0.53 | 23.22 | 1.42 | 33.37 | 2.12 | 1.39 |
| 2 | 52.82 | 1.05 | 11.38 | 0.59 | 22.37 | 1.12 | 32.26 | 1.75 | 1.13 | |
| AKR4 | 1 | 62.93 | 2.13 | 2.64 | 0.36 | 25.43 | 1.95 | 25.47 | 0.22 | 1.17 |
| AKR5 | 1 | 54.97 | 2.35 | 30.42 | 1.12 | 22.85 | 2.23 | 23.56 | 1.80 | 1.88 |
| 2 | 46.66 | 0.92 | 31.07 | 1.28 | 19.92 | 1.37 | 25.33 | 2.34 | 1.48 | |
| 3 | 51.13 | 1.69 | 28.61 | 0.68 | 22.56 | 2.14 | 20.01 | 0.72 | 1.31 | |
| AKR6 | 1 | 52.76 | 1.67 | 18.87 | 0.63 | 18.64 | 0.69 | 25.90 | 2.32 | 1.33 |
| HDAC1 | 1 | 54.25 | 1.57 | 33.14 | 1.85 | 18.90 | 1.35 | 31.50 | 1.91 | 1.67 |
| 2 | 57.87 | 2.60 | 30.91 | 1.23 | 17.18 | 0.36 | 32.16 | 2.09 | 1.57 | |
| 3 | 51.85 | 0.88 | 30.17 | 1.02 | 19.73 | 1.83 | 28.16 | 0.98 | 1.18 | |
| HDAC2 | 1 | 67.83 | 3.02 | 33.28 | 1.53 | 21.64 | 1.97 | 34.36 | 2.65 | 2.29 |
| 2 | 66.26 | 2.59 | 40.21 | 3.52 | 20.00 | 0.67 | 33.13 | 2.23 | 2.25 | |
| TRXR | 1 | 58.20 | 1.62 | 33.85 | 1.91 | 19.07 | 0.85 | 26.92 | 1.26 | 1.41 |
| 2 | 57.65 | 1.53 | 27.63 | 0.35 | 20.51 | 1.25 | 26.64 | 1.20 | 1.08 | |
| 3 | 50.72 | 0.32 | 33.48 | 1.82 | 18.79 | 0.77 | 26.62 | 1.20 | 1.03 | |
| GLUR | 1 | 43.93 | 1.45 | 38.20 | 1.17 | 9.93 | 0.41 | 20.16 | 1.99 | 1.25 |
| 2 | 44.38 | 1.55 | 35.82 | 0.78 | 12.64 | 1.01 | 16.41 | 1.35 | 1.17 | |
| 3 | 42.44 | 1.09 | 32.79 | 0.28 | 14.03 | 1.31 | 18.62 | 1.72 | 1.10 | |
| CCC3 | 1 | 42.91 | 1.25 | 31.55 | 0.86 | 12.49 | 0.73 | 18.77 | 1.51 | 1.09 |
| CCC2 | 1 | 51.24 | 2.05 | 42.66 | 2.29 | 15.44 | 1.10 | 19.54 | 1.40 | 1.71 |
| 2 | 48.60 | 1.56 | 34.18 | 0.85 | 16.51 | 1.51 | 18.79 | 1.22 | 1.29 | |
| KCC1A | 1 | 59.45 | 2.89 | 37.85 | 2.71 | 15.65 | −1.46 | 21.44 | 0.32 | 1.11 |
| 2 | 53.34 | 1.03 | 28.62 | 0.71 | 21.27 | 0.82 | 25.40 | 1.86 | 1.11 | |
| GSTE2 | 1 | 63.42 | 0.91 | 36.27 | 1.53 | 27.20 | 1.88 | 24.47 | 0.60 | 1.23 |
| GSTE4 | 1 | 64.51 | 1.68 | 30.14 | 0.06 | 25.20 | 1.79 | 24.30 | 1.72 | 1.31 |
| 2 | 63.20 | 1.31 | 34.97 | 1.26 | 24.36 | 1.43 | 22.21 | 0.92 | 1.23 | |
| 3 | 64.03 | 1.55 | 33.48 | 0.89 | 22.31 | 0.53 | 22.96 | 1.21 | 1.04 | |
| GSTD2 | 1 | 50.10 | 1.08 | 31.62 | 1.81 | 19.91 | 1.64 | 25.27 | 2.17 | 1.68 |
| 2 | 52.29 | 2.04 | 25.51 | 0.02 | 20.10 | 1.75 | 25.09 | 2.10 | 1.48 | |
| 3 | 49.98 | 1.03 | 32.08 | 1.95 | 20.32 | 1.87 | 22.45 | 1.05 | 1.47 | |
| 4 | 54.75 | 3.11 | 29.78 | 1.27 | 18.43 | 0.84 | 20.16 | 0.13 | 1.34 | |
| GSTT1 | 1 | 46.82 | 1.27 | 37.47 | 2.55 | 14.90 | 0.97 | 13.96 | 0.24 | 1.26 |
| 2 | 45.26 | 0.94 | 31.40 | 1.22 | 14.95 | 0.99 | 17.65 | 1.39 | 1.13 | |
| GSTX2 | 1 | 64.43 | 2.54 | 35.83 | 0.67 | 22.83 | 0.35 | 27.07 | 1.11 | 1.17 |
| GSTT2 | 1 | 55.10 | 1.41 | 35.79 | 1.03 | 19.27 | 1.42 | 22.70 | 1.26 | 1.28 |
| 2 | 55.08 | 1.40 | 36.45 | 1.17 | 18.77 | 1.13 | 22.13 | 1.01 | 1.18 | |
| 3 | 53.66 | 0.92 | 35.78 | 1.02 | 18.55 | 1.00 | 22.90 | 1.34 | 1.07 | |
| ACE | 1 | 66.49 | 2.03 | 38.42 | 1.51 | 27.23 | 1.26 | 40.77 | 1.71 | 1.63 |
| 2 | 63.40 | 1.07 | 35.51 | 0.71 | 26.21 | 0.69 | 40.82 | 1.72 | 1.05 | |
| Target | Conformer | MM-PBSA Component | ΔG TOTAL | ||||||
|---|---|---|---|---|---|---|---|---|---|
| VDWAALS | EEL | EPB | ENPOLAR | EDISPER | DELTA G Gas | DELTA G Solv | |||
| RHO | 1 | −29.83 | −10.07 | 23.41 | −23.48 | 36.28 | −39.90 | 36.20 | −3.70 |
| 2 | −25.21 | −4.55 | 18.20 | −20.40 | 31.68 | −29.76 | 29.47 | −0.29 | |
| 3 | −18.54 | −4.42 | 14.80 | −15.28 | 25.09 | −22.97 | 24.61 | 1.64 | |
| Cdc42 | 1 | −21.78 | −3.06 | 14.44 | −18.36 | 28.95 | −24.84 | 25.03 | 0.19 |
| 2 | −26.98 | −7.22 | 21.43 | −21.01 | 35.15 | −34.20 | 35.57 | 1.37 | |
| RAC | 1 | −28.43 | −10.29 | 25.03 | −23.27 | 35.58 | −38.72 | 37.34 | −1.38 |
| 2 | −31.63 | −2.24 | 20.23 | −24.42 | 37.46 | −33.88 | 33.27 | −0.61 | |
| RAC1 | 1 | −28.67 | −10.89 | 28.85 | −22.23 | 35.55 | −39.55 | 42.17 | 2.62 |
| 2 | −25.52 | −5.29 | 20.96 | −20.64 | 32.56 | −30.81 | 32.88 | 2.07 | |
| 3 | −30.26 | −10.77 | 26.13 | −24.38 | 37.11 | −41.03 | 38.85 | −2.18 | |
| LGUL | 1 | −24.40 | −25.51 | 35.21 | −24.56 | 39.18 | −49.90 | 49.83 | −0.07 |
| RHO1 | 1 | −25.42 | −6.01 | 19.92 | −20.62 | 32.41 | −31.43 | 31.71 | 0.28 |
| 2 | −23.94 | −4.63 | 17.11 | −19.43 | 31.08 | −28.57 | 28.76 | 0.19 | |
| 3 | −26.08 | −5.48 | 18.58 | −20.67 | 32.40 | −31.56 | 30.31 | −1.25 | |
| CA | 1 | −31.39 | −37.24 | 35.91 | −28.63 | 43.79 | −68.63 | 51.06 | −17.57 |
| 2 | −35.19 | −10.00 | 26.43 | −27.00 | 41.71 | −45.19 | 41.14 | −4.05 | |
| 3 | −29.70 | −38.59 | 39.81 | −27.36 | 43.77 | −68.29 | 56.22 | −12.07 | |
| AKR1 | 1 | −32.60 | −7.54 | 22.94 | −25.50 | 40.65 | −40.13 | 38.09 | −2.04 |
| 2 | −34.44 | −11.13 | 27.87 | −26.01 | 41.34 | −45.57 | 43.21 | −2.36 | |
| AKR2 | 1 | −25.62 | −4.71 | 20.90 | −20.50 | 35.55 | −30.33 | 35.95 | 5.62 |
| 2 | −27.29 | −3.71 | 16.04 | −21.99 | 36.13 | −30.99 | 30.17 | −0.82 | |
| AKR3 | 1 | −26.18 | −5.66 | 18.09 | −22.71 | 36.63 | −31.84 | 32.02 | 0.17 |
| 2 | −32.10 | −9.72 | 21.75 | −26.18 | 40.80 | −41.82 | 36.37 | −5.45 | |
| AKR4 | 1 | −35.96 | −8.29 | 24.18 | −28.56 | 45.11 | −44.25 | 40.74 | −3.51 |
| AKR5 | 1 | −28.66 | −3.77 | 17.40 | −23.05 | 37.38 | −32.44 | 31.74 | −0.70 |
| 2 | −28.63 | −6.59 | 22.85 | −22.50 | 38.13 | −35.22 | 38.48 | 3.26 | |
| 3 | −26.62 | −5.04 | 17.17 | −21.58 | 34.92 | −31.66 | 30.51 | −1.15 | |
| AKR6 | 1 | −31.54 | −8.85 | 19.78 | −26.51 | 41.95 | −40.38 | 35.21 | −5.17 |
| HDAC1 | 1 | −25.06 | −2.02 | 16.86 | −19.07 | 31.83 | −27.08 | 29.63 | 2.56 |
| 2 | −32.98 | −8.30 | 31.81 | −24.47 | 40.25 | −41.28 | 47.59 | 6.31 | |
| 3 | −32.33 | −12.10 | 26.99 | −23.74 | 38.74 | −44.43 | 41.98 | −2.45 | |
| HDAC2 | 1 | −28.79 | −47.26 | 47.24 | −26.80 | 43.29 | −76.05 | 63.73 | −12.32 |
| 2 | −33.84 | −4.59 | 23.46 | −24.07 | 39.96 | −38.43 | 39.35 | 0.92 | |
| TRXR | 1 | −28.27 | −3.60 | 18.81 | −22.35 | 36.89 | −31.87 | 33.35 | 1.47 |
| 2 | −26.53 | −6.41 | 20.67 | −20.50 | 34.85 | −32.94 | 35.02 | 2.07 | |
| 3 | −23.49 | −4.74 | 21.24 | −19.13 | 32.89 | −28.24 | 35.00 | 6.77 | |
| GLUR | 1 | −30.46 | −3.63 | 22.92 | −24.00 | 38.87 | −34.09 | 37.79 | 3.70 |
| 2 | −34.17 | −7.47 | 25.09 | −26.45 | 43.36 | −41.63 | 42.00 | 0.37 | |
| 3 | −37.26 | −9.08 | 35.26 | −27.81 | 44.66 | −46.34 | 52.10 | 5.76 | |
| CCC3 | 1 | −41.84 | −8.25 | 10.76 | −28.87 | 47.67 | −50.09 | 29.56 | −20.53 |
| CCC2 | 1 | −9.06 | −4.09 | 75.89 | −30.93 | 47.72 | −13.15 | 92.68 | 79.53 |
| 2 | −44.26 | 0.76 | 10.52 | −29.36 | 46.69 | −43.50 | 27.85 | −15.64 | |
| KCC1A | 1 | −38.70 | 38.29 | 10.86 | −27.73 | 46.16 | −0.41 | 29.29 | 28.87 |
| 2 | −38.64 | 3.52 | 10.86 | −27.86 | 46.84 | −35.12 | 29.84 | −5.28 | |
| GSTE2 | 1 | −24.34 | −1.90 | 13.17 | −20.38 | 33.97 | −26.24 | 26.76 | 0.52 |
| GSTE4 | 1 | −30.05 | −6.13 | 20.59 | −25.19 | 40.32 | −36.18 | 35.73 | −0.45 |
| 2 | −31.98 | −2.82 | 15.71 | −25.69 | 41.17 | −34.80 | 31.19 | −3.61 | |
| 3 | −27.34 | −0.81 | 13.45 | −22.34 | 36.93 | −28.15 | 28.05 | −0.10 | |
| GSTD2 | 1 | −20.18 | −2.36 | 11.48 | −16.94 | 27.99 | −22.55 | 22.52 | −0.02 |
| 2 | −17.26 | −1.94 | 9.18 | −14.83 | 24.47 | −19.20 | 18.82 | −0.38 | |
| 3 | −19.71 | −1.55 | 8.90 | −16.27 | 26.90 | −21.26 | 19.54 | −1.72 | |
| 4 | −17.93 | −0.89 | 9.04 | −14.96 | 24.69 | −18.82 | 18.76 | −0.06 | |
| GSTT1 | 1 | −19.24 | −2.73 | 10.98 | −16.64 | 27.32 | −21.97 | 21.66 | −0.31 |
| 2 | −25.45 | −4.02 | 14.12 | −21.37 | 34.20 | −29.46 | 26.95 | −2.51 | |
| GSTX2 | 1 | −29.27 | −3.04 | 15.38 | −24.22 | 37.99 | −32.31 | 29.15 | −3.16 |
| GSTT2 | 1 | −29.29 | −11.48 | 23.41 | −24.17 | 36.64 | −40.77 | 35.88 | −4.89 |
| 2 | −27.99 | −7.79 | 20.40 | −23.26 | 36.31 | −35.78 | 33.45 | −2.33 | |
| 3 | −30.40 | −8.32 | 23.34 | −25.04 | 39.16 | −38.72 | 37.46 | −1.26 | |
| ACE | 1 | −37.59 | −2.92 | 55.56 | −27.91 | 45.47 | −40.51 | 73.12 | 32.61 |
| 2 | −36.45 | −3.73 | 29.43 | −27.15 | 45.03 | −40.17 | 47.30 | 7.13 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Araújo, M.O.; Pérez-Castillo, Y.; Oliveira, L.H.G.; Nunes, F.C.; Sousa, D.P.d. Larvicidal Activity of Cinnamic Acid Derivatives: Investigating Alternative Products for Aedes aegypti L. Control. Molecules 2021, 26, 61. https://doi.org/10.3390/molecules26010061
Araújo MO, Pérez-Castillo Y, Oliveira LHG, Nunes FC, Sousa DPd. Larvicidal Activity of Cinnamic Acid Derivatives: Investigating Alternative Products for Aedes aegypti L. Control. Molecules. 2021; 26(1):61. https://doi.org/10.3390/molecules26010061
Chicago/Turabian StyleAraújo, Marianna O., Yunierkis Pérez-Castillo, Louise H. G. Oliveira, Fabíola C. Nunes, and Damião P. de Sousa. 2021. "Larvicidal Activity of Cinnamic Acid Derivatives: Investigating Alternative Products for Aedes aegypti L. Control" Molecules 26, no. 1: 61. https://doi.org/10.3390/molecules26010061
APA StyleAraújo, M. O., Pérez-Castillo, Y., Oliveira, L. H. G., Nunes, F. C., & Sousa, D. P. d. (2021). Larvicidal Activity of Cinnamic Acid Derivatives: Investigating Alternative Products for Aedes aegypti L. Control. Molecules, 26(1), 61. https://doi.org/10.3390/molecules26010061








