NMR Profiling of Ononis diffusa Identifies Cytotoxic Compounds against Cetuximab-Resistant Colon Cancer Cell Lines
Abstract
:1. Introduction
2. Results
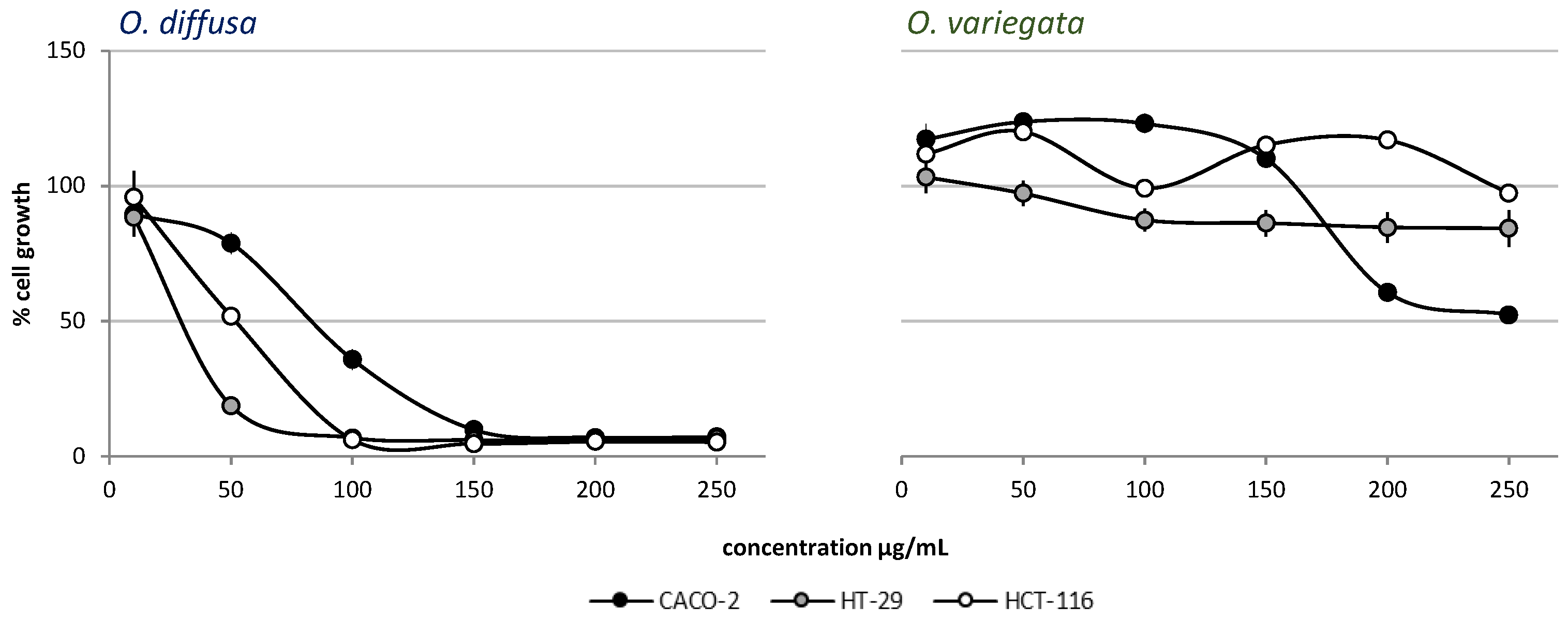
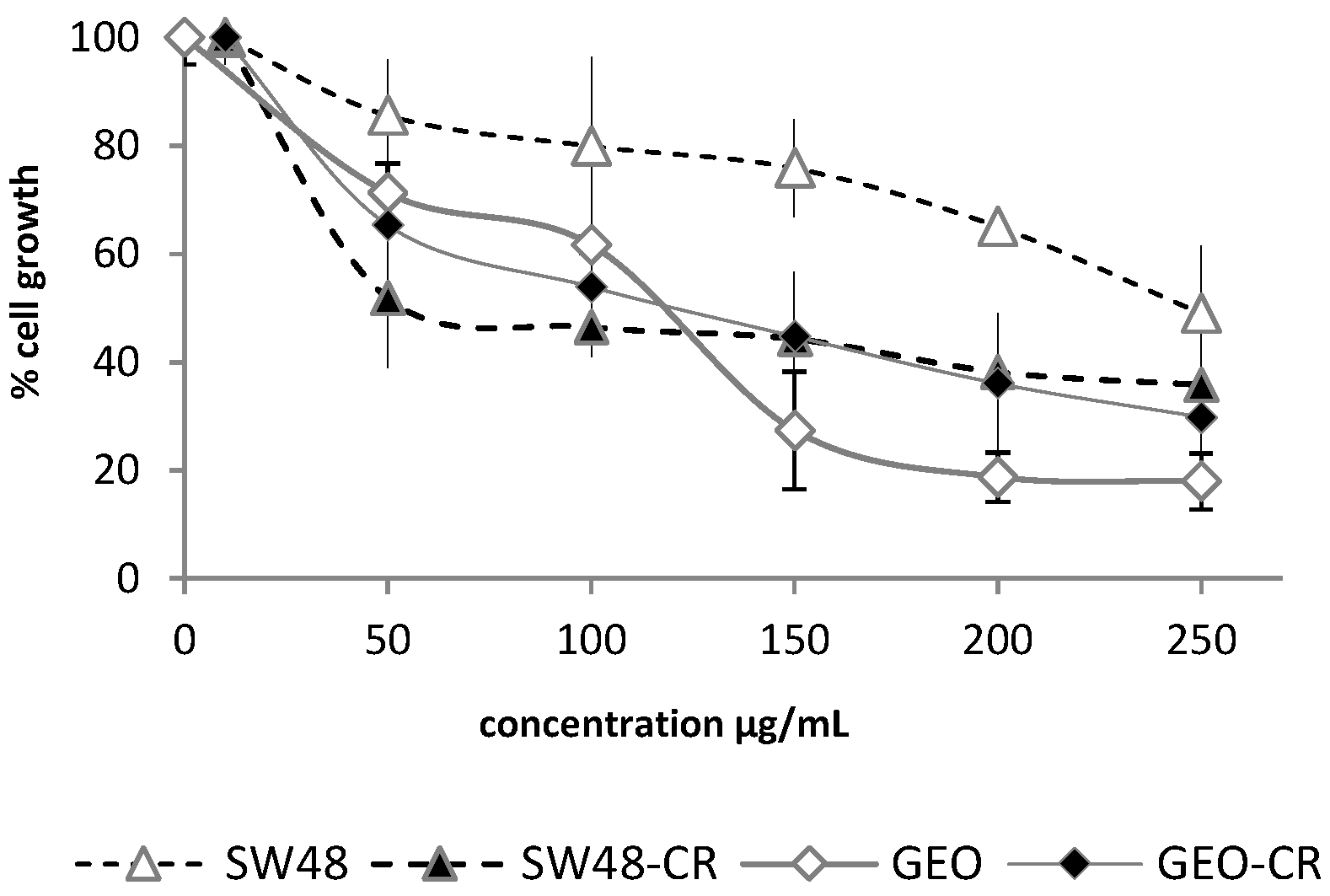
2.1. Cytotoxic Activity
2.1.1. Cytotoxic Activity of Crude Extracts
2.1.2. Cytotoxic Activity of the Crude Extract against Colon Cancer Cell Lines with Acquired Resistance to Cetuximab
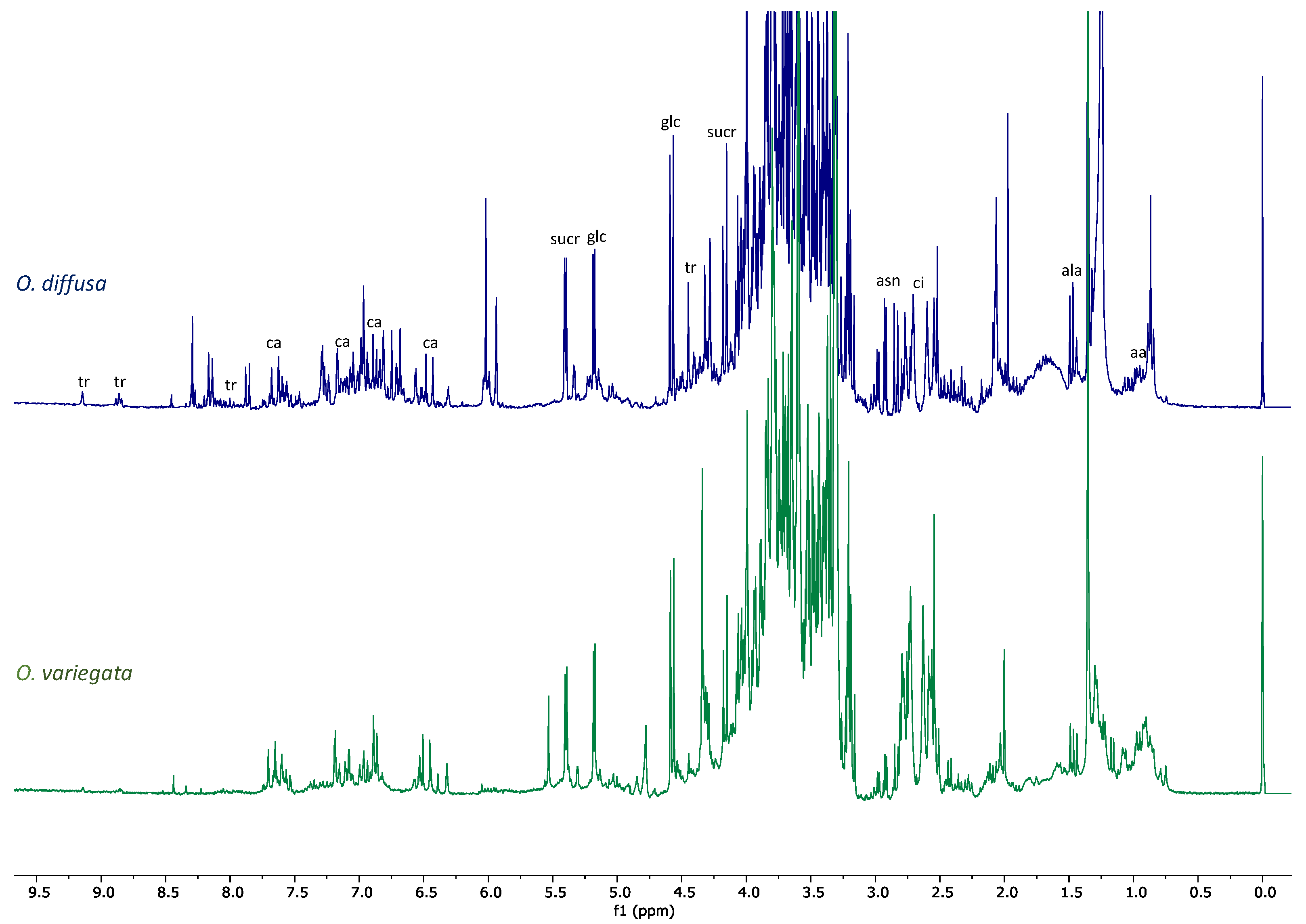
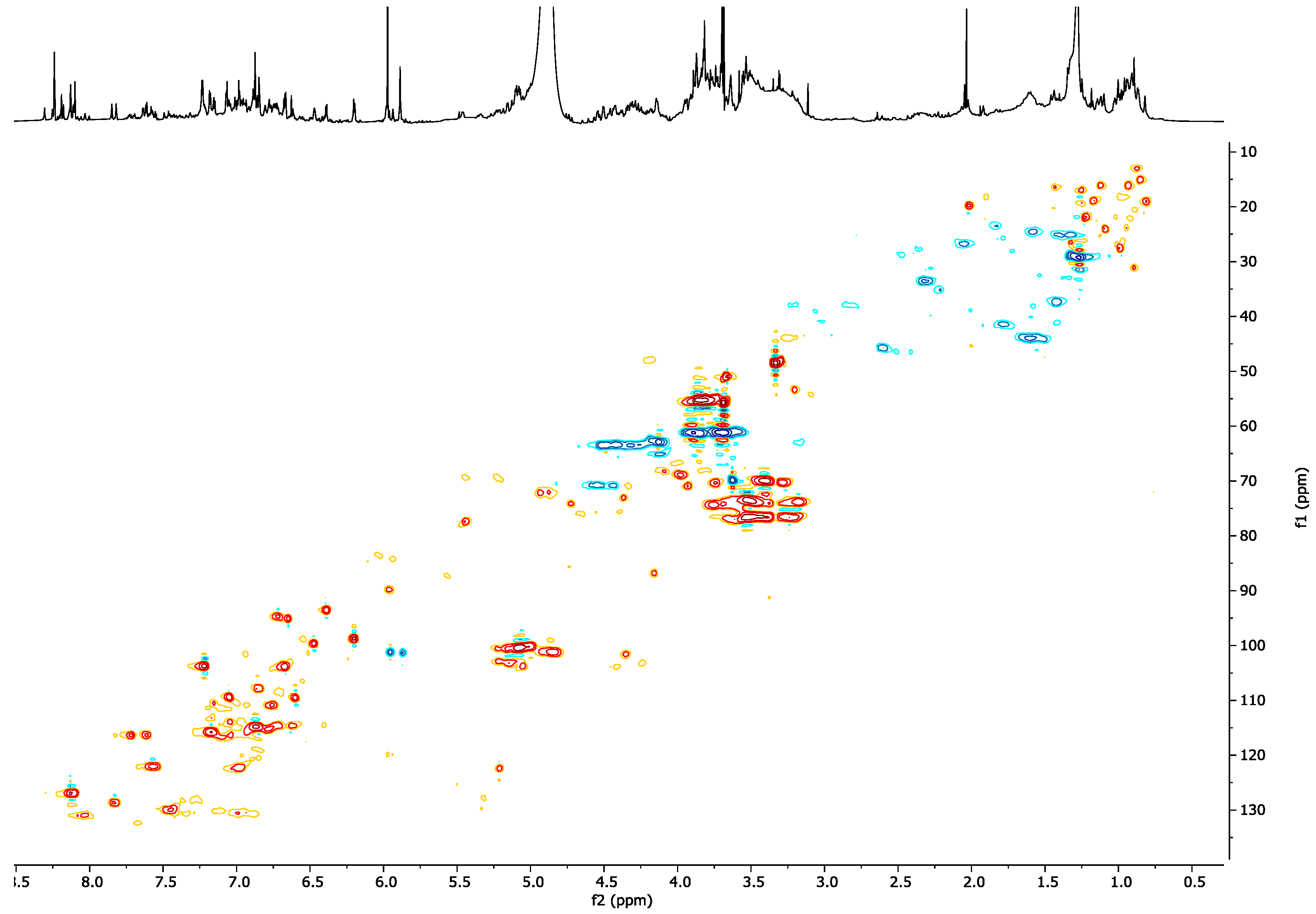
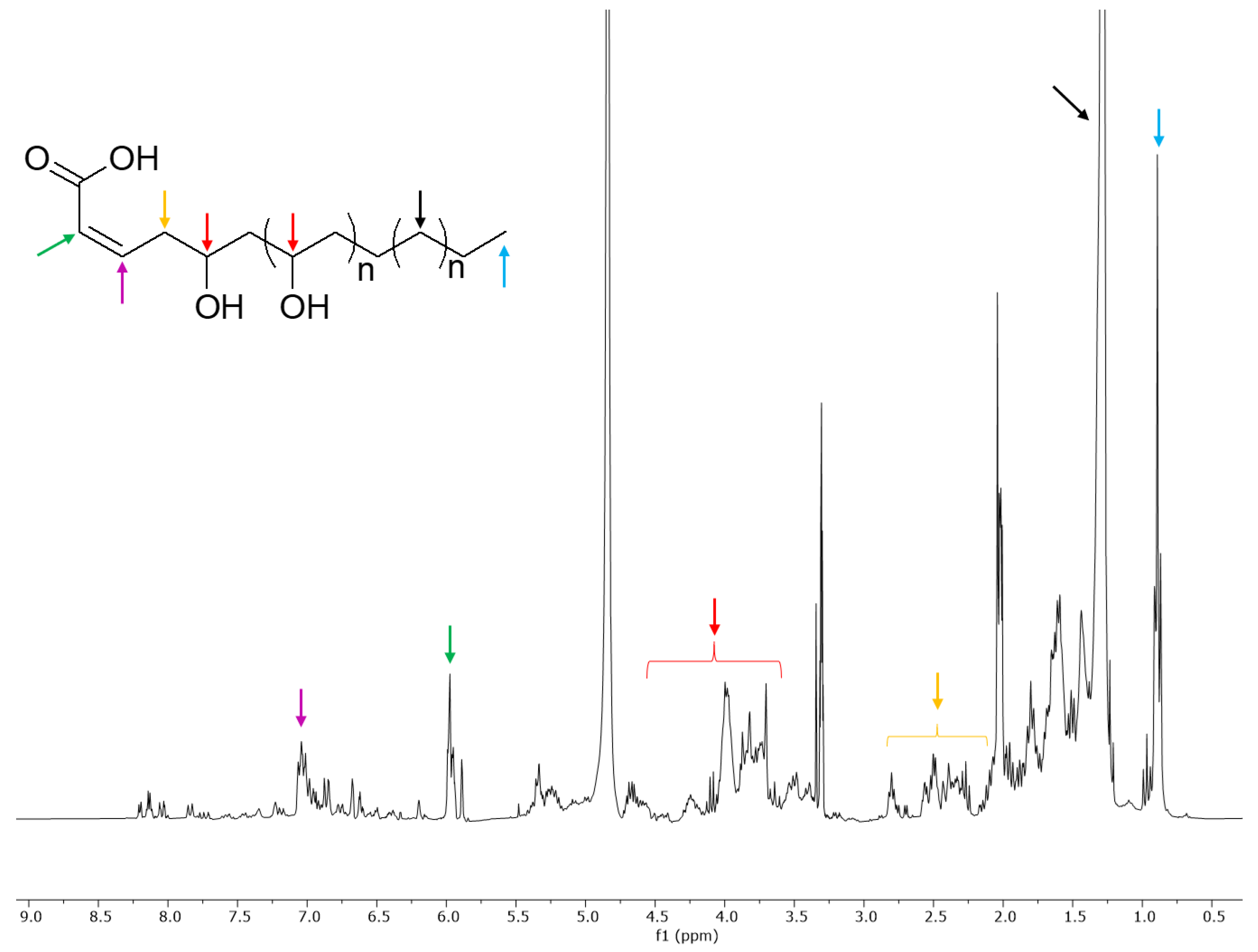
2.2. NMR Profiling of the Extracts
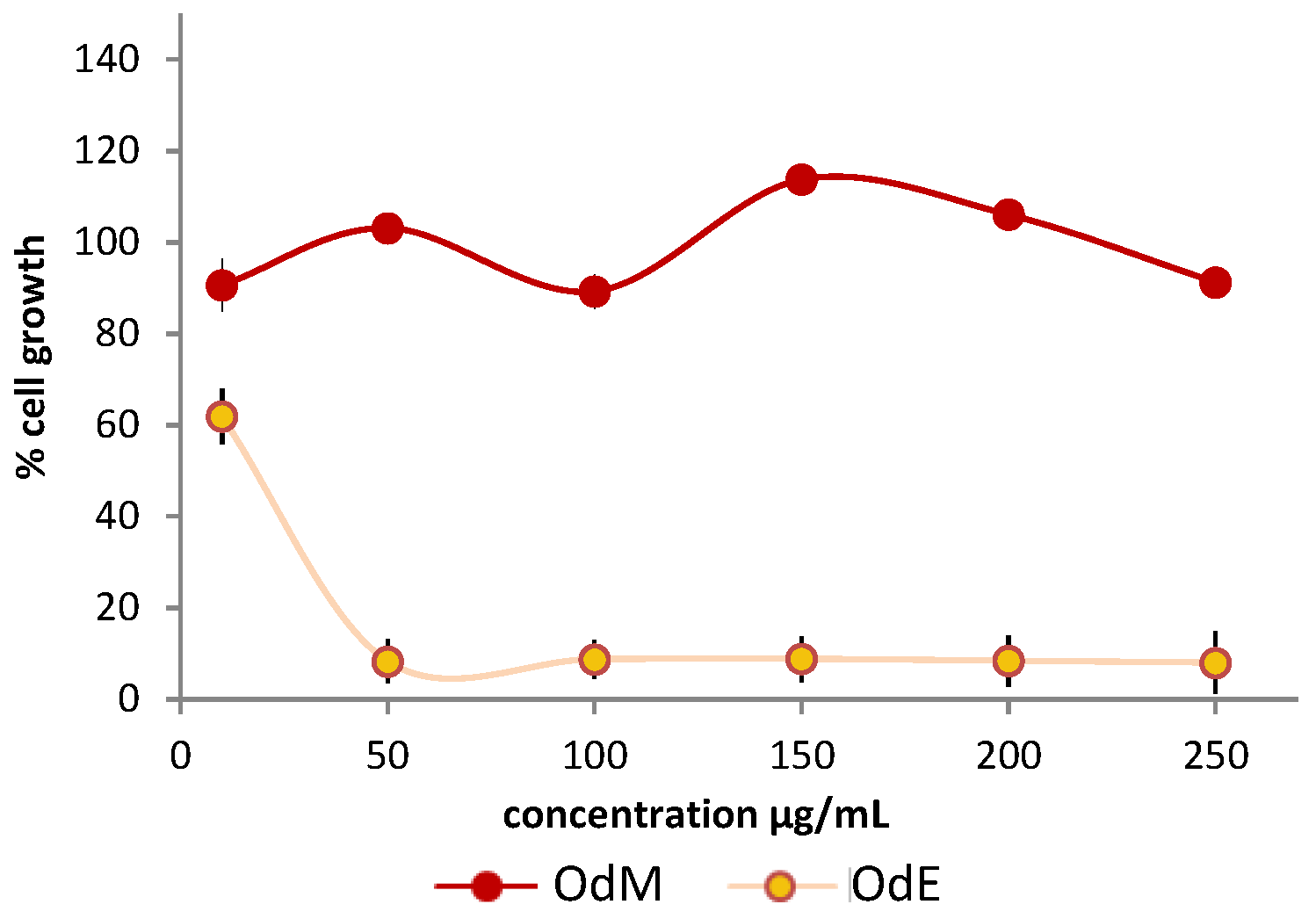
2.3. Partial Purification of the Extract and Biological Activity
3. Discussion
4. Materials and Methods
4.1. Plant Sampling
4.2. Plant Chemical Profile
4.2.1. Extraction
4.2.2. NMR Analysis
4.3. Partial Purification of O. diffusa Extract
4.4. Cell Lines
4.4.1. Cell Cultures
4.4.2. Proliferation Assay
4.4.3. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Yuliana, N.D.; Jahangir, M.; Verpoorte, R.; Choi, Y.H. Metabolomics for the rapid dereplication of bioactive compounds from natural sources. Phytochem. Rev. 2013, 12, 293–304. [Google Scholar] [CrossRef]
- Verpoorte, R. Exploration of nature’s chemodiversity: The role of secondary metabolites as leads in drug development. Drug Discov. Today 1998, 3, 232–238. [Google Scholar] [CrossRef]
- Paterson, I.; Anderson, E.A. Chemistry. The renaissance of natural products as drug candidates. Science 2005, 310, 451–453. [Google Scholar] [CrossRef] [PubMed]
- Pichersky, E.; Lewinsohn, E. Convergent evolution in plant specialized metabolism. Annu. Rev. Plant Biol. 2011, 62, 549–566. [Google Scholar] [CrossRef] [Green Version]
- Nakabayashi, R.; Saito, K. Integrated metabolomics for abiotic stress responses in plants. Curr. Opin. Plant Biol. 2015, 24, 10–16. [Google Scholar] [CrossRef] [Green Version]
- Verpoorte, R.; Choi, Y.H.; Kim, H.K. Ethnopharmacology and systems biology: A perfect holistic match. J. Ethnopharmacol. 2005, 100, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Malvezzi, M.; Carioli, G.; Bertuccio, P.; Rosso, T.; Boffetta, P.; Levi, F.; La Vecchia, C.; Negri, E. European cancer mortality predictions for the year 2016 with focus on leukaemias. Ann. Oncol. 2016, 27, 725–731. [Google Scholar] [CrossRef]
- Hashim, D.; Boffetta, P.; La Vecchia, C.; Rota, M.; Bertuccio, P.; Malvezzi, M.; Negri, E. The global decrease in cancer mortality: Trends and disparities. Ann. Oncol. 2016, 27, 926–933. [Google Scholar] [CrossRef]
- Giordano, G.; Remo, A.; Porras, A.; Pancione, M. Immune Resistance and EGFR Antagonists in Colorectal Cancer. Cancers 2019, 11, 1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciardiello, F.; Tortora, G. EGFR antagonists in cancer treatment. N. Engl. J. Med. 2008, 358, 1160–1174. [Google Scholar] [CrossRef] [Green Version]
- Napolitano, S.; Martini, G.; Rinaldi, B.; Martinelli, E.; Donniacuo, M.; Berrino, L.; Vitagliano, D.; Morgillo, F.; Barra, G.; De Palma, R.; et al. Primary and Acquired Resistance of Colorectal Cancer to Anti-EGFR Monoclonal Antibody Can Be Overcome by Combined Treatment of Regorafenib with Cetuximab. Clin. Cancer Res. 2015, 21, 2975–2983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Cutsem, E.; Kohne, C.H.; Lang, I.; Folprecht, G.; Nowacki, M.P.; Cascinu, S.; Shchepotin, I.; Maurel, J.; Cunningham, D.; Tejpar, S.; et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: Updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J. Clin. Oncol. 2011, 29, 2011–2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graziani, V.; Scognamiglio, M.; Belli, V.; Esposito, A.; D’Abrosca, B.; Chambery, A.; Russo, R.; Panella, M.; Russo, A.; Ciardiello, F.; et al. Metabolomic approach for a rapid identification of natural products with cytotoxic activity against human colorectal cancer cells. Sci. Rep. 2018, 8, 5309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mezrag, A.; Malafronte, N.; Bouheroum, M.; Travaglino, C.; Russo, D.; Milella, L.; Severino, L.; De Tommasi, N.; Braca, A.; Dal Piaz, F. Phytochemical and antioxidant activity studies on Ononis angustissima L. aerial parts: Isolation of two new flavonoids. Nat. Prod. Res. 2017, 31, 507–514. [Google Scholar] [CrossRef]
- Gampe, N.; Darcsi, A.; Nagyne Nedves, A.; Boldizsar, I.; Kursinszki, L.; Beni, S. Phytochemical analysis of Ononis arvensis L. by liquid chromatography coupled with mass spectrometry. J. Mass Spectrom. 2019, 54, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Stojkovic, D.; Dias, M.I.; Drakulic, D.; Barros, L.; Stevanovic, M.; Ferreira, I.C.F.R.; Sokovic, M.D. Methanolic Extract of the Herb Ononis spinosa L. Is an Antifungal Agent with no Cytotoxicity to Primary Human Cells. Pharmaceuticals 2020, 13, 78. [Google Scholar] [CrossRef]
- Ghribi, L.; Waffo-Teguo, P.; Cluzet, S.; Marchal, A.; Marques, J.; Merillon, J.M.; Ben Jannet, H. Isolation and structure elucidation of bioactive compounds from the roots of the Tunisian Ononis angustissima L. Bioorg. Med. Chem. Lett. 2015, 25, 3825–3830. [Google Scholar] [CrossRef]
- Wang, G.; Huang, Y.; Wu, Z.; Zhao, C.; Cong, H.; Ju, S.; Wang, X. KRAS-mutant colon cancer cells respond to combined treatment of ABT263 and axitinib. Biosci. Rep. 2019, 39, 3. [Google Scholar] [CrossRef] [Green Version]
- Scognamiglio, M.; Fiumano, V.; D’Abrosca, B.; Esposito, A.; Choi, Y.H.; Verpoorte, R.; Fiorentino, A. Chemical interactions between plants in Mediterranean vegetation: The influence of selected plant extracts on Aegilops geniculata metabolome. Phytochemistry 2014, 106, 69–85. [Google Scholar] [CrossRef]
- Scognamiglio, M.; D’Abrosca, B.; Esposito, A.; Fiorentino, A. Chemical Composition and Seasonality of Aromatic Mediterranean Plant Species by NMR-Based Metabolomics. J. Anal. Methods Chem. 2015, 2015, 258570. [Google Scholar] [CrossRef]
- Scognamiglio, M.; Schneider, B. Identification of Potential Allelochemicals From Donor Plants and Their Synergistic Effects on the Metabolome of Aegilops geniculata. Front. Plant Sci. 2020, 11, 1046. [Google Scholar] [CrossRef]
- Sigismund, S.; Avanzato, D.; Lanzetti, L. Emerging functions of the EGFR in cancer. Mol. Oncol. 2018, 12, 3–20. [Google Scholar] [CrossRef]
- Blick, S.K.; Scott, L.J. Cetuximab: A review of its use in squamous cell carcinoma of the head and neck and metastatic colorectal cancer. Drugs 2007, 67, 2585–2607. [Google Scholar] [CrossRef] [PubMed]
- Leto, S.M.; Trusolino, L. Primary and acquired resistance to EGFR-targeted therapies in colorectal cancer: Impact on future treatment strategies. J. Mol. Med. 2014, 92, 709–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blee, E. Impact of phyto-oxylipins in plant defense. Trends Plant Sci. 2002, 7, 315–322. [Google Scholar] [CrossRef]
- Howe, G.A.; Schilmiller, A.L. Oxylipin metabolism in response to stress. Curr. Opin. Plant Biol. 2002, 5, 230–236. [Google Scholar] [CrossRef]
- Farmer, E.E.; Almeras, E.; Krishnamurthy, V. Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Curr. Opin. Plant Biol. 2003, 6, 372–378. [Google Scholar] [CrossRef]
- Caldwell, G.S. The influence of bioactive oxylipins from marine diatoms on invertebrate reproduction and development. Mar. Drugs 2009, 7, 367–400. [Google Scholar] [CrossRef] [Green Version]
- Christensen, L.P. Bioactive C17 and C18 Acetylenic Oxylipins from Terrestrial Plants as Potential Lead Compounds for Anticancer Drug Development. Molecules 2020, 25, 2568. [Google Scholar] [CrossRef]
- D’Abrosca, B.; Ciaramella, V.; Graziani, V.; Papaccio, F.; Della Corte, C.M.; Potenza, N.; Fiorentino, A.; Ciardiello, F.; Morgillo, F. Urtica dioica L. inhibits proliferation and enhances cisplatin cytotoxicity in NSCLC cells via Endoplasmic Reticulum-stress mediated apoptosis. Sci. Rep. 2019, 9, 4986. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.K.; Verpoorte, R. Sample preparation for plant metabolomics. Phytochem. Anal. 2010, 21, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Tun, J.O.; Salvador-Reyes, L.A.; Velarde, M.C.; Saito, N.; Suwanborirux, K.; Concepcion, G.P. Synergistic cytotoxicity of renieramycin M and doxorubicin in MCF-7 breast cancer cells. Mar. Drugs 2019, 17, 536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Graziani, V.; Potenza, N.; D’Abrosca, B.; Troiani, T.; Napolitano, S.; Fiorentino, A.; Scognamiglio, M. NMR Profiling of Ononis diffusa Identifies Cytotoxic Compounds against Cetuximab-Resistant Colon Cancer Cell Lines. Molecules 2021, 26, 3266. https://doi.org/10.3390/molecules26113266
Graziani V, Potenza N, D’Abrosca B, Troiani T, Napolitano S, Fiorentino A, Scognamiglio M. NMR Profiling of Ononis diffusa Identifies Cytotoxic Compounds against Cetuximab-Resistant Colon Cancer Cell Lines. Molecules. 2021; 26(11):3266. https://doi.org/10.3390/molecules26113266
Chicago/Turabian StyleGraziani, Vittoria, Nicoletta Potenza, Brigida D’Abrosca, Teresa Troiani, Stefania Napolitano, Antonio Fiorentino, and Monica Scognamiglio. 2021. "NMR Profiling of Ononis diffusa Identifies Cytotoxic Compounds against Cetuximab-Resistant Colon Cancer Cell Lines" Molecules 26, no. 11: 3266. https://doi.org/10.3390/molecules26113266
APA StyleGraziani, V., Potenza, N., D’Abrosca, B., Troiani, T., Napolitano, S., Fiorentino, A., & Scognamiglio, M. (2021). NMR Profiling of Ononis diffusa Identifies Cytotoxic Compounds against Cetuximab-Resistant Colon Cancer Cell Lines. Molecules, 26(11), 3266. https://doi.org/10.3390/molecules26113266









