Use of Silicon Nanowire Sensors for Early Cancer Diagnosis
Abstract
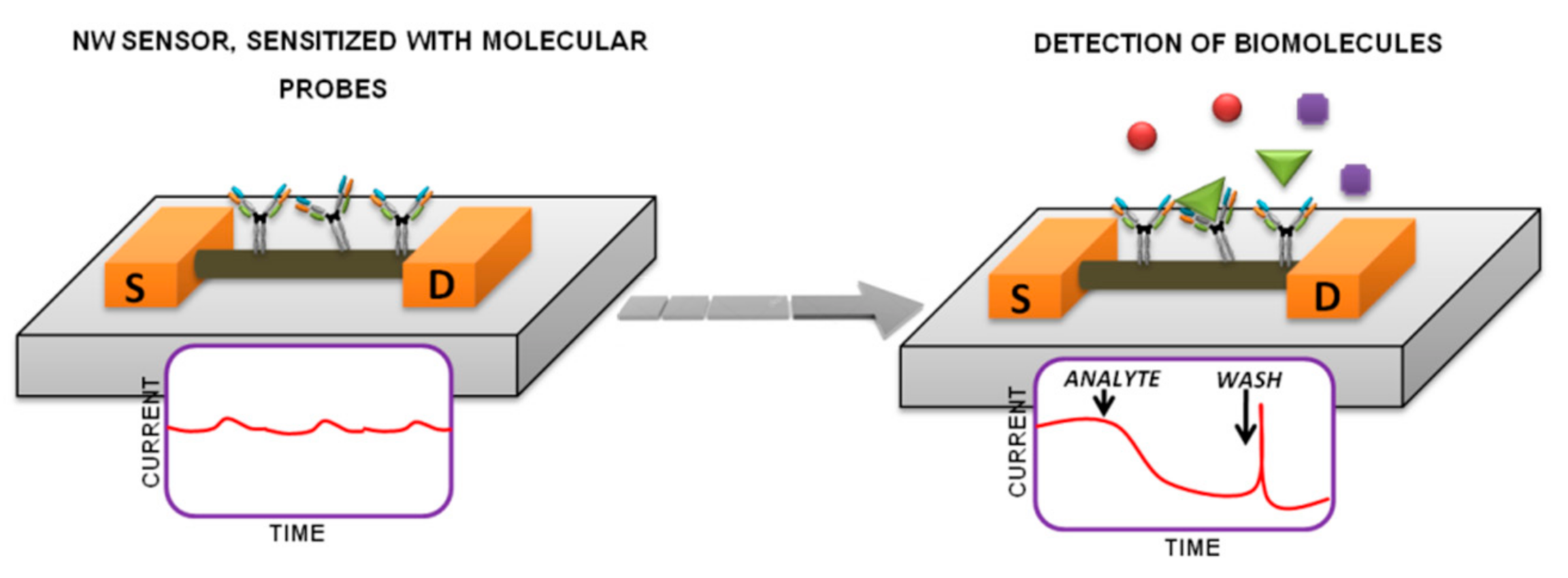
:1. Introduction
2. Detecting Protein Markers with the Use of Si-NW-Nanosensors
3. Detection of Specific Nucleic Scids with the Use of Si-NW-Nanosensors
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- World Health Organization. Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 1 June 2021).
- Kaprin, A.D.; Starinskiy, V.V.; Petrov, V.G. The State of Cancer Care for the Population of Russia in 2018; MNIOI Them. P.A. Herzen: Branch of the Federal State Budgetary Institution “National Medical Research Center of Radiology” Ministry of Health of Russia: Moscow, Russia, 2019; p. 250. ISBN 978-5-85502-251-3. [Google Scholar]
- Malsagova, K.A.; Pleshakova, T.O.; Kozlov, A.F.; Shumov, I.D.; Ilnitskii, M.A.; Miakonkikh, A.V.; Popov, V.P.; Rudenko, K.V.; Glukhov, A.V.; Kupriyanov, I.N.; et al. Micro-Raman Spectroscopy for Monitoring of Deposition Quality of High-k Stack Protective Layer onto Nanowire FET Chips for Highly Sensitive miRNA Detection. Biosensors 2018, 8, 72. [Google Scholar] [CrossRef] [Green Version]
- Rissin, D.M.; Kan, C.W.; Campbell, T.G.; Howes, S.C.; Fournier, D.R.; Song, L.; Piech, T.; Patel, P.P.; Chang, L.; Rivnak, A.J.; et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 2010, 28, 595–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malsagova, K.A.; Ivanov, Y.D.; Pleshakova, T.O.; Kaysheva, A.L.; Shumov, I.D.; Kozlov, A.F.; Archakov, A.I.; Popov, V.; Fomin, B.I.; Latyshev, A.V. A SOI-nanowire biosensor for the multiple detection of D-NFATc1 protein in the serum. Anal. Methods 2015, 7, 8078–8085. [Google Scholar] [CrossRef]
- Ivanov, Y.D.; Pleshakova, T.; Malsagova, K.; Kozlov, A.; Kaysheva, A.; Shumov, I.; Galiullin, R.; Kurbatov, L.; Popov, V.; Naumova, O.; et al. Detection of marker miRNAs in plasma using SOI-NW biosensor. Sens. Actuators B Chem. 2018, 261, 566–571. [Google Scholar] [CrossRef]
- Malsagova, K.A.; Pleshakova, T.O.; Galiullin, R.A.; Shumov, I.D.; Kozlov, A.F.; Romanova, T.S.; Popov, V.P.; Glukhov, A.V.; Konev, V.A.; Archakov, A.I.; et al. Nanowire Aptamer-Sensitized Biosensor Chips with Gas Plasma-Treated Surface for the Detection of Hepatitis C Virus Core Antigen. Coatings 2020, 10, 753. [Google Scholar] [CrossRef]
- Malsagova, K.A.; Pleshakova, T.O.; Galiullin, R.A.; Kaysheva, A.L.; Shumov, I.D.; Ilnitskii, M.A.; Popov, V.P.; Glukhov, A.V.; Archakov, A.I.; Ivanov, Y.D. Ultrasensitive nanowire-based detection of HCVcoreAg in the serum using a microwave generator. Anal. Methods 2018, 10, 2740–2749. [Google Scholar] [CrossRef]
- Wang, H.; Han, X.; Ou, X.; Lee, C.-S.; Zhang, X.; Lee, S.-T. Silicon nanowire based single-molecule SERS sensor. Nanoscale 2013, 5, 8172–8176. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zhang, G.-J. Silicon nanowire-transistor biosensor for study of molecule-molecule interactions. Rev. Anal. Chem. 2014, 33. [Google Scholar] [CrossRef]
- He, B.; Morrow, T.J.; Keating, C.D. Nanowire sensors for multiplexed detection of biomolecules. Curr. Opin. Chem. Biol. 2008, 12, 522–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leonardi, A.A.; Faro, M.J.L.; Irrera, A. Biosensing platforms based on silicon nanostructures: A critical review. Anal. Chim. Acta 2021, 1160, 338393. [Google Scholar] [CrossRef]
- Tran, D.P.; Pham, T.T.T.; Wolfrum, B.; Offenhäusser, A.; Thierry, B. CMOS-Compatible Silicon Nanowire Field-Effect Transistor Biosensor: Technology Development toward Commercialization. Materials 2018, 11, 785. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Chen, R.; Xu, L.; Ning, Y.; Xie, S.; Zhang, G.-J. Silicon Nanowire Biosensor for Highly Sensitive and Multiplexed Detection of Oral Squamous Cell Carcinoma Biomarkers in Saliva. Anal. Sci. 2015, 31, 73–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, G.; Patolsky, F.; Cui, Y.; Wang, W.U.; Lieber, C.M. Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nat. Biotechnol. 2005, 23, 1294–1301. [Google Scholar] [CrossRef]
- Lichtenstein, A.; Havivi, E.; Shacham, R.; Hahamy, E.; Leibovich, R.; Pevzner, A.; Krivitsky, V.; Davivi, G.; Presman, I.; Elnathan, R.; et al. Supersensitive fingerprinting of explosives by chemically modified nanosensors arrays. Nat. Commun. 2014, 5, 4195. [Google Scholar] [CrossRef] [Green Version]
- Doucey, M.-A.; Carrara, S. Nanowire Sensors in Cancer. Trends Biotechnol. 2019, 37, 86–99. [Google Scholar] [CrossRef] [Green Version]
- Noor, M.O.; Krull, U.J. Silicon nanowires as field-effect transducers for biosensor development: A review. Anal. Chim. Acta 2014, 825, 1–25. [Google Scholar] [CrossRef]
- Gao, A.; Dai, P.; Lu, N.; Li, T.; Wang, Y.; Hemmila, S.; Kallio, P. Integration of microfluidic system with silicon nanowires biosensor for multiplexed detection. In Proceedings of the 2013 International Conference on Manipulation, Manufacturing and Measurement on the Nanoscale, Suzhou, China, 26–30 August 2013; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA, 2013; pp. 333–336. [Google Scholar]
- Shehada, N.; Cancilla, J.C.; Torrecilla, J.S.; Pariente, E.S.; Brönstrup, G.; Christiansen, S.; Johnson, D.W.; Leja, M.; Davies, M.P.A.; Liran, O.; et al. Silicon Nanowire Sensors Enable Diagnosis of Patients via Exhaled Breath. ACS Nano 2016, 10, 7047–7057. [Google Scholar] [CrossRef] [Green Version]
- Tran, T.T.; Mulchandani, A. Carbon nanotubes and graphene nano field-effect transistor-based biosensors. TrAC Trends Anal. Chem. 2016, 79, 222–232. [Google Scholar] [CrossRef] [Green Version]
- Wasik, D.; Mulchandani, A.; Yates, M.V. A heparin-functionalized carbon nanotube-based affinity biosensor for dengue virus. Biosens. Bioelectron. 2017, 91, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hodak, M.; Lu, W.; Bernholc, J. Selective sensing of ethylene and glucose using carbon-nanotube-based sensors: An ab initio investigation. Nanoscale 2017, 9, 1687–1698. [Google Scholar] [CrossRef] [PubMed]
- Majd, S.M.; Salimi, A. Ultrasensitive flexible FET-type aptasensor for CA 125 cancer marker detection based on carboxylated multiwalled carbon nanotubes immobilized onto reduced graphene oxide film. Anal. Chim. Acta 2018, 1000, 273–282. [Google Scholar] [CrossRef]
- Ramnani, P.; Gao, Y.; Ozsoz, M.; Mulchandani, A. Electronic Detection of MicroRNA at Attomolar Level with High Specificity. Anal. Chem. 2013, 85, 8061–8064. [Google Scholar] [CrossRef]
- Popov, V.P.; Antonova, A.I.; Frantsuzov, A.A.; Safronov, L.N.; Feofanov, G.N.; Naumova, O.V.; Kilanov, D.V. Properties of silicon-on-insulator structures and devices. Semiconductor 2001, 35, 1030–1037. [Google Scholar] [CrossRef]
- Naumova, O.V.; Fomin, B.I.; Nasimov, D.A.; Dudchenko, N.V.; Devyatova, S.F.; Zhanaev, E.D.; Popov, V.; Latyshev, A.V.; Aseev, A.L.; Ivanov, Y.D.; et al. SOI nanowires as sensors for charge detection. Semicond. Sci. Technol. 2010, 25, 055004. [Google Scholar] [CrossRef]
- Patolsky, F.; Zheng, G.; Hayden, O.; Lakadamyali, M.; Zhuang, X.; Lieber, C.M. Electrical detection of single viruses. Proc. Natl. Acad. Sci. 2004, 101, 14017–14022. [Google Scholar] [CrossRef] [Green Version]
- Vu, C.-A.; Chen, W.-Y. Field-Effect Transistor Biosensors for Biomedical Applications: Recent Advances and Future Prospects. Sensors 2019, 19, 4214. [Google Scholar] [CrossRef] [Green Version]
- Hu, P.; Zhang, J.; Li, L.; Wang, Z.; O’Neill, W.; Estrela, P. Carbon Nanostructure-Based Field-Effect Transistors for Label-Free Chemical/Biological Sensors. Sensors 2010, 10, 5133–5159. [Google Scholar] [CrossRef] [Green Version]
- Lu, N.; Gao, A.; Dai, P.; Mao, H.; Zuo, X.; Fan, C.; Wang, Y.; Yuelin, W. Ultrasensitive Detection of Dual Cancer Biomarkers with Integrated CMOS-Compatible Nanowire Arrays. Anal. Chem. 2015, 87, 11203–11208. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Hideshima, S.; Kuroiwa, S.; Nakanishi, T.; Osaka, T. Label-free detection of tumor markers using field effect transistor (FET)-based biosensors for lung cancer diagnosis. Sens. Actuators B Chem. 2015, 212, 329–334. [Google Scholar] [CrossRef]
- Zhu, K.; Zhang, Y.; Li, Z.; Zhou, F.; Feng, K.; Dou, H.; Wang, T. Simultaneous Detection of α-Fetoprotein and Carcinoembryonic Antigen Based on Si Nanowire Field-Effect Transistors. Sensors 2015, 15, 19225–19236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Fan, Y.; Wu, Z.; Liu, C. A Silicon Nanowire Array Biosensor Fabricated by Complementary Metal Oxide Semiconductor Technique for Highly Sensitive and Selective Detection of Serum Carcinoembryonic Antigen. Micromachines 2019, 10, 764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, A.; Yang, X.; Tong, J.; Zhou, L.; Wang, Y.; Zhao, J.; Mao, H.; Li, T. Multiplexed detection of lung cancer biomarkers in patients serum with CMOS-compatible silicon nanowire arrays. Biosens. Bioelectron. 2017, 91, 482–488. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Gao, A.; Chen, S.; Wang, Y.; Li, T. Highly Sensitive Juntionless Nanowire Transistor Biosensor in Detecting Breast Tumor Marker. In Proceedings of the 2018 IEEE Sensors Applications Symposium (SAS), Seoul, Korea, 12–14 March 2018; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA, 2018; pp. 1–4. [Google Scholar]
- Chen, H.-C.; Chen, Y.-T.; Tsai, R.-Y.; Chen, M.-C.; Chen, S.-L.; Xiao, M.-C.; Chen, C.-L.; Hua, M.-Y. A sensitive and selective magnetic graphene composite-modified polycrystalline-silicon nanowire field-effect transistor for bladder cancer diagnosis. Biosens. Bioelectron. 2015, 66, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Pham, V.B.; Le, T.T.T.; Phan, T.N.K.; Nguyen, T.T.; Dang, C.M. Application of silicon nanowire for detection and quantitative analysis of alpha-fetoprotein biomarker. Int. J. Nanotechnol. 2018, 15, 210. [Google Scholar] [CrossRef]
- Yang, C.-Y.; Chiang, H.-C.; Kuo, C.-J.; Hsu, C.-W.; Chan, S.-F.; Lin, Z.-Y.; Lin, C.-H.; Chen, Y.-T. Hepatocellular Carcinoma Diagnosis by Detecting α-Fucosidase with a Silicon Nanowire Field-Effect Transistor Biosensor. ECS J. Solid State Sci. Technol. 2018, 7, Q3153–Q3158. [Google Scholar] [CrossRef]
- Zida, S.I.; Yang, C.-C.; Khung, Y.L.; Lin, Y.-D. Fabrication and Characterization of an Aptamer-Based N-type Silicon Nanowire FET Biosensor for VEGF Detection. J. Med. Biol. Eng. 2020, 40, 601–609. [Google Scholar] [CrossRef]
- Puppo, F.; Doucey, M.-A.; Delaloye, J.-F.; Moh, T.S.Y.; Pandraud, G.; Sarro, P.M.; De Micheli, G.; Carrara, S. SiNW-FET in-Air Biosensors for High Sensitive and Specific Detection in Breast Tumor Extract. IEEE Sens. J. 2015, 16, 3374–3381. [Google Scholar] [CrossRef] [Green Version]
- Kruchinina, M.V.; Prudnikova, Y.I.; Kurilovich, S.A.; Gromov, A.A.; Kruchinin, V.N.; Atuchin, V.V.; Naumova, O.V.; Spesivtsev, E.V.; Volodin, V.A.; Peltek, S.E.; et al. Ellipsometry, raman spectroscopy and soi-nanowire biosensor in diagnosis of colorectalcancer. Sib. J. Oncol. 2017, 16, 32–41. [Google Scholar] [CrossRef] [Green Version]
- Tran, D.P.; Wolfrum, B.; Stockmann, R.; Pai, J.-H.; Pourhassan-Moghaddam, M.; Offenhäusser, A.; Thierry, B. Complementary Metal Oxide Semiconductor Compatible Silicon Nanowires-on-a-Chip: Fabrication and Preclinical Validation for the Detection of a Cancer Prognostic Protein Marker in Serum. Anal. Chem. 2015, 87, 1662–1668. [Google Scholar] [CrossRef]
- Hung, J.-Y.; Manga, Y.B.; Chen, Y.-J.; Huang, H.-M.; Yang, W.-L.; Wu, C.-C. P16INK4a detection using an ultra-sensitive silicon nanowire field effect transistor. In Proceedings of the 2018 7th International Symposium on Next Generation Electronics (ISNE), Taipei, Taiwan, 7–9 May 2018; pp. 1–2. [Google Scholar] [CrossRef]
- Elfström, N.; Juhasz, R.; Sychugov, I.; Engfeldt, T.; Karlström, A.E.; Linnros, J. Surface Charge Sensitivity of Silicon Nanowires: Size Dependence. Nano Lett. 2007, 7, 2608–2612. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Rajendran, B.; Kamins, T.; Li, X.; Chen, Y.; Williams, R.S. Silicon nanowires for sequence-specific DNA sensing: Device fabrication and simulation. Appl. Phys. A 2005, 80, 1257–1263. [Google Scholar] [CrossRef]
- Giardina, M.G.; Matarazzo, M.; Varriale, A.; Morante, R.; Napoli, A.; Martino, R. Serum Alpha-L-Fucosidase. A Useful Marker in the Diagnosis of Hepatocellular Carcinoma. Cancer 1992, 70, 1044–1048. [Google Scholar] [CrossRef]
- Bukharina, N.S.; Ivanov, Y.D.; Pleshakova, T.O.; Frantsuzov, P.A.; Andreeva, E.Y.; Kaysheva, A.L.; Izotov, A.A.; Pavlova, T.I.; Ziborov, V.S.; Radko, S.P.; et al. Atomic force microscopy fishing of GP120 on immobilized aptamers and its mass spectrometry identification. Biochem. (Moscow) Suppl. Ser. B Biomed. Chem. 2014, 8, 115–124. [Google Scholar] [CrossRef]
- Kulbachinskiy, A.V. Methods for selection of aptamers to protein targets. Biochem. (Moscow) 2007, 72, 1505–1518. [Google Scholar] [CrossRef]
- Gold, L.; Ayers, D.; Bertino, J.; Bock, C.; Bock, A.; Brody, E.N.; Carter, J.; Dalby, A.B.; Eaton, B.E.; Fitzwater, T.; et al. Aptamer-Based Multiplexed Proteomic Technology for Biomarker Discovery. PLoS ONE 2010, 5, e15004. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Yang, W.; Lai, R.Y. A folding-based electrochemical aptasensor for detection of vascular endothelial growth factor in human whole blood. Biosens. Bioelectron. 2011, 26, 2442–2447. [Google Scholar] [CrossRef]
- Gasparini, G. Prognostic Value of Vascular Endothelial Growth Factor in Breast Cancer. Oncology 2000, 5, 37–44. [Google Scholar] [CrossRef]
- Kakeji, Y.; Koga, T.; Sumiyoshi, Y.; Shibahara, K.; Oda, S.; Maehara, Y.; Sugimachi, K. Clinical significance of vascular endothelial growth factor expression in gastric cancer. J. Exp. Clin. Cancer Res. 2002, 21, 125–129. [Google Scholar]
- Smedbakken, L.; Jensen, J.K.; Hallén, J.; Atar, D.; Januzzi, J.L.; Halvorsen, B.; Aukrust, P.; Ueland, T. Activated Leukocyte Cell Adhesion Molecule and Prognosis in Acute Ischemic Stroke. Stroke 2011, 42, 2453–2458. [Google Scholar] [CrossRef] [Green Version]
- Stern, E.; Vacic, A.; Rajan, N.K.; Criscione, J.M.; Park, J.; Ilic, B.R.; Mooney, D.; Reed, M.A.; Fahmy, T.M. Label-free biomarker detection from whole blood. Nat. Nanotechnol. 2009, 5, 138–142. [Google Scholar] [CrossRef] [Green Version]
- Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clin. Pharmacol. Ther. 2001, 69, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Moyer, V.A. Screening for Prostate Cancer: U.S. Preventive Services Task Force Recommendation Statement. Ann. Intern. Med. 2012, 157, 120–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wittmann, J.; Jäck, H.-M. Serum microRNAs as powerful cancer biomarkers. Biochim. Biophys. Acta (BBA) Bioenerg. 2010, 1806, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Shimomura, A.; Shiino, S.; Kawauchi, J.; Takizawa, S.; Sakamoto, H.; Matsuzaki, J.; Ono, M.; Takeshita, F.; Niida, S.; Shimizu, C.; et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci. 2016, 107, 326–334. [Google Scholar] [CrossRef]
- De Planell-Saguer, M.; Rodicio, M.C. Detection methods for microRNAs in clinic practice. Clin. Biochem. 2013, 46, 869–878. [Google Scholar] [CrossRef]
- Jiang, J.; Lee, E.J.; Gusev, Y.; Schmittgen, T.D. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005, 33, 5394–5403. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Lee, E.J.; Jiang, J.; Sarkar, A.; Yang, L.; Elton, T.S.; Chen, C. Real-time PCR quantification of precursor and mature microRNA. Methods 2008, 44, 31–38. [Google Scholar] [CrossRef] [Green Version]
- Válóczi, A. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004, 32, e175. [Google Scholar] [CrossRef] [Green Version]
- Yao, X.; Huang, H.; Xu, L. In Situ Detection of Mature miRNAs in Plants Using LNA-Modified DNA Probes. Adv. Struct. Saf. Stud. 2012, 883, 143–154. [Google Scholar] [CrossRef]
- Liu, C.-G.; Calin, G.A.; Volinia, S.; Croce, C.M. MicroRNA expression profiling using microarrays. Nat. Protoc. 2008, 3, 563–578. [Google Scholar] [CrossRef]
- Creighton, C.J.; Reid, J.G.; Gunaratne, P.H. Expression profiling of microRNAs by deep sequencing. Brief. Bioinform. 2009, 10, 490–497. [Google Scholar] [CrossRef]
- Lu, N.; Gao, A.; Dai, P.; Song, S.; Fan, C.; Wang, Y.; Li, T. CMOS-Compatible Silicon Nanowire Field-Effect Transistors for Ultrasensitive and Label-Free MicroRNAs Sensing. Small 2014, 10, 2022–2028. [Google Scholar] [CrossRef]
- Zhang, G.-J.; Chua, J.H.; Chee, R.-E.; Agarwal, A.; Wong, S.M. Label-free direct detection of MiRNAs with silicon nanowire biosensors. Biosens. Bioelectron. 2009, 24, 2504–2508. [Google Scholar] [CrossRef]
- Lu, N.; Gao, A.; Dai, P.; Li, T.; Wang, Y.; Gao, X.; Song, S.; Fan, C.; Wang, Y. Ultra-sensitive nucleic acids detection with electrical nanosensors based on CMOS-compatible silicon nanowire field-effect transistors. Methods 2013, 63, 212–218. [Google Scholar] [CrossRef]
- Gao, A.; Lu, N.; Wang, Y.; Dai, P.; Li, T.; Gao, X.; Wang, Y.; Fan, C. Enhanced Sensing of Nucleic Acids with Silicon Nanowire Field Effect Transistor Biosensors. Nano Lett. 2012, 12, 5262–5268. [Google Scholar] [CrossRef]
- Zhang, G.-J. Silicon Nanowire Biosensor for Ultrasensitive and Label-Free Direct Detection of miRNAs. In Advanced Structural Safety Studies; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2010; Volume 676, pp. 111–121. [Google Scholar]
- He, J.; Zhu, J.; Gong, C.; Qi, J.; Xiao, H.; Jiang, B.; Zhao, Y. Label-Free Direct Detection of miRNAs with Poly-Silicon Nanowire Biosensors. PLoS ONE 2015, 10, e0145160. [Google Scholar] [CrossRef]
- Malsagova, K.A.; Pleshakova, T.O.; Galiullin, R.A.; Kozlov, A.F.; Romanova, T.S.; Shumov, I.D.; Popov, V.P.; Tikhonenko, F.V.; Glukhov, A.V.; Smirnov, A.Y.; et al. SOI-Nanowire Biosensor for the Detection of Glioma-Associated miRNAs in Plasma. Chemosensors 2020, 8, 95. [Google Scholar] [CrossRef]
- Ahmad, R.; Mahmoudi, T.; Ahn, M.-S.; Hahn, Y.-B. Recent advances in nanowires-based field-effect transistors for biological sensor applications. Biosens. Bioelectron. 2018, 100, 312–325. [Google Scholar] [CrossRef]
| Analyte | Medium | Detection Limit * | Method | Ref. |
|---|---|---|---|---|
| PSA | buffer | 1.7 × 10−15 M | Si-NW | [15] |
| serum | 3.13 × 10−14 M | |||
| buffer | 3.48 × 10−17 M | Si-NW PDMS | [31] | |
| serum | 3.48 × 10−16 M | |||
| buffer | 3.48 × 10−17 M | [32] | ||
| serum | 3.48 × 10−16 M | |||
| CEA | buffer | 6.51 × 10−16 M | Si-NW | [15] |
| serum | 1.17 × 10−14 M | |||
| serum | 6.51 × 10−16 M | Si-NW PDMS | [33] | |
| serum | 1.3 × 10−16 M | Si-NW | [34] | |
| buffer | 1.3 × 10−17 M | [35] | ||
| serum | 1.3 × 10−16 M | |||
| buffer | 1.3 × 10−17 M | JNT | [36] | |
| MUC1 | buffer | 4.09 × 10−16 M | Si-NW | [15] |
| serum | 7.37 × 10−15 M | |||
| APOA2 | urina | 3.8 × 10−13 M | Poly-Si-NW | [37] |
| D-NFATc1 | buffer | 2.5 × 10−15 M | Si-NW | [5] |
| serum | 2.5 × 10−14 M | |||
| CYFRA21-1 | buffer | 3.33 × 10−17 M | Si-NW PDMS | [31] |
| serum | 3.33 × 10−16 M | |||
| buffer | 3.33 × 10−17 M | Si-NW | [32] | |
| serum | 3.33×10−16 M | |||
| AFP | buffer | 1.46 × 10−13 M | Si-NW | [38] |
| serum | 7.28 × 10−15 M | Si-NW PDMS | [33] | |
| α-fucosidase | buffer | 1.3 × 10−12 M | Si-NW PDMS | [39] |
| VEGF | buffer | 2.59 × 10−9 M | Si-NW | [40] |
| tissue | 5.0 × 10−15 M | [41] | ||
| Tumor M2-PK | buffer | 10−13–10−15 M | Si-NW | [42] |
| ALCAM | serum | 2.38 × 10−13 M | Si-NW | [43] |
| p16 INK4a | buffer | 6.48 × 10−15 M | Poly-Si-NW | [44] |
| Analyte | Medium | Detection Limit * | Method | Ref. |
|---|---|---|---|---|
| DNA | buffer | 1.0 × 10−15 M | Si-NW-FET | [69] |
| buffer | 1.0 × 10−15 M | [70] | ||
| buffer | 1.0 × 10−15 M | |||
| miRNA | buffer | 1.0 × 10−17 M | Si-NW | [71] |
| U6 snRNA | cells | ~2.2 × 10−5 M * | poly-Si-NW FET | [72] |
| miRNA | buffer | 1.0 × 10−15 M | ||
| miRNA-363 | buffer | 1.0 × 10−17 M | Si-NW | [73] |
| plasma | no data |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivanov, Y.D.; Romanova, T.S.; Malsagova, K.A.; Pleshakova, T.O.; Archakov, A.I. Use of Silicon Nanowire Sensors for Early Cancer Diagnosis. Molecules 2021, 26, 3734. https://doi.org/10.3390/molecules26123734
Ivanov YD, Romanova TS, Malsagova KA, Pleshakova TO, Archakov AI. Use of Silicon Nanowire Sensors for Early Cancer Diagnosis. Molecules. 2021; 26(12):3734. https://doi.org/10.3390/molecules26123734
Chicago/Turabian StyleIvanov, Yuri D., Tatyana S. Romanova, Kristina A. Malsagova, Tatyana O. Pleshakova, and Alexander I. Archakov. 2021. "Use of Silicon Nanowire Sensors for Early Cancer Diagnosis" Molecules 26, no. 12: 3734. https://doi.org/10.3390/molecules26123734
APA StyleIvanov, Y. D., Romanova, T. S., Malsagova, K. A., Pleshakova, T. O., & Archakov, A. I. (2021). Use of Silicon Nanowire Sensors for Early Cancer Diagnosis. Molecules, 26(12), 3734. https://doi.org/10.3390/molecules26123734







