Phenolics-Rich Extracts of Dietary Plants as Regulators of Fructose Uptake in Caco-2 Cells via GLUT5 Involvement
Abstract
:1. Introduction
2. Results
2.1. The Effect of Extracts on Cell Metabolic Activity
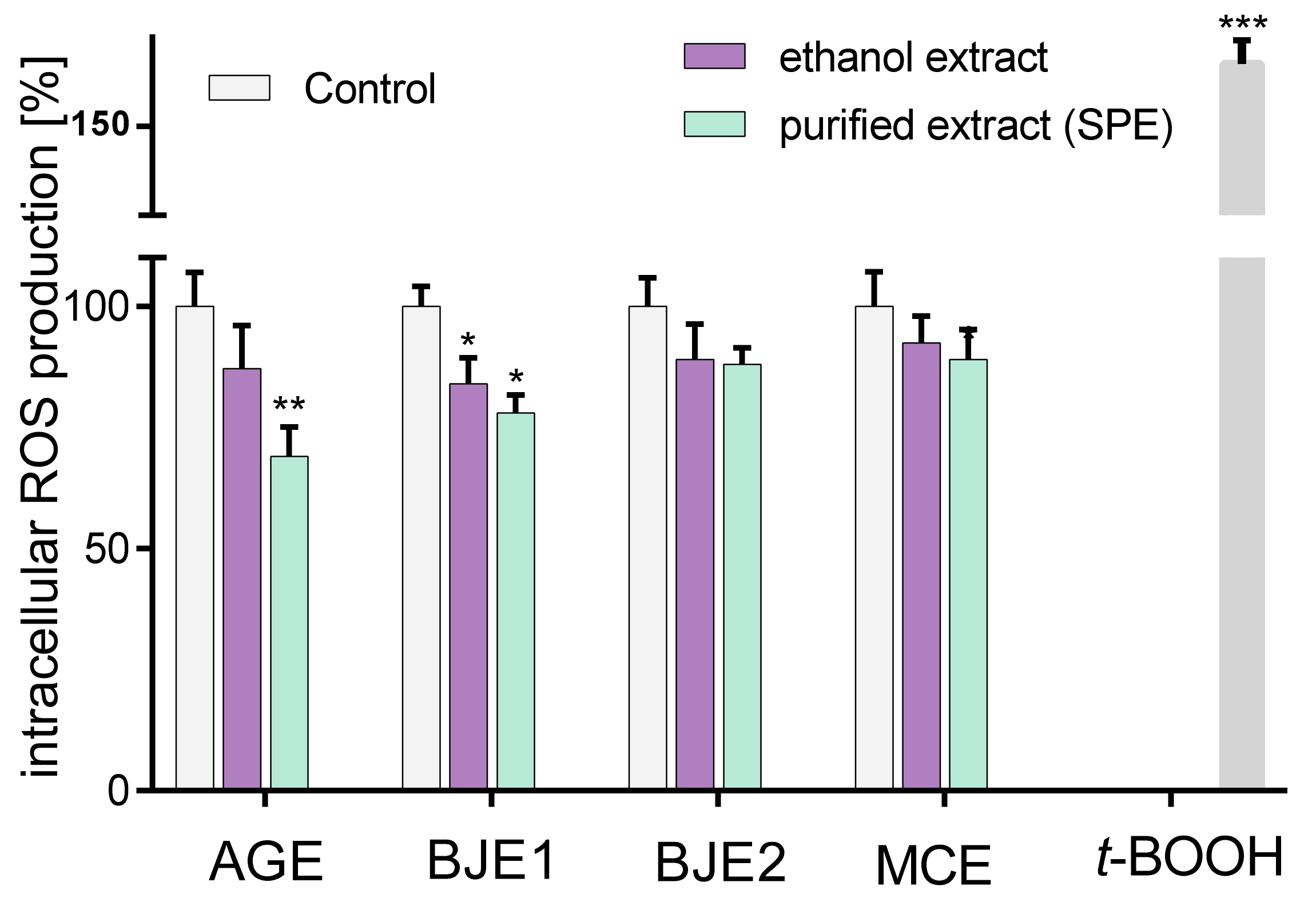
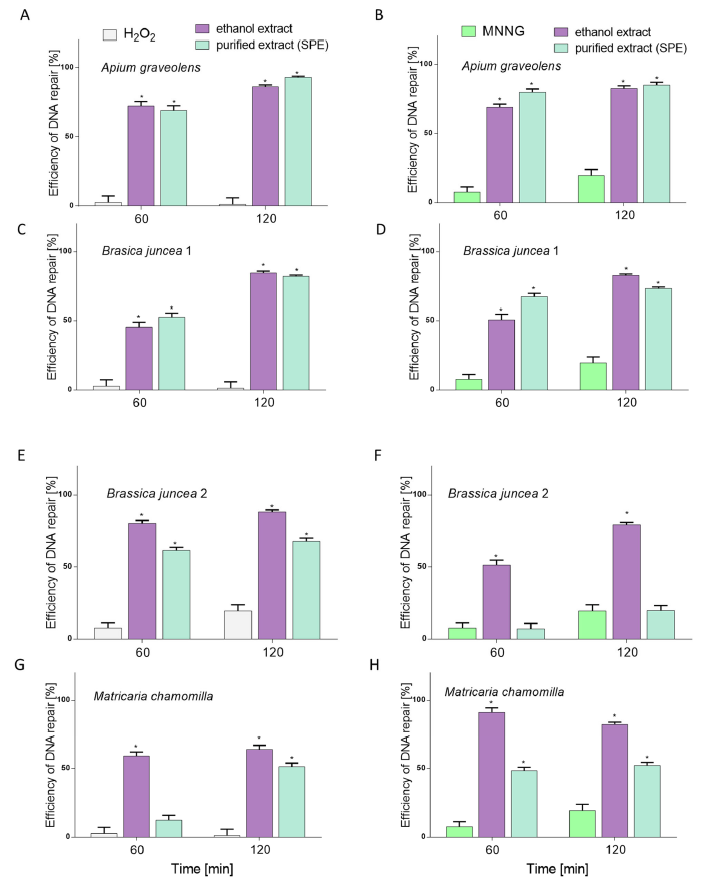
2.2. The Effect of Extracts on Intracellular Reactive Oxygen Species Production and DNA Repair
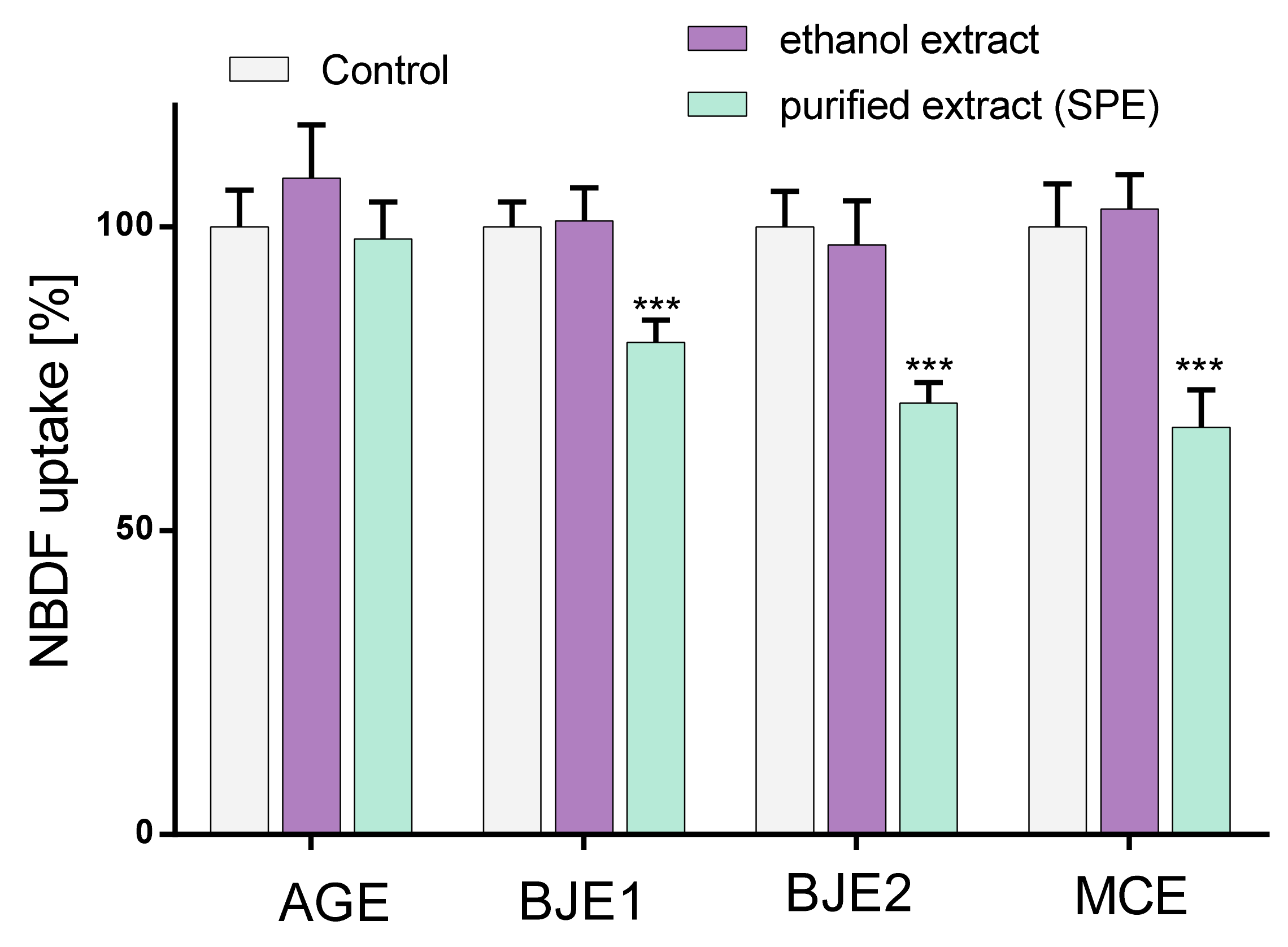
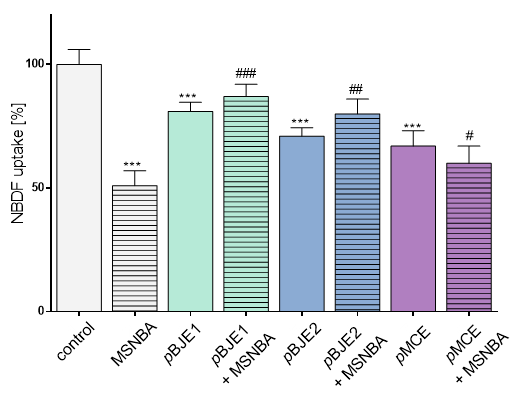
2.3. The Influence of Extracts on Fructose Uptake
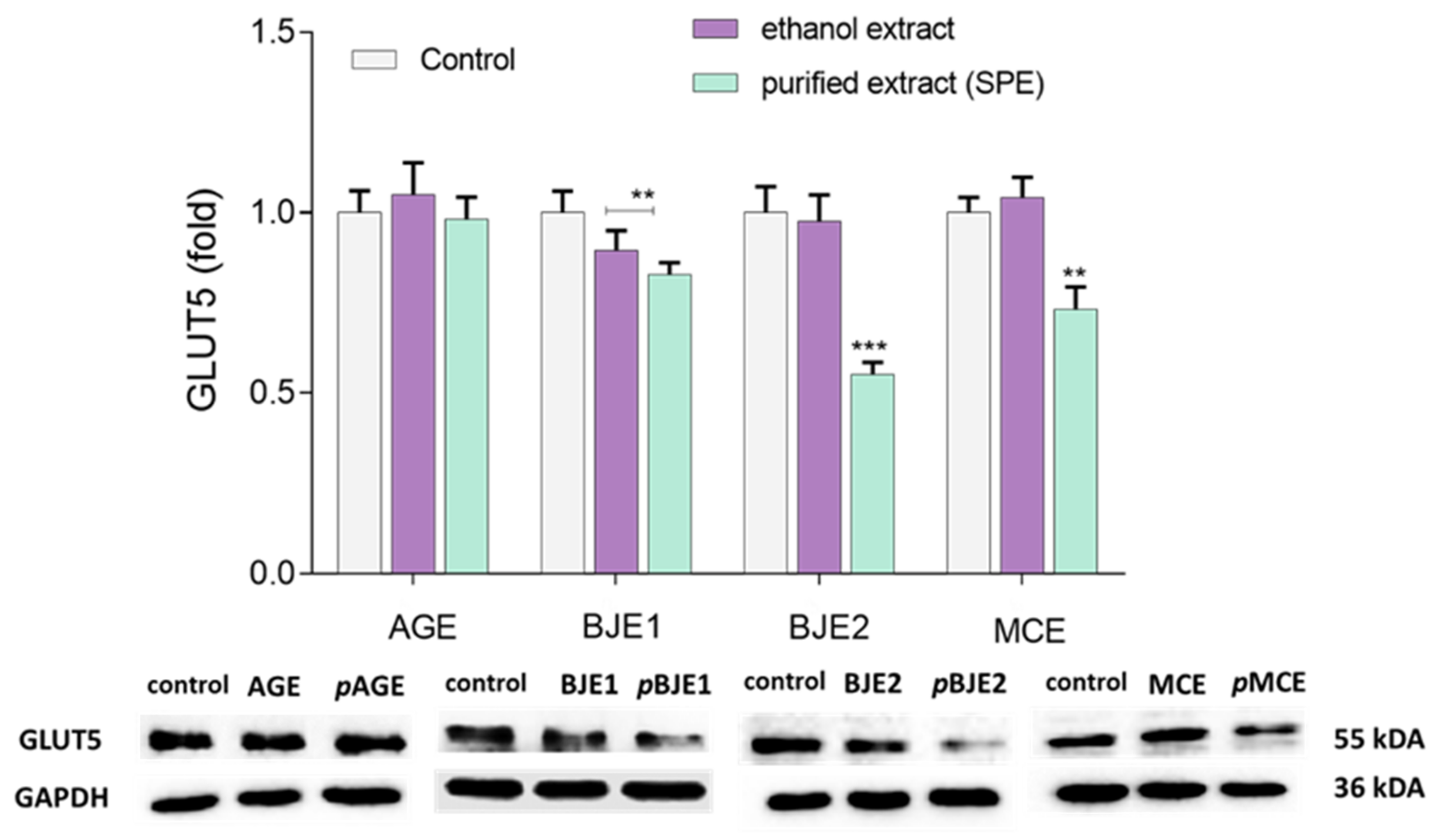
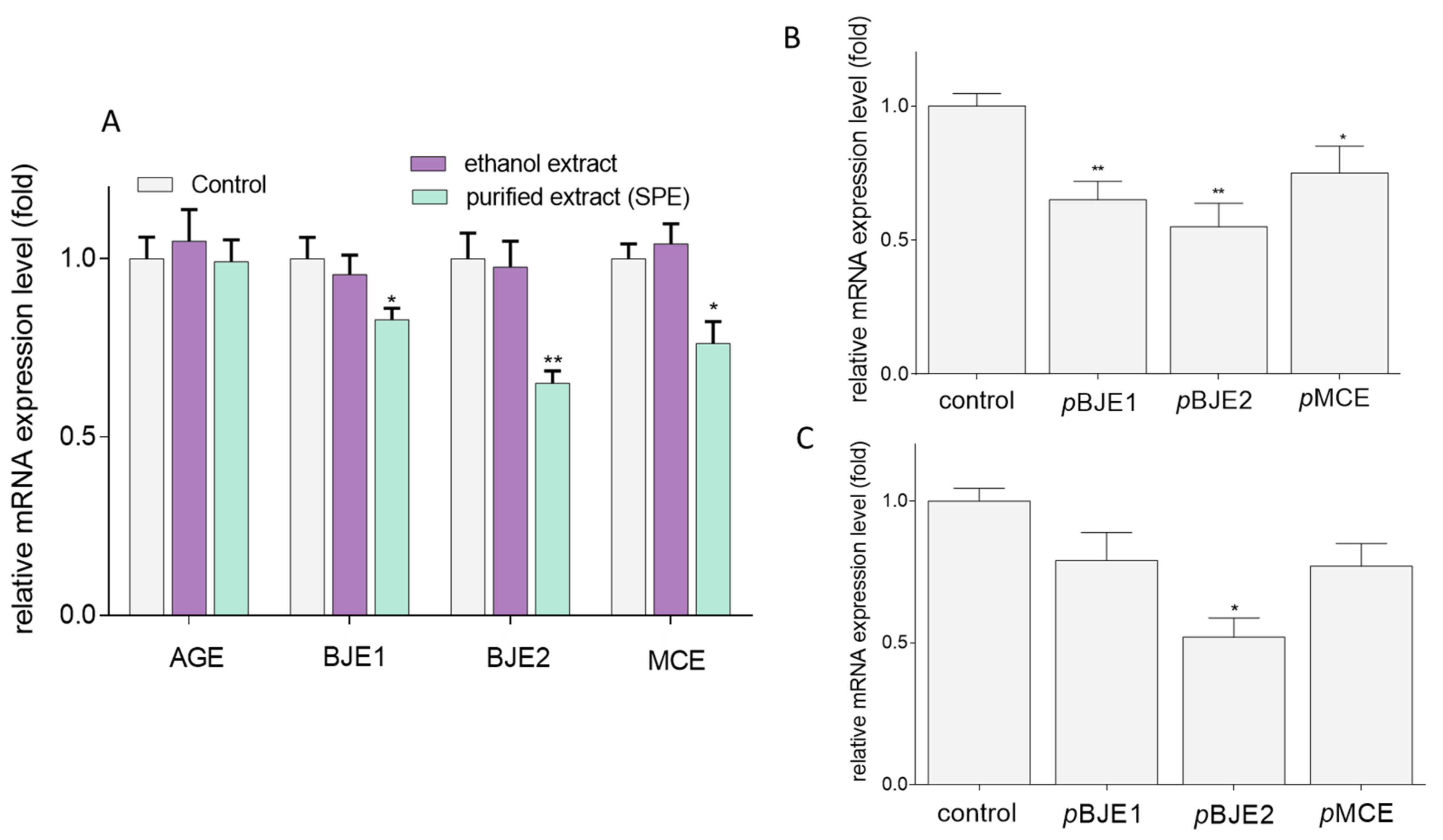
2.4. The Effects of Extracts on GLUT5 Level
2.5. Phenolic Compounds Profile and Content Determined by UPLC Method
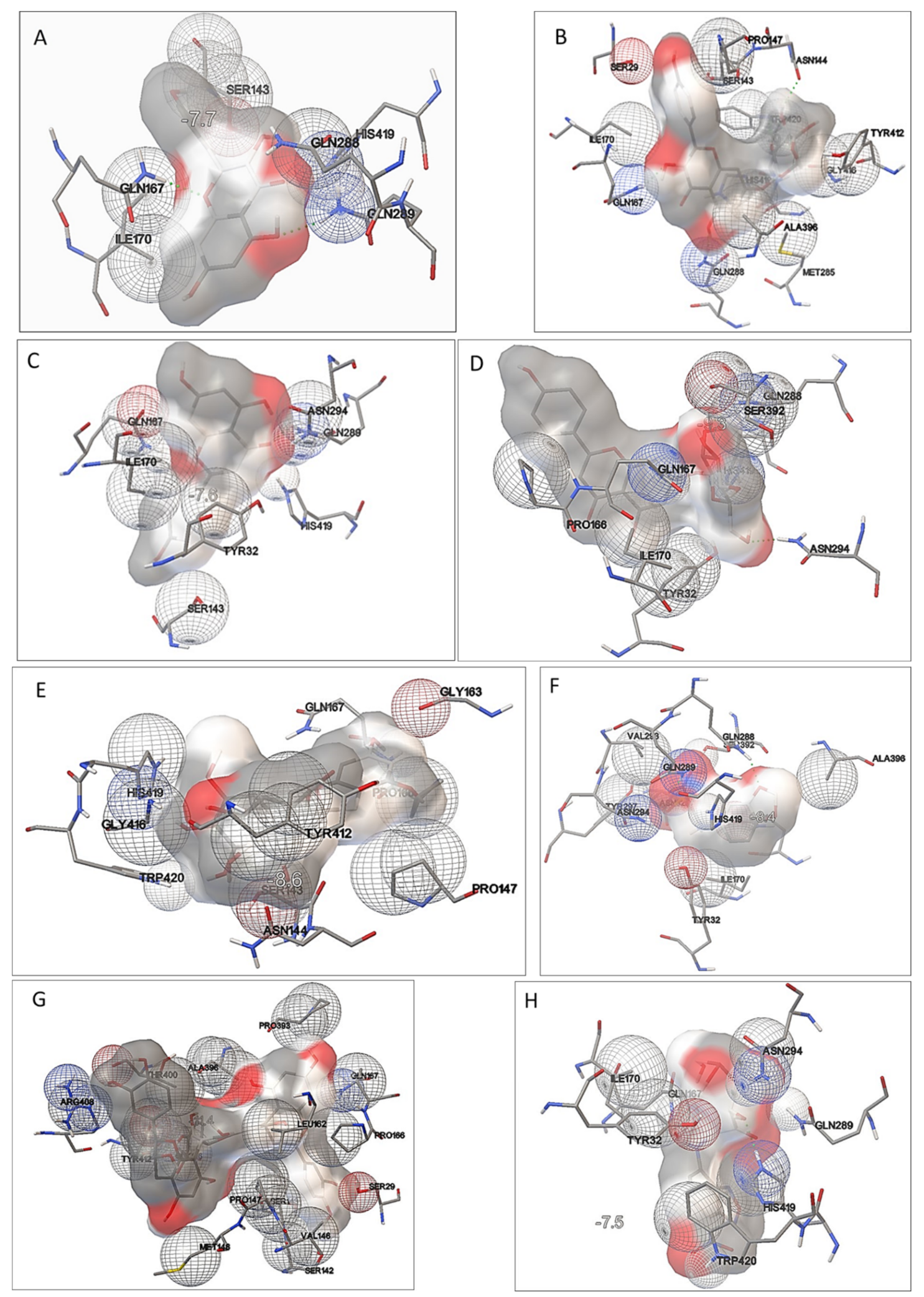
2.6. Determination of Phenolic Constituents’ Binding Site in GLUT5 Model
3. Discussion
4. Materials and Methods
4.1. Preparation of Plant Extracts
4.2. Determination of Phenolic Compounds Composition by UPLC Method
4.3. Cell Culture and Exposure Conditions
4.4. Cell Viability
4.5. Detection of Intracellular Reactive Oxygen Species Generation
4.6. Western Blot Analysis
4.7. DNA Damage and Repair
4.8. Gene Expression Analysis
4.9. NBDF Uptake
4.10. Molecular Docking Simulations
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Douard, V.; Ferraris, R.P. The role of fructose transporters in diseases linked to excessive fructose intake. J. Physiol. 2013, 591, 401–414. [Google Scholar] [CrossRef]
- Hannou, S.A.; Haslam, D.E.; McKeown, N.M.; Herman, M.A. Fructose metabolism and metabolic disease. J. Clin. Investig. 2018, 128, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Mai, B.H.; Yan, L.J. The negative and detrimental effects of high fructose on the liver, with special reference to metabolic disorders. Diabetes Metab. Syndr. Obes. Targets Ther. 2019, 12, 821–826. [Google Scholar] [CrossRef] [Green Version]
- Douard, V.; Ferraris, R.P. Regulation of the fructose transporter GLUT5 in health and disease. Am. J. Physiol. Endocrinol. Metab. 2008, 295, E227–E237. [Google Scholar] [CrossRef] [Green Version]
- Iizuka, K. The role of carbohydrate response element binding protein in intestinal and hepatic fructose metabolism. Nutrients 2017, 9, 181. [Google Scholar] [CrossRef] [Green Version]
- Chyau, C.C.; Wang, H.F.; Zhang, W.J.; Chen, C.C.; Huang, S.H.; Chang, C.C.; Peng, R.Y. Antrodan alleviates high-fat and high-fructose diet-induced fatty liver disease in C57BL/6 mice model via AMPK/Sirt1/SREBP-1c/PPARγ pathway. Int. J. Mol. Sci. 2020, 21, 360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gugliucci, A. Fructose surges damage hepatic adenosyl-monophosphate-dependent kinase and lead to increased lipogenesis and hepatic insulin resistance. Med. Hypotheses 2016, 93, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Nomura, N.; Verdon, G.; Kang, H.J.; Shimamura, T.; Nomura, Y.; Sonoda, Y.; Hussien, S.A.; Qureshi, A.A.; Coincon, M.; Sato, Y.; et al. Structure and mechanism of the mammalian fructose transporter GLUT5. Nature 2015, 526, 397–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gauer, J.S.; Tumova, S.; Lippiat, J.D.; Kerimi, A.; Williamson, G. Differential patterns of inhibition of the sugar transporters GLUT2, GLUT5 and GLUT7 by flavonoids. Biochem. Pharmacol. 2018, 152, 11–20. [Google Scholar] [CrossRef]
- Patel, C.; Douard, V.; Yu, S.; Gao, N.; Ferraris, R.P. Transport, metabolism, and endosomal traffickingdependent regulation of intestinal fructose absorption. FASEB J. 2015, 29, 4046–4058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ter Horst, K.W.; Serlie, M.J. Fructose consumption, lipogenesis, and non-alcoholic fatty liver disease. Nutrients 2017, 9, 981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, T.; Abdelmalek, M.F.; Sullivan, S.; Nadeau, K.J.; Green, M.; Roncal, C.; Nakagawa, T.; Kuwabara, M.; Sato, Y.; Kang, D.H.; et al. Fructose and sugar: A major mediator of non-alcoholic fatty liver disease. J. Hepatol. 2018, 68, 1063–1075. [Google Scholar] [CrossRef] [Green Version]
- Patel, C.; Douard, V.; Yu, S.; Tharabenjasin, P.; Gao, N.; Ferraris, R.P. Fructose-induced increases in expression of intestinal fructolytic and gluconeogenic genes are regulated by GLUT5 and KHK. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2015, 309, R499–R509. [Google Scholar] [CrossRef] [Green Version]
- Zwarts, I.; van Zutphen, T.; Kruit, J.K.; Liu, W.; Oosterveer, M.H.; Verkade, H.J.; Uhlenhaut, N.H.; Jonker, J.W. Identification of the fructose transporter GLUT5 (SLC2A5) as a novel target of nuclear receptor LXR. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Gugliucci, A. Formation of fructose-mediated advanced glycation end products and their roles in metabolic and inflammatory diseases. Adv. Nutr. 2017, 8, 54–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, A.M.G.; Ursu, O.; Babkin, P.; Iancu, C.V.; Whang, A.; Oprea, T.I.; Choe, J.Y. Discovery of a specific inhibitor of human GLUT5 by virtual screening and in vitro transport evaluation. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, R.S.; Pons, J.L.; Labesse, G. Insights into substrate and inhibitor selectivity among human GLUT transporters through comparative modeling and molecular docking. ACS Omega 2019, 4, 4748–4760. [Google Scholar] [CrossRef]
- Thorens, B.; Mueckler, M. The SLC2 (GLUT) family of membrane transporters. Mol. Asp. Med. 2014, 34, 121–138. [Google Scholar]
- Yan, N. Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem. Sci. 2013, 38, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Ebert, K.; Ludwig, M.; Geillinger, K.E.; Schoberth, G.C.; Essenwanger, J.; Stolz, J.; Daniel, H.; Witt, H. Reassessment of GLUT7 and GLUT9 as putative fructose and glucose transporters. J. Membr. Biol. 2017, 250, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Lim, Y.; Kwon, O. Selected phytochemicals and culinary plant extracts inhibit fructose uptake in caco-2 cells. Molecules 2015, 20, 17393–17404. [Google Scholar] [CrossRef] [Green Version]
- Villa-Rodriguez, J.A.; Aydin, E.; Gauer, J.S.; Pyner, A.; Williamson, G.; Kerimi, A. Green and Chamomile Teas, but not acarbose, attenuate glucose and fructose transport via inhibition of GLUT2 and GLUT5. Mol. Nutr. Food Res. 2017, 61, 1–14. [Google Scholar] [CrossRef]
- Andrade, N.; Araújo, J.R.; Correia-Branco, A.; Carletti, J.V.; Martel, F. Effect of dietary polyphenols on fructose uptake by human intestinal epithelial (Caco-2) cells. J. Funct. Foods 2017, 36, 429–439. [Google Scholar] [CrossRef]
- Loureiro, G.; Martel, F. The effect of dietary polyphenols on intestinal absorption of glucose and fructose: Relation with obesity and type 2 diabetes. Food Rev. Int. 2019, 35, 390–406. [Google Scholar] [CrossRef]
- Schreck, K.; Melzig, M.F. Fructose transporters and their inhibition by natural plant extracts in Caco-2 cells. Molecules 2018, 23, 2544. [Google Scholar] [CrossRef] [Green Version]
- Satsu, H.; Awara, S.; Unno, T.; Shimizu, M. Suppressive effect of nobiletin and epicatechin gallate on fructose uptake in human intestinal epithelial Caco-2 cells. Biosci. Biotechnol. Biochem. 2018, 82, 636–646. [Google Scholar] [CrossRef] [Green Version]
- Kooti, W.; Daraei, N. A Review of the antioxidant activity of aelery (Apium graveolens L). J. Evid.-Based Complement. Altern. Med. 2017, 22, 1029–1034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emad, A.M.; Ali, S.F.; Abdel-Rahman, E.A.; Meselhy, M.R.; Farag, M.A.; Ali, S.S.; Abdel-Sattar, E.A. Anti-inflammatory and antioxidant effects of Apium graveolens L. extracts mitigate against fatal acetaminophen-induced acute liver toxicity. J. Food Biochem. 2020, 44, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.K.; Xu, C.C.; Zhang, L.; Ma, H.; Chen, X.J.; Sui, Y.C.; Zhang, H.Z. Evaluation of bioactive components and antioxidant capacity of four celery (Apium graveolens L.) leaves and petioles. Int. J. Food Prop. 2020, 23, 1097–1109. [Google Scholar] [CrossRef]
- Parikh, H.; Pandita, N.; Khanna, A. Phytoextract of Indian mustard seeds acts by suppressing the generation of ROS against acetaminophen-induced hepatotoxicity in HepG2 cells. Pharm. Biol. 2015, 53, 975–984. [Google Scholar] [CrossRef] [Green Version]
- Golubkina, N.; Kekina, H.; Caruso, G. Yield, quality and antioxidant properties of indian mustard (Brassica juncea L.) in response to foliar biofortification with selenium and iodine. Plants 2018, 7, 80. [Google Scholar] [CrossRef] [Green Version]
- Park, S.Y.; Jang, H.L.; Lee, J.H.; Choi, Y.; Kim, H.; Hwang, J.; Seo, D.; Kim, S.; Nam, J.S. Changes in the phenolic compounds and antioxidant activities of mustard leaf (Brassica juncea) kimchi extracts during different fermentation periods. Food Sci. Biotechnol. 2017, 26, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Villa-Rodriguez, J.A.; Kerimi, A.; Abranko, L.; Tumova, S.; Ford, L.; Blackburn, R.S.; Rayner, C.; Williamson, G. Acute metabolic actions of the major polyphenols in chamomile: An in vitro mechanistic study on their potential to attenuate postprandial hyperglycaemia. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Alpers, D. In vivo versus cell culture models: The example of lipid biosynthesis in the enterocyte in vivo versus cell culture models: The example of lipid biosynthesis in the enterocyte. Future Lipidol. 2017, 3, 367–370. [Google Scholar] [CrossRef]
- Kwon, O.; Eck, P.; Chen, S.; Corpe, C.P.; Lee, J.; Kruhlak, M.; Levine, M. Inhibition of the intestinal glucose transporter GLUT2 by flavonoids. FASEB J. 2007, 21, 366–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levi, J.; Cheng, Z.; Gheysens, O.; Patel, M.; Chan, C.T.; Wang, Y.; Namavari, M.; Gambhir, S.S. Fluorescent fructose derivatives for imaging breast cancer cells. Bioconjug. Chem. 2007, 18, 628–634. [Google Scholar] [CrossRef] [Green Version]
- Zakłos-Szyda, M.; Pietrzyk, N.; Szustak, M.; Podsędek, A. Viburnum opulus l. Juice phenolics inhibit mouse 3T3-L1 cells adipogenesis and pancreatic lipase activity. Nutrients 2020, 12, 2003. [Google Scholar] [CrossRef] [PubMed]
- Koch, W. Dietary polyphenols-important non-nutrients in the prevention of chronic noncommunicable diseases. A systematic review. Nutrients 2019, 11, 1039. [Google Scholar] [CrossRef] [Green Version]
- Williamson, G. Possible effects of dietary polyphenols on sugar absorption and digestion. Mol. Nutr. Food Res. 2013, 57, 48–57. [Google Scholar] [CrossRef]
- Mahraoui, L.; Rodolosse, A.; Barbat, A.; Dussaulx, E.; Zweibaum, A.; Rousset, M.; Brot-Laroche, E. Presence and differential expression of SGLT1, GLUT1, GLUT2, GLUT3 and GLUT5 hexose-transporter mRNAs in Caco-2 cell clones in relation to cell growth and glucose consumption. Biochem. J. 1994, 298 Pt 3, 629–633. [Google Scholar] [CrossRef] [Green Version]
- Pietrzyk, N.; Zakłos-Szyda, M.; Koziołkiewicz, M.; Podsędek, A. Viburnum opulus L. fruit phenolic compounds protect against FFA-induced steatosis of HepG2 cells via AMPK pathway. J. Funct. Foods 2021, 80, 104437. [Google Scholar] [CrossRef]
- Zakłos-Szyda, M.; Pawlik, N.; Polka, D.; Nowak, A.; Koziołkiewicz, M.; Podsędek, A. Viburnum opulus fruit phenolic compounds as cytoprotective agents able to decrease free fatty acids and glucose uptake by Caco-2 cells. Antioxidants 2019, 8, 262. [Google Scholar] [CrossRef] [Green Version]
- Zakłos-Szyda, M.; Kowalska-Baron, A.; Pietrzyk, N.; Drzazga, A. Evaluation of Viburnum opulus L. fruit phenolics cytoprotective potential on insulinoma MIN6 cells relevant for diabetes mellitus and obesity. Antioxidants 2020, 9, 433. [Google Scholar] [CrossRef]
- Zakłos-Szyda, M.; Nowak, A.; Pietrzyk, N.; Podsędek, A. Viburnum opulus L. juice phenolic compounds influence osteogenic differentiation in human osteosarcoma Saos-2 cells. Int. J. Mol. Sci. 2020, 21, 4909. [Google Scholar] [CrossRef] [PubMed]
- Kulig, D.; Matysiak, M.; Baldovská, S.; Štefániková, J.; Maruniaková, N.; Mňahončáková, E.; árvay, J.; Galbavý, D.; Kolesárová, A. Screening of polyphenolic compounds from traditional medicinal herbs. J. Microbiol. Biotechnol. Food Sci. 2019, 9, 487–491. [Google Scholar] [CrossRef]
- Priecina, L.; Karklina, D.; Kince, T. The impact of steam-blanching and dehydration on phenolic, organic acid composition, and total carotenoids in celery roots. Innov. Food Sci. Emerg. Technol. 2018, 49, 192–201. [Google Scholar] [CrossRef]
- Yao, Y.; Sang, W.; Zhou, M.; Ren, G. Phenolic composition and antioxidant activities of 11 celery cultivars. J. Food Sci. 2010, 75, C9–C13. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.Z.; Lu, S.; Harnly, J.M. Detection and quantification of glycosylated flavonoid malonates in celery, chinese celery, and celery seed by LC-DAD-ESI/MS. J. Agric. Food Chem. 2007, 55, 1321–1326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, Y.; Ren, G. Effect of thermal treatment on phenolic composition and antioxidant activities of two celery cultivars. LWT-Food Sci. Technol. 2011, 44, 181–185. [Google Scholar] [CrossRef]
- Haghi, G.; Hatami, A.; Safaei, A.; Mehran, M. Analysis of phenolic compounds in Matricaria chamomilla and its extracts by UPLC-UV. Res. Pharm. Sci. 2014, 9, 31–37. [Google Scholar] [PubMed]
- Nováková, L.; Vildová, A.; Mateus, J.P.; Gonalves, T.; Solich, P. Development and application of UHPLC-MS/MS method for the determination of phenolic compounds in Chamomile flowers and Chamomile tea extracts. Talanta 2010, 82, 1271–1280. [Google Scholar] [CrossRef]
- Cvetanović, A.; Švarc-Gajić, J.; Zeković, Z.; Jerković, J.; Zengin, G.; Gašić, U.; Tešić, Ž.; Mašković, P.; Soares, C.; Fatima Barroso, M.; et al. The influence of the extraction temperature on polyphenolic profiles and bioactivity of chamomile (Matricaria chamomilla L.) subcritical water extracts. Food Chem. 2019, 271, 328–337. [Google Scholar] [CrossRef] [Green Version]
- Sotiropoulou, N.S.; Megremi, S.F.; Tarantilis, P. Evaluation of antioxidant activity, toxicity, and phenolic profile of aqueous extracts of chamomile (Matricaria chamomilla L.) and sage (Salvia ocinalis L.) prepared at different temperatures. Appl. Sci. 2020, 10, 2270. [Google Scholar] [CrossRef] [Green Version]
- Đorđević, B.S.; Todorović, Z.B.; Troter, D.Z.; Stanojević, L.P.; Stojanović, G.S.; Đalović, I.G.; Mitrović, P.M.; Veljković, V.B. Extraction of phenolic compounds from black mustard (Brassica nigra L.) seed by deep eutectic solvents. J. Food Meas. Charact. 2021, 15, 1931–1938. [Google Scholar] [CrossRef]
- Aziz, S.; El-Zayat, M.; El-Khateeb, A. Phytochemical characterization, antioxidant and antimicrobial activities of Brassica juncea (L.) mustard seeds aqueous and ethanolic extracts. J. Plant Prod. 2020, 11, 85–88. [Google Scholar] [CrossRef]
- Fang, Z.; Hu, Y.; Liu, D.; Chen, J.; Ye, X. Changes of phenolic acids and antioxidant activities during potherb mustard (Brassica juncea, Coss.) pickling. Food Chem. 2008, 108, 811–817. [Google Scholar] [CrossRef]
- Lin, L.Z.; Harnly, J.M. Phenolic component profiles of mustard greens, yu choy, and 15 other Brassica vegetables. J. Agric. Food Chem. 2010, 58, 6850–6857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, L.Z.; Sun, J.; Chen, P.; Harnly, J. UHPLC-PDA-ESI/HRMS/MSn analysis of anthocyanins, flavonol glycosides, and hydroxycinnamic acid derivatives in red mustard greens (Brassica juncea Coss variety). J. Agric. Food Chem. 2011, 59, 12059–12072. [Google Scholar] [CrossRef] [Green Version]
- Häkkinen, S.H.; Törrönen, A.R. Content of flavonols and selected phenolic acids in strawberries and Vaccinium species: Influence of cultivar, cultivation site and technique. Food Res. Int. 2000, 33, 517–524. [Google Scholar] [CrossRef]
- Amaral, J.S.; Valentão, P.; Andrade, P.B.; Martins, R.C.; Seabra, R.M. Phenolic composition of hazelnut leaves: Influence of cultivar, geographical origin and ripening stage. Sci. Hortic. 2010, 126, 306–313. [Google Scholar] [CrossRef]
- López-Yerena, A.; Lozano-Castellón, J.; Olmo-Cunillera, A.; Tresserra-Rimbau, A.; Quifer-Rada, P.; Jiménez, B.; Pérez, M.; Vallverdú-Queralt, A. Effects of organic and conventional growing systems on the phenolic profile of extra-virgin olive oil. Molecules 2019, 24, 1986. [Google Scholar] [CrossRef] [Green Version]
- Rafraf, M.; Zemestani, M.; Asghari-Jafarabadi, M. Effectiveness of chamomile tea on glycemic control and serum lipid profile in patients with type 2 diabetes. J. Endocrinol. Investig. 2015, 38, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Zemestani, M.; Rafraf, M.; Asghari-Jafarabadi, M. Chamomile tea improves glycemic indices and antioxidants status in patients with type 2 diabetes mellitus. Nutrition 2016, 32, 66–72. [Google Scholar] [CrossRef]
- Moser, S.; Lim, J.; Chegeni, M.; Wightman, J.L.D.; Hamaker, B.R.; Ferruzzi, M.G. Concord and niagara grape juice and their phenolics modify intestinal glucose transport in a coupled in vitro digestion/Caco-2 human intestinal model. Nutrients 2016, 8, 414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Tan, J.; Zhang, L.; Liu, Y.; Cheng, Y.; Zhang, Q.; Ding, H. Apigenin ameliorates vascular injury in rats with high fructose-induced metabolic disturbance by inhibiting PI3K/AKT/GLUT1. RSC Adv. 2018, 8, 24470–24476. [Google Scholar] [CrossRef] [Green Version]
- Johnston, K.L.; Clifford, M.N.; Morgan, L.M. Coffee acutely modifies gastrointestinal hormone secretion and glucose tolerance in humans: Glycemic effects of chlorogenic acid and caffeine. Am. J. Clin. Nutr. 2003, 78, 728–733. [Google Scholar] [CrossRef]
- Cao, H.; Ou, J.; Chen, L.; Zhang, Y.; Szkudelski, T.; Delmas, D.; Daglia, M.; Xiao, J. Dietary polyphenols and type 2 diabetes: Human study and clinical trial. Crit. Rev. Food Sci. Nutr. 2019, 59, 3371–3379. [Google Scholar] [CrossRef]
- Shah, A.; Dagdeviren, S.; Lewandowski, J.P.; Schmider, A.B.; Ricci-Blair, E.M.; Natarajan, N.; Hundal, H.; Noh, H.L.; Friedline, R.H.; Vidoudez, C.; et al. Thioredoxin interacting protein is required for a chronic energy-rich diet to promote intestinal fructose absorption. iScience 2020, 23, 101521. [Google Scholar] [CrossRef]
- Zhang, W.; Tang, R.; Ba, G.; Li, M.; Lin, H. Anti-allergic and anti-inflammatory effects of resveratrol via inhibiting TXNIP-oxidative stress pathway in a mouse model of allergic rhinitis. World Allergy Organ. J. 2020, 13, 100473. [Google Scholar] [CrossRef]
- Oh, A.; Sohn, S.; Lee, J.; Park, J.; Nam, K.T.; Kim, Y.; Lee, H.; Cha, J.; Gil, L.; Cancer, Y.; et al. ChREBP deficiency leads to diarrhea-predominant irritable bowel syndrome. Metabolism 2020, 85, 286–297. [Google Scholar] [CrossRef]
- Noblet, B.; Benhamed, F.; O-Sullivan, I.S.; Zhang, W.; Filhoulaud, G.; Montagner, A.; Polizzi, A.; Marmier, S.; Burnol, A.F.; Guilmeau, S.; et al. Dual regulation of TxNIP by ChREBP and FoxO1 in liver. iScience 2021, 24, 102218. [Google Scholar] [CrossRef]
- Samantha, R.; Almalik, D. Loss of intestinal ChREBP impairs absorption of dietary sugars and prevents glycemic excursion curves. bioRxiv 2021, 444615. [Google Scholar]
- Koepsell, H. Glucose transporters in the small intestine in health and disease. Pflug. Arch. Eur. J. Physiol. 2020, 472, 1207–1248. [Google Scholar] [CrossRef]
- Viscarra, J.; Sul, H.S. Epigenetic regulation of hepatic lipogenesis: Role in hepatosteatosis and diabetes. Diabetes 2020, 69, 525–531. [Google Scholar] [CrossRef]
- Honma, K.; Mochizuki, K.; Goda, T. Induction by fructose force-feeding of histone H3 and H4 acetylation at their lysine residues around the Slc2a5 gene and its expression in mice. Biosci. Biotechnol. Biochem. 2013, 77, 2188–2191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanwal, R.; Datt, M.; Liu, X.; Gupta, S. Dietary flavones as dual inhibitors of DNA methyltransferases and histone methyltransferases. PLoS ONE 2016, 11, 1–19. [Google Scholar]
- Scafuri, B.; Bontempo, P.; Altucci, L.; De Masi, L.; Facchiano, A. Molecular docking simulations on histone deacetylases (Hdac)-1 and-2 to investigate the flavone binding. Biomedicines 2020, 8, 1. [Google Scholar] [CrossRef]
- Pandey, M.; Kaur, P.; Shukla, S.; Abbas, A.; Fu, P.; Gupta, S. Plant flavone apigenin inhibits HDAC and remodels chromatin to induce growth arrest and apoptosis in human prostate cancer cells: In vitro and in vivo study. Mol. Carcinog. 2012, 51, 952–962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paredes-Gonzalez, X.; Fuentes, F.; Su, Z.Y.; Kong, A.N.T. Apigenin reactivates Nrf2 anti-oxidative stress signaling in mouse skin epidermal JB6 P + cells through epigenetics modifications. AAPS J. 2014, 16, 727–735. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Jiang, Z.H.; Li, C.G.; Zhu, Y.J.; Li, Z.; Tang, Y.Z.; Ni, C.L. Apigenin prevents metabolic syndrome in high-fructose diet-fed mice by Keap1-Nrf2 pathway. Biomed. Pharmacother. 2018, 105, 1283–1290. [Google Scholar] [CrossRef]
- Holman, G.D. Structure, function and regulation of mammalian glucose transporters of the SLC2 family. Pflug. Arch. Eur. J. Physiol. 2020, 472, 1155–1175. [Google Scholar] [CrossRef] [PubMed]
- Pantsar, T.; Poso, A. Binding affinity via docking: Fact and fiction. Molecules 2018, 23, 1899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mobley, D.L.; Dill, K.A. Binding of small-molecule ligands to proteins: “what you see” is not always “what you get”. Structure 2009, 17, 489–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ju, J.; Wei, S.J.; Savira, F.; Suharsono, Y.; Aragão, R.; Linsi, L.; Editor, B.; Reeger, U.; Sievers, W.; Michalopoulou, C.; et al. Improved software with new docking methods, expanded force field, and Python bindings. J. Chem. Inf. Model. 2020, 43, 7728. [Google Scholar]
- Trott, O.; Olson, A.J. Autodock vina: Improving the speed and accuracy of docking. J. Comput. Chem. 2019, 31, 455–461. [Google Scholar]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09. Revision, E.01; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Morris, G.M.; Ruth, H.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Software news and updates AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
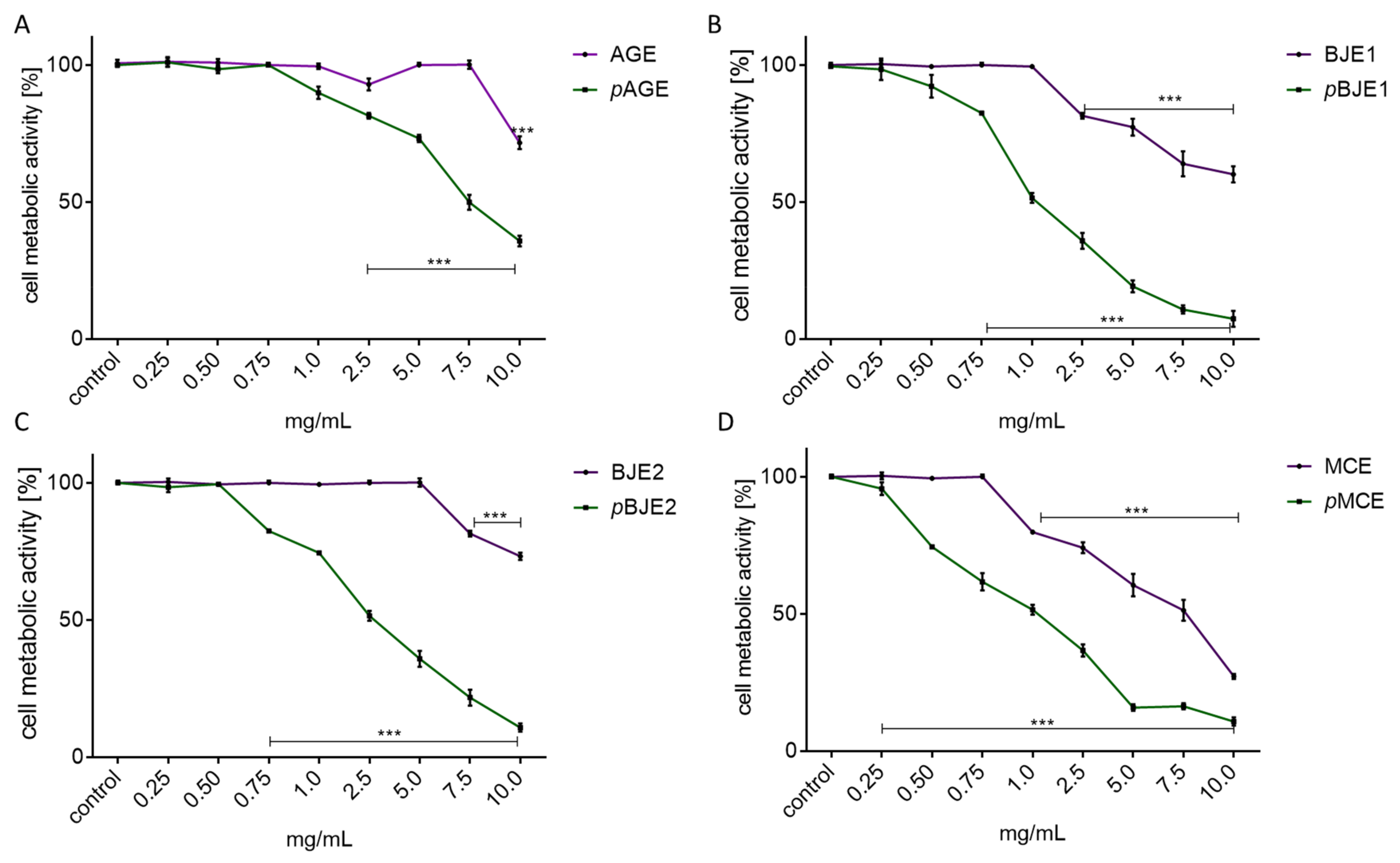


| Extract | AGE | pAGE | BJE1 | pBJE1 | BJE2 | pBJ2 | MCE | pMCE |
|---|---|---|---|---|---|---|---|---|
| IC0 [mg/mL] | >2.50 | 0.75 | 2.00 | 0.25 | 2.50 | 0.75 | >2.50 | 0.25 |
| The Compound Identified | λmax [nm] | Rt | Quantification [mg/g of Extract] | |||
|---|---|---|---|---|---|---|
| pAGE | pMCE | pBJE2 | pBJE1 | |||
| HYDROXYBENZOIC AND HYDROXYCINNAMIC ACIDS | ||||||
| Gallic acid | 270 | 2.0 | 12.68 ± 0.00 b | n.d. | n.d. | 3.96 ± 0.10 a |
| Chlorogenic acid | 324 | 4.88 | n.d. | 0.96 ± 0.01 | n.d | n.d. |
| Caffeic acid | 324 | 6.50 | n.d. | 0.81 ± 0.00 a | 2.02 ± 0.04 b | n.d. |
| 3-Coumaric acid | 277 | 9.24 | 6.24 ± 0.30 b | 0.17 ± 0.01 a | n.d. | n.d. |
| Salicylic acid | 303 | 10.70 | 41.88 ± 3.19 c | 28.00 ± 0.18 b | 24.63 ± 2.17 a | n.d. |
| 3,5-Dicaffeoylquinic acid | 326 | 10.85 | 6.23 ± 0.02 a | n.d. | 12.85 ± 0.31 b | n.d. |
| Ferulic acid | 315 | 11.90 | 5.19 ± 0.01 a | 36.20 ± 0.08 c | 17.78 ± 1.41 d | 7.25 ± 0.04 b |
| Sinapic acid | 326 | 12.20 | n.d. | n.d. | 6.40 ± 0.22 | n.d. |
| FLAVANOLS | ||||||
| Procyanidin B2 | 286 | 6.51 | 7.11 ± 0.67 | n.d. | n.d. | n.d. |
| (-)-Epicatechin | 284 | 7.00 | 6.27 ± 0.04 | n.d. | n.d. | n.d. |
| Procyanidin C1 | 278 | 7.87 | 256.76 ± 6.74 c | n.d. | 2.32 ± 0.10 a | 70.43 ± 9.94 b |
| Epigallocatechin-3-gallate | 274 | 9.50 | 6.30 ± 0.01 b | 4.94 ± 0.17 a | 51.62 ± 0.18 c | n.d. |
| Epicatechin-3-gallate | 276 | 11.23 | 6.51 ± 0.24 | n.d. | n.d. | n.d. |
| FLAVONOLS | ||||||
| Rutin | 355 | 9.70 | n.d. | n.d. | 25.28 ± 1.53 b | 4.98 ± 0.02 a |
| Quercetin-3-O-glucoside | 350 | 9.91 | n.d. | 1.90 ± 0.03 b | 27.70 ± 0.47 c | 0.45 ± 0.13 a |
| Quercetin-3-O-glucuronide | 350 | 9.98 | n.d. | n.d. | 22.20 ± 0.64 | n.d. |
| Kaempferol-3-O-glucoside | 353 | 10.75 | n.d. | 6.21 ± 0.04 b | 18.56 ± 0.53 c | 10.40 ± 0.08 a |
| Isorhamnetin-3-O-rutinoside | 345 | 12.20 | n.d. | n.d. | n.d. | 4.72 ± 0.22 |
| Isorhamnetin-3-O-glucoside | 352 | 12.42 | n.d. | 20.17 ± 0.44 c | 1.05 ± 0.12 a | 9.83 ± 0.26 b |
| Kaempferol | 352 | 12.98 | n.d. | n.d. | n.d. | 30.97 ± 0.14 |
| FLAVONS | ||||||
| Apigenin-7-O-glucoside | 336 | 13.00 | 302.01 ± 14.43 b | n.d. | 43.66 ± 1.65 a | n.d. |
| Luteolin | 369 | 13.18 | n.d. | 6.45 ± 0.46 | n.d. | n.d. |
| Apigenin | 336 | 13.20 | n.d. | 40.47 ± 1.01 a | 199.12 ± 0.85 c | 165.81 ± 0.45 b |
| TOTAL PHENOLICS | - | 680.20 ± 22.42 d | 153.07 ± 0.71 a | 455.19 ± 2.61 c | 308.79 ± 9.79 b | |
| Ligand | Binding Affinity | Residues Interacting with a Ligand | |
|---|---|---|---|
| kcal/mol | kJ/mol | ||
| Kaempferol | −7.7 | −32.2 | Gln288 *, Gln289, Ile170, Ser143, His419, Gln167 * |
| Kaempferol-3-O-glycoside | −9.4 | −39.3 | Pro147, Asn144, Ser143, Ser29, Ile170, Gln167 *, Trp420, His419, Tyr412, Gly416, Met285, Gln288 *, Ala396 |
| Apigenin | −7.6 | −31.8 | Gln289 *, Ile170, Ser143, His419, Gln167, Tyr32, Asn294 |
| Apigenin-7-O-glucoside | −9.3 | −38.9 | Ser392, Gln167, Tyr32, Ile170, Pro166, His419, Gln288 *, Asn294 * |
| Epigallocatechin-3-gallate (ECGC) | −8.6 | −35.9 | Ser143, Asn144, Tyr412, Gly416, Pro147, Trp420, Pro166, His419,Gln167, Gly163 |
| MSNBA | −8.4 | −35.1 | Tyr297, Asn325 *, Val293, Asn294, Gln289 *, Gln288 *, Ala396, Gln167, Ile170, Ser392, His419, Tyr32 |
| Procyanidin C1 | −11.4 | −47.7 | Gly416, Tyr412, Arg408, Ala396, Ser143, Ser142, Ser29, Pro147, Pro166, Leu162, Pro393, Gln167, Met148, Thr400, Val146 |
| NBDF | −7.5 | −31.4 | Gln167, His419 *, Ile170, Tyr32, Trp420,Asn294,Gln289 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zakłos-Szyda, M.; Pietrzyk, N.; Kowalska-Baron, A.; Nowak, A.; Chałaśkiewicz, K.; Ratajewski, M.; Budryn, G.; Koziołkiewicz, M. Phenolics-Rich Extracts of Dietary Plants as Regulators of Fructose Uptake in Caco-2 Cells via GLUT5 Involvement. Molecules 2021, 26, 4745. https://doi.org/10.3390/molecules26164745
Zakłos-Szyda M, Pietrzyk N, Kowalska-Baron A, Nowak A, Chałaśkiewicz K, Ratajewski M, Budryn G, Koziołkiewicz M. Phenolics-Rich Extracts of Dietary Plants as Regulators of Fructose Uptake in Caco-2 Cells via GLUT5 Involvement. Molecules. 2021; 26(16):4745. https://doi.org/10.3390/molecules26164745
Chicago/Turabian StyleZakłos-Szyda, Małgorzata, Nina Pietrzyk, Agnieszka Kowalska-Baron, Adriana Nowak, Katarzyna Chałaśkiewicz, Marcin Ratajewski, Grażyna Budryn, and Maria Koziołkiewicz. 2021. "Phenolics-Rich Extracts of Dietary Plants as Regulators of Fructose Uptake in Caco-2 Cells via GLUT5 Involvement" Molecules 26, no. 16: 4745. https://doi.org/10.3390/molecules26164745








