Donkey Milk Fermentation by Lactococcus lactis subsp. cremoris and Lactobacillus rhamnosus Affects the Antiviral and Antibacterial Milk Properties
Abstract
:1. Introduction
2. Results and Discussion
2.1. Microbial Characterization of the Donkey Milk
2.2. Screening of the LAB for Peptide Production
2.3. Antioxidant and Metal-Chelating Activity
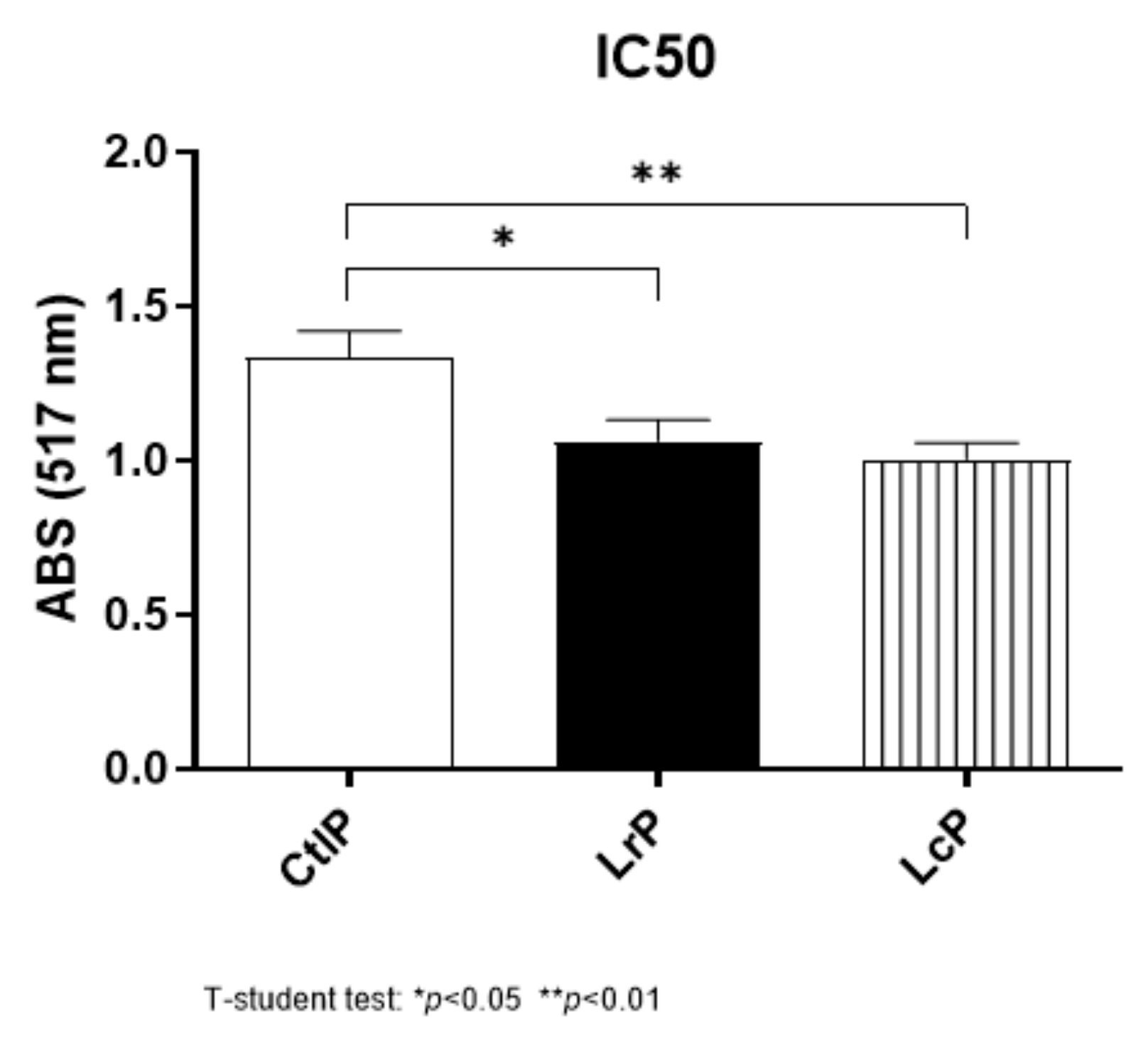
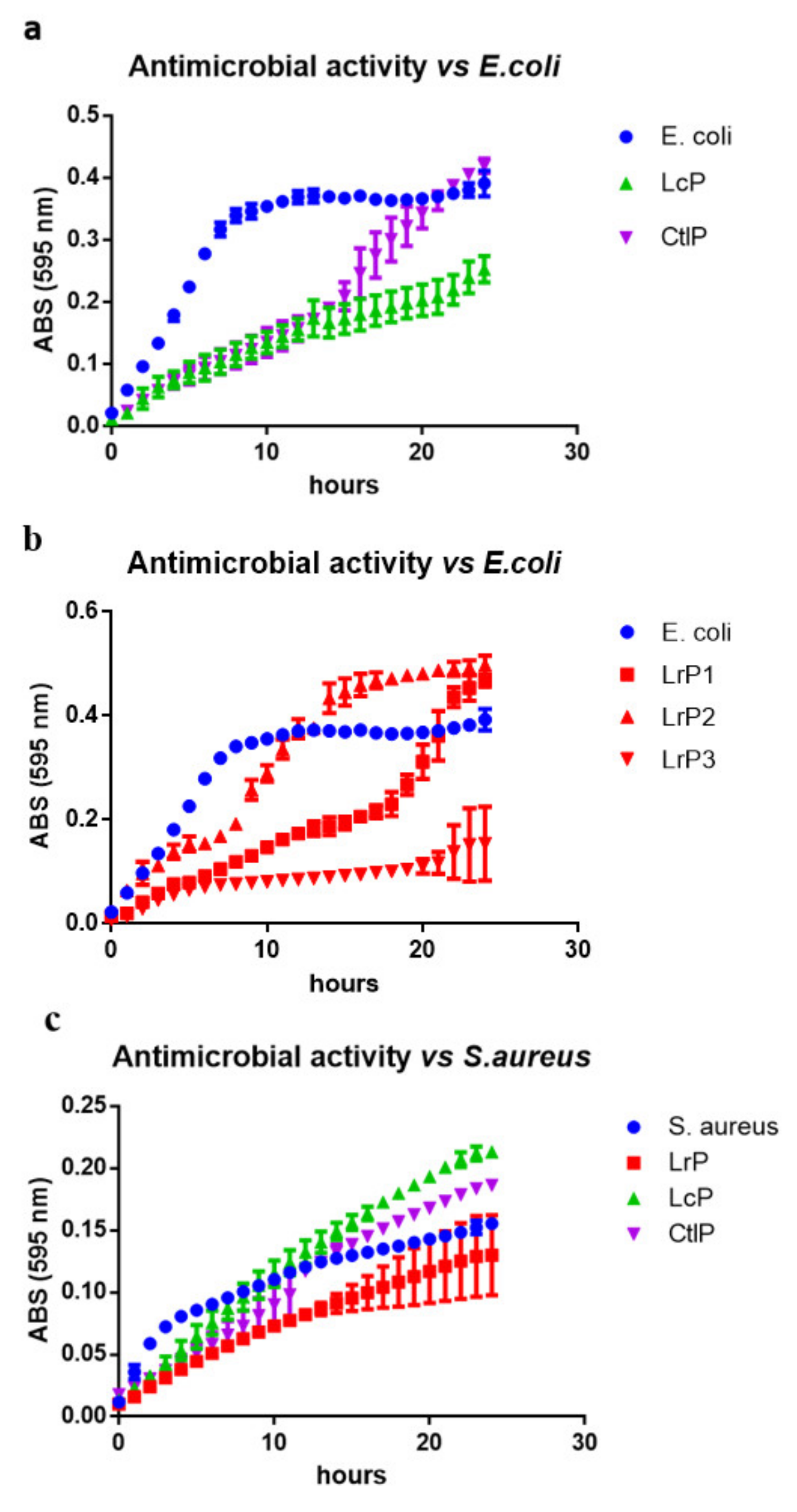
2.4. Antibacterial Activity
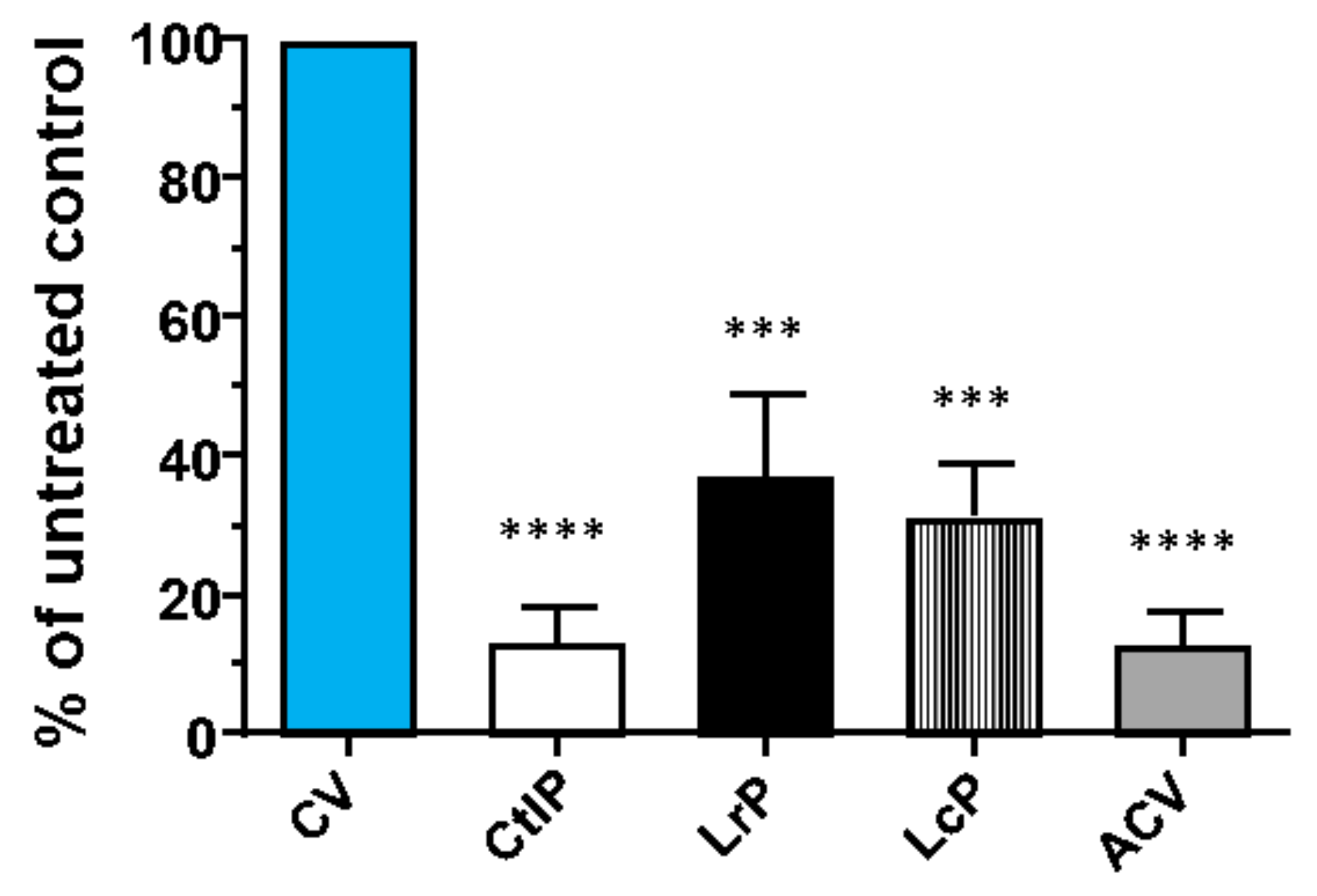
2.5. Antiviral Activity
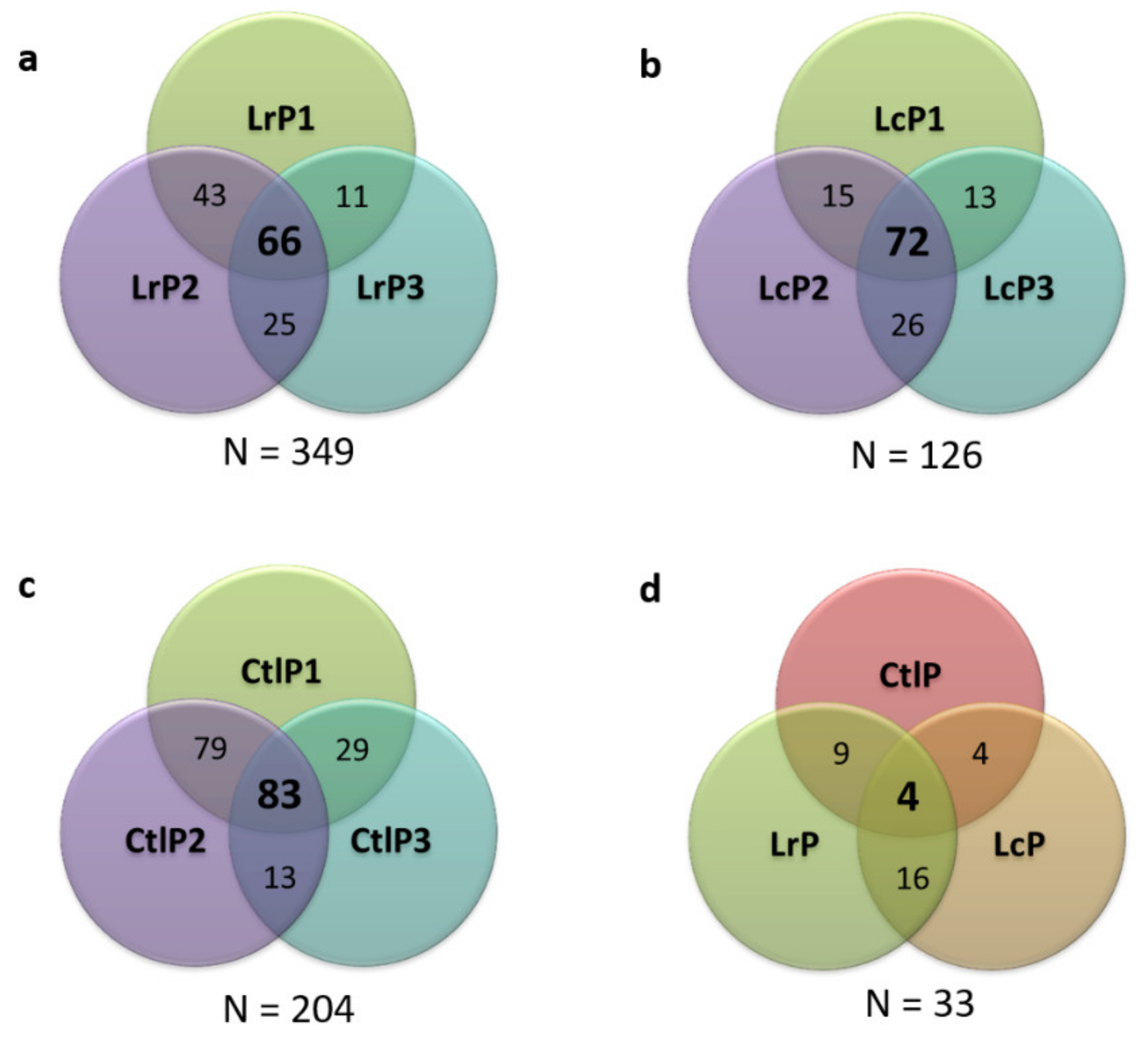
2.6. Characterization of the Peptide Profile of the Donkey Milk Samples
3. Materials and Methods
3.1. Donkey Milk
3.2. Microbial Strains and Growth Conditions
3.3. Characterization of the Endogenous Microflora of the Donkey Milk
3.4. Selection of the LAB Strains for the Proteolytic Activity
3.5. Peptide Production from Donkey Milk by Selected Strains
3.6. Peptide Extraction and Purification
3.7. Evaluation of the Peptide Bioactivities
3.7.1. Antioxidant Activity
3.7.2. Metal-Chelating Activity
3.7.3. Antibacterial Activity
3.7.4. Antiviral Activity
3.8. Peptide Identification by Means of LC-MS/MS Analysis
3.9. Peptide Data Searc
3.10. Peptide in Silico Analyses
3.11. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Hartmann, R.; Meisel, H. Food-derived peptides with biological activity: From research to food applications. Curr. Opin. Biotechnol. 2007, 18, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Lemes, A.C.; Sala, L.; Ores, J.D.C.; Braga, A.R.C.; Egea, M.B.; Fernandes, K.F. A review of the latest advances in encrypted bioactive peptides from protein-richwaste. Int. J. Mol. Sci. 2016, 17, 950. [Google Scholar] [CrossRef] [Green Version]
- Korhonen, H. Milk-derived bioactive peptides: From science to applications. J. Funct. Foods 2009, 1, 177–187. [Google Scholar] [CrossRef]
- Meisel, H. Multifunctional peptides encrypted in milk proteins. BioFactors 2004, 21, 55–61. [Google Scholar] [CrossRef]
- Hafeez, Z.; Cakir-Kiefer, C.; Roux, E.; Perrin, C.; Miclo, L.; Dary-Mourot, A. Strategies of producing bioactive peptides from milk proteins to functionalize fermented milk products. Food Res. Int. 2014, 63, 71–80. [Google Scholar] [CrossRef]
- Elfahri, K.R.; Vasiljevic, T.; Yeager, T.; Donkor, O.N. Anti-colon cancer and antioxidant activities of bovine skim milk fermented by selected Lactobacillus helveticus strains. J. Dairy Sci. 2016, 99, 31–40. [Google Scholar] [CrossRef]
- Savijoki, K.; Ingmer, H.; Varmanen, P. Proteolytic systems of lactic acid bacteria. Appl. Microbiol. Biotechnol. 2006, 71, 394–406. [Google Scholar] [CrossRef]
- Silva, S.V.; Malcata, F.X. Caseins as source of bioactive peptides. Int. Dairy J. 2005, 15, 1–15. [Google Scholar] [CrossRef]
- Pessione, E. Lactic acid bacteria contribution to gut microbiota complexity: Lights and shadows. Front. Cell. Infect. Microbiol. 2012, 2, 86. [Google Scholar] [CrossRef] [Green Version]
- Gobbetti, M.; Stepaniak, L.; De Angelis, M.; Corsetti, A.; Di Cagno, R. Latent bioactive peptides in milk proteins: Proteolytic activation and significance in dairy processing. Crit. Rev. Food Sci. Nutr. 2002, 42, 223–239. [Google Scholar] [CrossRef] [PubMed]
- Bruni, N.; Capucchio, M.T.; Biasibetti, E.; Pessione, E.; Cirrincione, S.; Giraudo, L.; Corona, A.; Dosio, F. Antimicrobial activity of lactoferrin-related peptides and applications in human and veterinary medicine. Molecules 2016, 21, 752. [Google Scholar] [CrossRef] [PubMed]
- Lemes, A.C.; De Paula, L.C.; Filho, J.G.O. Bioactive Peptides with Antihypertensive Property Obtained from Agroindustrial Byproducts—Mini Review. Austin J. Nutr. Metab. 2020, 7, 1082. [Google Scholar]
- Pessione, E.; Cirrincione, S. Bioactive molecules released in food by lactic acid bacteria: Encrypted peptides and biogenic amines. Front. Microbiol. 2016, 7, 876. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Ledesma, B.; Dávalos, A.; Bartolomé, B.; Amigo, L. Preparation of antioxidant enzymatic hydrolysates from α-lactalbumin and β-lactoglobulln. Identification of active peptides by HPLC-MS/MS. J. Agric. Food Chem. 2005, 53, 588–593. [Google Scholar] [CrossRef]
- Prioult, G.; Pecquet, S.; Fliss, I. Stimulation of Interleukin-10 Production by Acidic -Lactoglobulin-Derived Peptides Hydrolyzed with Lactobacillus paracasei NCC2461 Peptidases. Clin. Vaccine Immunol. 2004, 11, 266–271. [Google Scholar] [CrossRef] [Green Version]
- Daliri, E.; Oh, D.; Lee, B. Bioactive Peptides. Foods 2017, 6, 32. [Google Scholar] [CrossRef]
- Barlowska, J.; Szwajkowska, M.; Litwińczuk, Z.; Król, J. Nutritional Value and Technological Suitability of Milk from Various Animal Species Used for Dairy Production. Compr. Rev. Food Sci. Food Saf. 2011, 10, 291–302. [Google Scholar] [CrossRef]
- Flom, J.D.; Sicherer, S.H. Epidemiology of Cow ’s Milk Allergy Gastrointestinal Symptoms. Nutrients 2019, 11, 1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Claeys, W.L.; Verraes, C.; Cardoen, S.; De Block, J.; Huyghebaert, A.; Raes, K.; Dewettinck, K.; Herman, L. Consumption of raw or heated milk from different species: An evaluation of the nutritional and potential health benefits. Food Control 2014, 42, 188–201. [Google Scholar] [CrossRef]
- Tidona, F.; Sekse, C.; Criscione, A.; Jacobsen, M.; Bordonaro, S.; Marletta, D.; Vegarud, G.E. Antimicrobial effect of donkeys’ milk digested in vitro with human gastrointestinal enzymes. Int. Dairy J. 2011, 21, 158–165. [Google Scholar] [CrossRef]
- Piovesana, S.; Capriotti, A.L.; Cavaliere, C.; La Barbera, G.; Samperi, R.; Zenezini Chiozzi, R.; Laganà, A. Peptidome characterization and bioactivity analysis of donkey milk. J. Proteom. 2015, 119, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Aspri, M.; Leni, G.; Galaverna, G.; Papademas, P. Bioactive properties of fermented donkey milk, before and after in vitro simulated gastrointestinal digestion. Food Chem. 2018, 268, 476–484. [Google Scholar] [CrossRef]
- Martins, M.L.; Pinto, C.L.O.; Rocha, R.B.; de Araújo, E.F.; Vanetti, M.C.D. Genetic diversity of Gram-negative, proteolytic, psychrotrophic bacteria isolated from refrigerated raw milk. Int. J. Food Microbiol. 2006, 111, 144–148. [Google Scholar] [CrossRef] [PubMed]
- De Jonghe, V.; Coorevits, A.; De Block, J.; Van Coillie, E.; Grijspeerdt, K.; Herman, L.; De Vos, P.; Heyndrickx, M. Toxinogenic and spoilage potential of aerobic spore-formers isolated from raw milk. Int. J. Food Microbiol. 2010, 136, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Zhao, L.; Jiang, L.; Dong, M.L.; Ren, F.Z. The antimicrobial activity of donkey milk and its microflora changes during storage. Food Control 2008, 19, 1191–1195. [Google Scholar] [CrossRef]
- Leroy, F.; De Vuyst, L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 2004, 15, 67–78. [Google Scholar] [CrossRef]
- Brady, C.; Cleenwerck, I.; Venter, S.; Coutinho, T.; De Vos, P. Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): Proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov. Syst. Appl. Microbiol. 2013, 36, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Xin, L.; Meng, Z.; Zhang, L.; Cui, Y.; Han, X.; Yi, H. The diversity and proteolytic properties of psychrotrophic bacteria in raw cows’ milk from North China. Int. Dairy J. 2017, 66, 34–41. [Google Scholar] [CrossRef]
- Wang, J.; Jonkers, H.M.; Boon, N.; De Belie, N. Bacillus sphaericus LMG 22257 is physiologically suitable for self-healing concrete. Appl. Microbiol. Biotechnol. 2017, 101, 5101–5114. [Google Scholar] [CrossRef] [PubMed]
- Barghouthi, S.A. A Universal Method for the Identification of Bacteria Based on General PCR Primers. Indian J. Microbiol. 2011, 51, 430–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, S. Bacteria, viruses, membrane-enclosed microentities and fungi as the environmental evolutionary entities coexisting in non-human mammalian milk. J. Theor. Fimpology. 2014, 2, 1–16. [Google Scholar]
- Machado, S.G.; da Silva, F.L.; Bazzolli, D.M.S.; Heyndrickx, M.; Costa, P.M.D.A.; Vanetti, M.C.D. Pseudomonas spp. and Serratia liquefaciens as Predominant Spoilers in Cold Raw Milk. J. Food Sci. 2015, 80, M1842–M1849. [Google Scholar] [CrossRef] [PubMed]
- Zadoks, R.N.; Middleton, J.R.; McDougall, S.; Katholm, J.; Schukken, Y.H. Molecular epidemiology of mastitis pathogens of dairy cattle and comparative relevance to humans. J. Mammary Gland Biol. Neoplasia 2011, 16, 357–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zenezini Chiozzi, R.; Capriotti, A.L.; Cavaliere, C.; La Barbera, G.; Piovesana, S.; Samperi, R.; Laganà, A. Purification and identification of endogenous antioxidant and ACE-inhibitory peptides from donkey milk by multidimensional liquid chromatography and nanoHPLC-high resolution mass spectrometry. Anal. Bioanal. Chem. 2016, 408, 5657–5666. [Google Scholar] [CrossRef]
- Virtanen, T.; Pihlanto, A.; Akkanen, S.; Korhonen, H. Development of antioxidant activity in milk whey during fermentation with lactic acid bacteria. J. Appl. Microbiol. 2007, 102, 106–115. [Google Scholar] [CrossRef]
- Rizzello, C.G.; Lorusso, A.; Russo, V.; Pinto, D.; Marzani, B.; Gobbetti, M. Improving the antioxidant properties of quinoa flour through fermentation with selected autochthonous lactic acid bacteria. Int. J. Food Microbiol. 2017, 241, 252–261. [Google Scholar] [CrossRef]
- Maryati, Y.; Susilowati, A.; Melanie, H.; Lotulung, P.D. Characteristic of phenolic compound and antioxidant activity of fermented broccoli (Brassica oleracea L. ssp.) beverage by lactic acid bacteria (LAB). AIP Conf. Proc. 2017, 1803. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Vorland, L.H.; Ulvatne, H.; Rekdal, Ø.; Svendsen, J.S. Initial binding sites of antimicrobial peptides in Staphylococcus aureus and Escherichia coli. Scand. J. Infect. Dis. 1999, 31, 467–473. [Google Scholar] [CrossRef]
- Poole, C.L.; James, S.H. Antiviral Therapies for Herpesviruses: Current Agents and New Directions. Clin. Ther. 2018, 40, 1282–1298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Álvarez, D.M.; Castillo, E.; Duarte, L.F.; Arriagada, J.; Corrales, N.; Farías, M.A.; Henríquez, A.; Agurto-Muñoz, C.; González, P.A. Current Antivirals and Novel Botanical Molecules Interfering with Herpes Simplex Virus Infection. Front. Microbiol. 2020, 11, 139. [Google Scholar] [CrossRef]
- Brumini, D.; Furlund, C.B.; Comi, I.; Devold, T.G.; Marletta, D.; Vegarud, G.E.; Jonassen, C.M. Antiviral activity of donkey milk protein fractions on echovirus type 5. Int. Dairy J. 2013, 28, 109–111. [Google Scholar] [CrossRef]
- Martini, M.; Altomonte, I.; Licitra, R.; Salari, F. Nutritional and Nutraceutical Quality of Donkey Milk. J. Equine Vet. Sci. 2018, 65, 33–37. [Google Scholar] [CrossRef]
- Wang, X.; Sun, Y.; Wang, F.; You, L.; Cao, Y.; Tang, R.; Wen, J.; Cui, X. A novel endogenous antimicrobial peptide CAMP211-225 derived from casein in human milk. Food Funct. 2020, 11, 2291–2298. [Google Scholar] [CrossRef] [PubMed]
- Sedaghati, M.; Ezzatpanah, H.; Mashhadi Akbar Boojar, M.; Tajabadi Ebrahimi, M.; Kobarfard, F. Isolation and identification of some antibacterial peptides in the plasmin-digest of β-casein. LWT Food Sci. Technol. 2016, 68, 217–225. [Google Scholar] [CrossRef]
- Liu, Y.; Eichler, J.; Pischetsrieder, M. Virtual screening of a milk peptide database for the identification of food-derived antimicrobial peptides. Mol. Nutr. Food Res. 2015, 59, 2243–2254. [Google Scholar] [CrossRef]
- Amarowicz, R.; Estrella, I.; Troszyn, A. Its Fractions. J. Food Lipids 2007, 15, 119–136. [Google Scholar] [CrossRef]
- Durak, A.; Baraniak, B.; Jakubczyk, A.; Świeca, M. Biologically active peptides obtained by enzymatic hydrolysis of Adzuki bean seeds. Food Chem. 2013, 141, 2177–2183. [Google Scholar] [CrossRef] [PubMed]
- Luganini, A.; Nicoletto, S.F.; Pizzuto, L.; Pirri, G.; Giuliani, A.; Landolfo, S.; Gribaudo, G. Inhibition of herpes simplex virus type 1 and type 2 infections by peptide-derivatized dendrimers. Antimicrob. Agents Chemother. 2011, 55, 3231–3239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bovio, E.; Garzoli, L.; Poli, A.; Luganini, A.; Villa, P.; Musumeci, R.; Mccormack, G.P.; Cocuzza, C.E.; Gribaudo, G.; Mehiri, M.; et al. Marine Fungi from the sponge grantia compressa: Biodiversity, chemodiversity, and biotechnological potential. Mar. Drugs 2019, 17, 220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luganini, A.; Sibille, G.; Mognetti, B.; Sainas, S.; Pippione, A.C.; Giorgis, M.; Boschi, D.; Lolli, M.L.; Gribaudo, G. Effective deploying of a novel DHODH inhibitor against herpes simplex type 1 and type 2 replication. Antivir. Res. 2021, 189, 105057. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.D.; Beverly, R.L.; Qu, Y.; Dallas, D.C. Milk bioactive peptide database: A comprehensive database of milk protein-derived bioactive peptides and novel visualization. Food Chem. 2017, 232, 673–682. [Google Scholar] [CrossRef] [PubMed]

| Peptide (mg mL−1) | L-Lactic Acid (mg mL−1) | pH | |
|---|---|---|---|
| L. reuteri Lb2 BM-DSM | 0.082 ± 0.005 | 0.568 ± 0.033 | 6.27 ± 0.29 |
| L. helveticus 6E8 | 0.107 ± 0.014 | 0.813 ± 0.019 | 5.21 ± 0.31 |
| L. rhamnosus 17D10 | 0.283 ± 0.008 | 4.501 ± 0.107 | 3.86 ± 0.19 |
| L. lactis subsp cremoris 40FEL3 | 0.221 ± 0.015 | 3.390 ± 0.099 | 4.24 ± 0.28 |
| L. lactis subsp lactis MG1363 | 0.170 ± 0.006 | 0.219 ± 0.023 | 6.33 ± 0.25 |
| Experimental Peptide | Sample | % Similarity (Identity/Blosum 62) | Peptide from Database | Specie | Protein | AA Intervals | Activity |
|---|---|---|---|---|---|---|---|
| DPATQPIVPVHNP | LrP | 60/60 | YPVTQPLAPVHNPIS | Homo sapiens | β-casein | 211–225 | Antimicrobial |
| FDPATQPIVPVHNPV | LcP | 60/70 | YPVTQPLAPVHNPIS | Homo sapiens | β-casein | 211–225 | Antimicrobial |
| FIAIPPKK | CtlP | 70/80 | MAIPPKK | Bos taurus | κ-casein | 127–133 | Antithrombin ACE-inhibitory |
| 60/60 | IAIPP | Homo sapiens | κ-casein | 118–122 | ACE-inhibitory | ||
| 60/60 | AIPPKKNQD | Bos taurus | κ-casein | 128–136 | ACE-inhibitory | ||
| FIAIPPKKLQ | LrP | 70/70 | AIPPKKNQD | Bos taurus | κ-casein | 128–136 | ACE-inhibitory |
| 60/70 | MAIPPKK | Bos taurus | κ-casein | 127–133 | Antithrombin | ||
| KVAPFPQPVVPYPQ | LrP, LcP | 70/70 | VAPFPQPVVP | Equus asinus | β-casein | 176–185 | ACE-inhibitory |
| 60/60 | VLPVPQKAVPYPQR | Bos taurus | β-casein | 185–198 | Antimicrobial | ||
| LPSQPVLSPPQSKVAPFP | LrP | 60/60 | PPQSVLSLSQSKVLPVPQ | Bos taurus | β-casein | 173–190 | ACE-inhibitory |
| PATQPIVPVHNPV | LrP | 60/70 | YPVTQPLAPVHNPIS | Homo sapiens | β-casein | 211–225 | Antimicrobial |
| PATQPIVPVHNPVI | LrP | 60/70 | YPVTQPLAPVHNPIS | Homo sapiens | β-casein | 211–225 | Antimicrobial |
| PATQPIVPVHNPVIV | LrP, LcP, CtlP | 60/70 | YPVTQPLAPVHNPIS | Homo sapiens | β-casein | 211–225 | Antimicrobial |
| QPVVPYPQRDTPVQAF | CtlP | 60/70 | VPYPQRDMPIQAFL | Bos taurus | β-casein | 193–206 | Antimicrobial |
| QSKVAPFPQPVVPYPQ | LrP | 60/70 | VAPFPQPVVP | Equus asinus | β-casein | 176–185 | ACE-inhibitory |
| RDTPVQAFLL | LrP | 60/70 | RDMPIQAF | Bos taurus | β-casein | 198–205 | ACE-inhibitory |
| RDTPVQAFLLYQDPQLGLT | LrP | 60/70 | DMPIQAFLLYQEPVLGPVR | Bos taurus | β-casein | 199–217 | Anti-inflammatory |
| SKVAPFPQPVVPYPQ | LrP, LcP | 60/60 | VLPVPQKAVPYPQR | Bos taurus | β-casein | 185–198 | Antimicrobial |
| VAPFPQPVVP | Equus asinus | β-casein | 176–185 | ACE-inhibitory | |||
| VAPFPQPVVPYPQ | LcP | 70/70 | VAPFPQPVVP | Equus asinus | β-casein | 176–185 | ACE-inhibitory |
| 60/70 | VLPVPQKAVPYPQR | Bos taurus | β-casein | 185–198 | Antimicrobial | ||
| VAPFPQPVVPYPQR | CtlP | 70/70 | VLPVPQKAVPYPQR | Bos taurus | β-casein | 185–198 | Antimicrobial |
| 70/70 | VAPFPQPVVP | Equus asinus | β-casein | 176–185 | ACE-inhibitory | ||
| VVPYPQRDTPVQAF | CtlP | 70/80 | VPYPQRDMPIQAFL | Bos taurus | β-casein | 193–206 | Antimicrobial |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cirrincione, S.; Luganini, A.; Lamberti, C.; Manfredi, M.; Cavallarin, L.; Giuffrida, M.G.; Pessione, E. Donkey Milk Fermentation by Lactococcus lactis subsp. cremoris and Lactobacillus rhamnosus Affects the Antiviral and Antibacterial Milk Properties. Molecules 2021, 26, 5100. https://doi.org/10.3390/molecules26165100
Cirrincione S, Luganini A, Lamberti C, Manfredi M, Cavallarin L, Giuffrida MG, Pessione E. Donkey Milk Fermentation by Lactococcus lactis subsp. cremoris and Lactobacillus rhamnosus Affects the Antiviral and Antibacterial Milk Properties. Molecules. 2021; 26(16):5100. https://doi.org/10.3390/molecules26165100
Chicago/Turabian StyleCirrincione, Simona, Anna Luganini, Cristina Lamberti, Marcello Manfredi, Laura Cavallarin, Maria Gabriella Giuffrida, and Enrica Pessione. 2021. "Donkey Milk Fermentation by Lactococcus lactis subsp. cremoris and Lactobacillus rhamnosus Affects the Antiviral and Antibacterial Milk Properties" Molecules 26, no. 16: 5100. https://doi.org/10.3390/molecules26165100






