NLIP and HAD-like Domains of Pah1 and Lipin 1 Phosphatidate Phosphatases Are Essential for Their Catalytic Activities
Abstract
:1. Introduction
2. Results and Discussion
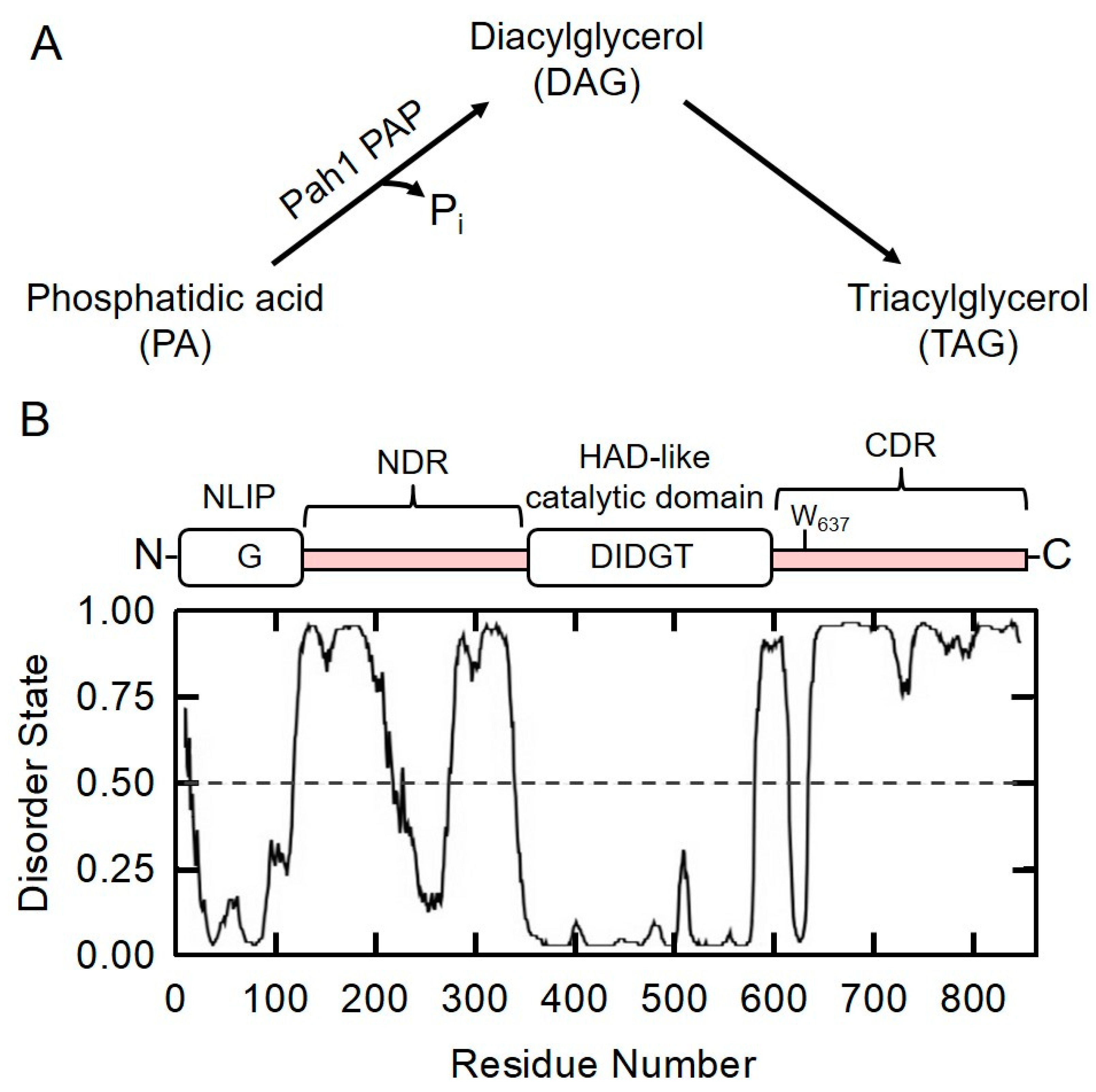
2.1. Prediction of Intrinsically Disordered Regions in Pah1 by DISPRED3 Algorithm
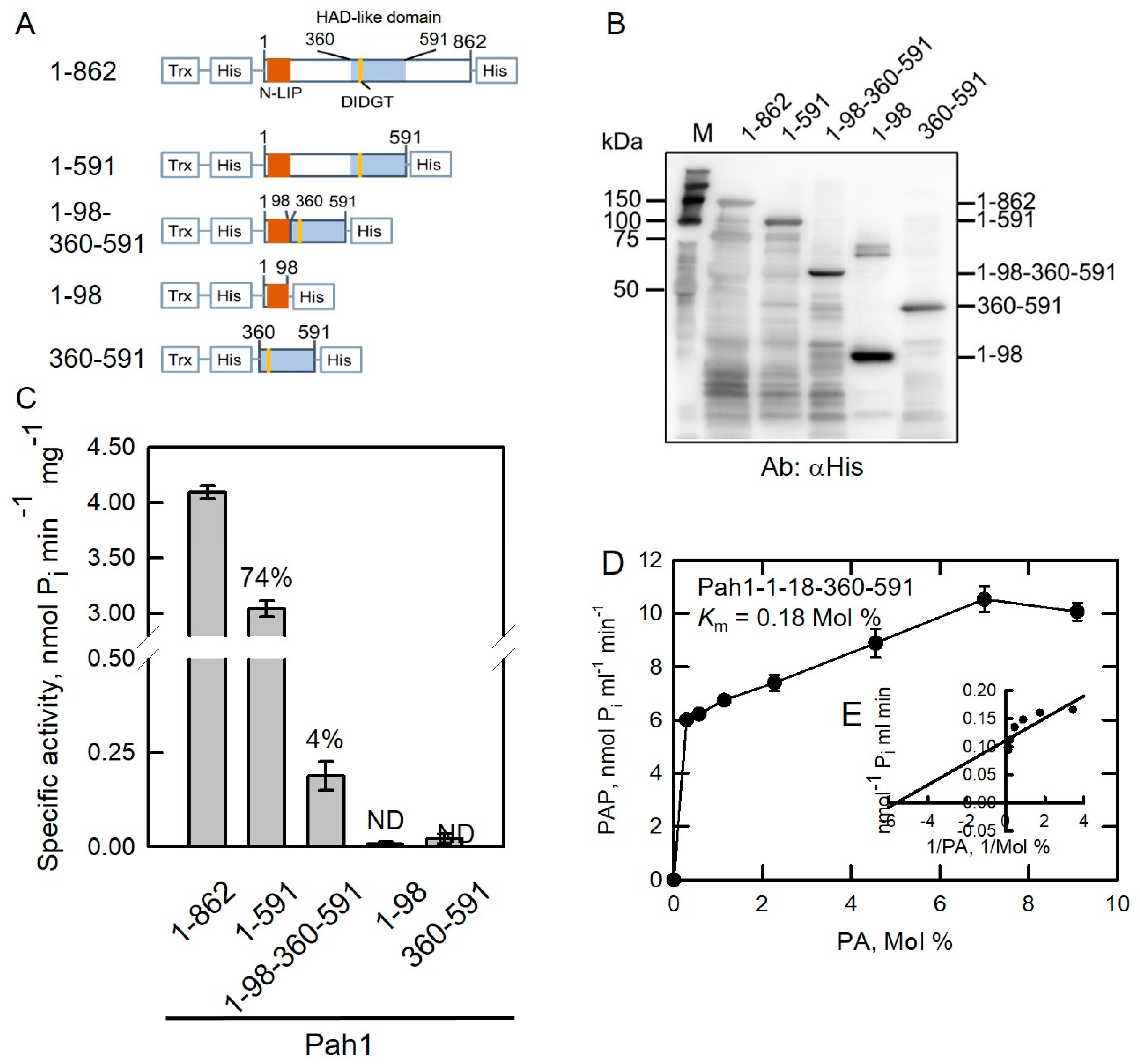
2.2. HAD-like Domain Is Essential for Pah1 Catalytic Activity
2.3. Disordered Regions Are Responsible for the Solubility of Pah1
2.4. Prediction of Intrinsically Disordered Regions in Human Lipin 1-α by DISPRED3 Algorithm
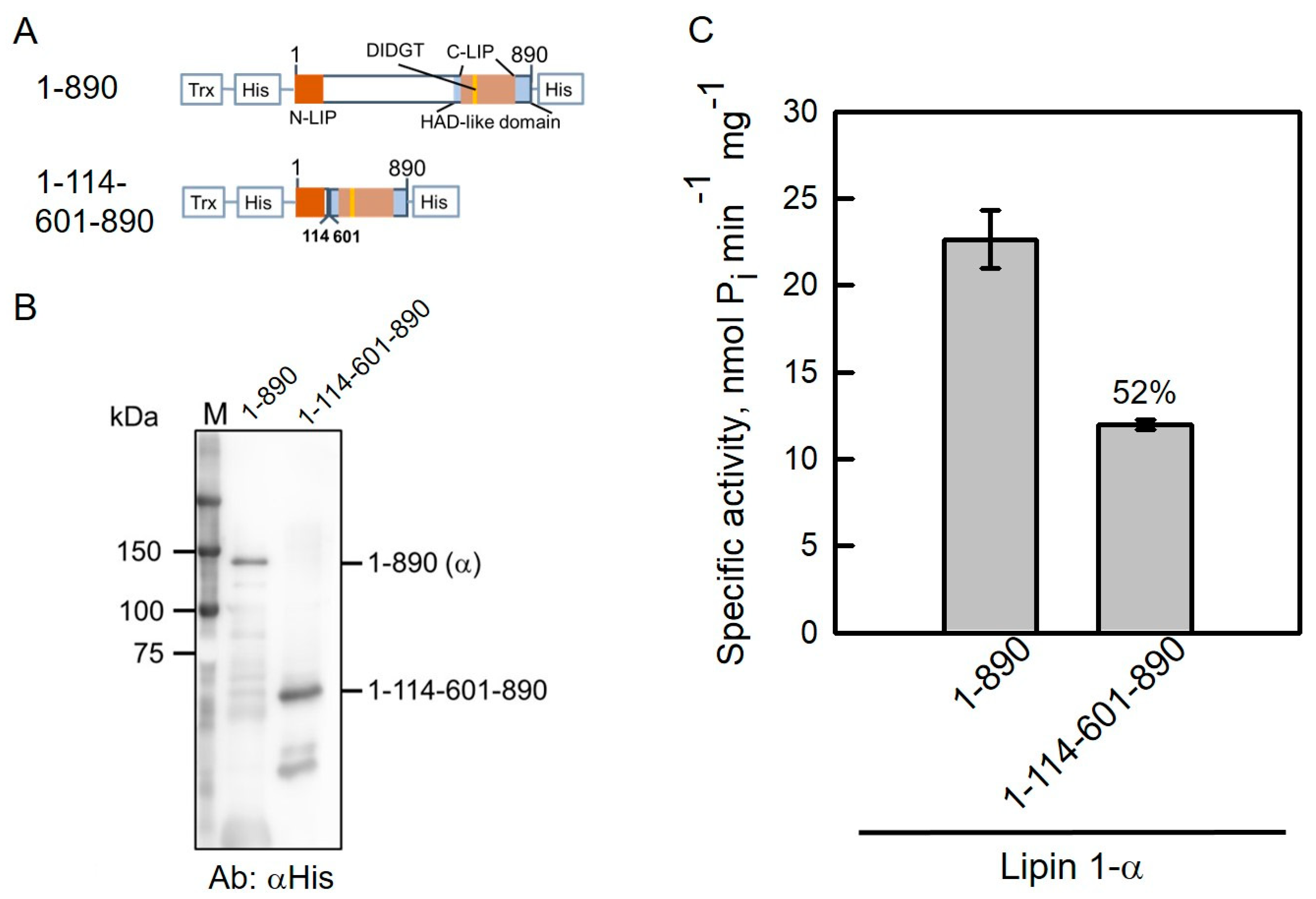
2.5. NLIP–HAD-like Truncation of Human Lipin 1 Is Active
3. Materials and Methods
3.1. Reagents
3.2. Molecular Manipulations and Construction of Plasmids
3.3. Escherichia coli Strains and Protein Expression Conditions
3.4. Purification of Recombinant Proteins by Affinity Chromatography
3.5. SDS-Polyacrylamide Gel Electrophoresis and Western Blotting Analysis
3.6. PAP Activity Assay
3.7. PAP Enzyme Kinetic
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Smith, S.W.; Weiss, S.B.; Kennedy, E.P. The enzymatic dephosphorylation of phosphatidic acids. J. Biol. Chem. 1957, 228, 915–922. [Google Scholar] [CrossRef]
- Carman, G.M.; Han, G.S. Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu. Rev. Biochem. 2011, 80, 859–883. [Google Scholar] [CrossRef] [PubMed]
- Henry, S.A.; Kohlwein, S.; Carman, G.M. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 2012, 190, 317–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chae, M.; Han, G.S.; Carman, G.M. The Saccharomyces cerevisiae actin patch protein App1p is a phosphatidate phosphatase enzyme. J. Biol. Chem. 2012, 287, 40186–40196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toke, D.A.; Bernnett, W.L.; Dillon, D.A.; Wu, W.I.; Chen, X.; Ostrander, D.B.; Oshiro, J.; Cremesti, A.; Voelker, D.R.; Fischl, A.S.; et al. Isolation and characterization of the Saccharomyces cerevisiae DPP1 gene encoding for diacylglycerol pyrophosphate phosphatase. J. Biol. Chem. 1998, 273, 3278–3284. [Google Scholar] [CrossRef] [Green Version]
- Toke, D.A.; Bernnett, W.L.; Oshiro, J.; Wu, W.I.; Voelker, D.R.; Carman, G.M. Isolation and characterization of the Saccharomyces cerevisiae LPP1 gene encoding a Mg2+-independent phosphatidate phosphatase. J. Biol. Chem. 1998, 273, 14331–14338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, G.-S.; Wu, W.-I.; Carman, G.M. The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 2006, 281, 9210–9218. [Google Scholar] [CrossRef] [Green Version]
- Han, G.S.; Siniossoglou, S.; Carman, G.M. The cellular functions of the yeast Lipin homolog Pah1p are dependent on its phosphatidate phosphatase activity. J. Biol. Chem. 2007, 282, 37026–37035. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.; Han, G.S.; Carman, G.M. A conserved tryptophan within the WRDPLVDID domain of yeast Pah1 phosphatidate phosphatase is required for its in vivo function in lipid metabolism. J. Biol. Chem. 2017, 292, 19580–19589. [Google Scholar] [CrossRef] [Green Version]
- Adeyo, O.; Horn, P.J.; Lee, S.; Binns, D.D.; Chandrahas, A.; Chapman, K.D.; Goodman, J.M. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 2011, 192, 1046–1055. [Google Scholar] [CrossRef] [Green Version]
- Karanasios, E.; Barbosa, A.D.; Sembongi, H.; Mari, M.; Han, G.S.; Reggiori, F.; Carman, G.M.; Siniossoglou, S. Regulation of lipid droplet and membrane biogenesis by the acidic tail of the phosphatidate phosphatase Pah1p. Mol. Biol. Cell 2013, 24, 2124–2133. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Pillai, A.N.; Rahaman, A. A putative NEM1 homologue regulates lipid droplet biogenesis via PAH1 in Tetrahymena thermophila. J. Biosci. 2018, 43, 693–706. [Google Scholar] [CrossRef]
- Carman, G.M.; Han, G.S. Fat-regulating phosphatidic acid phosphatase: A review of its roles and regulation in lipid homeostasis. J. Lipid Res. 2019, 60, 2–6. [Google Scholar] [CrossRef]
- Sasser, T.; Qiu, Q.S.; Karunakaran, S.; Padolina, M.; Reyes, A.; Flood, B.; Smith, S.; Gonzales, C.; Fratti, R.A. The yeast lipin 1 orthologue Pah1p regulates vacuole homeostasis and membrane fusion. J. Biol. Chem. 2012, 287, 2221–2236. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Zhang, Y.; Nielsen, J.; Liu, Z. Production of β-carotene in Saccharomyces cerevisiae through altering yeast lipid metabolism. Biotechnol. Bioeng. 2021, 118, 2046–2052. [Google Scholar] [CrossRef]
- Zhang, J.L.; Bai, Q.Y.; Peng, Y.Z.; Fan, J.; Jin, C.C.; Cao, Y.X.; Yuan, Y.J. High production of triterpenoids in Yarrowia lipolytica through manipulation of lipid components. Biotechnol. Biofuels 2020, 13, 133. [Google Scholar] [CrossRef]
- Guerfal, M.; Claes, K.; Knittelfelder, O.; De Rycke, R.; Kohlwein, S.D.; Callewaerrt, N. Enhanced membrane protein expression by engineering increased intracellular membrane production. Microb. Cell Fact. 2013, 12, 122. [Google Scholar] [CrossRef] [Green Version]
- Carman, G.M. Lipid metabolism has been good to me. J. Biol. Chem. 2021, 297, 100786. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.S.; Su, W.M.; Han, G.S.; Plote, D.; Xu, Z.; Carman, G.M. Pho85p-Pho80p phosphorylation of yeast Pah1p phosphatidate phosphatase regulates its activity, location, abundance, and function in lipid metabolism. J. Biol. Chem. 2012, 287, 11290–11310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsieh, L.S.; Su, W.M.; Han, G.S.; Carman, G.M. Phosphorylation regulates the ubiquitin-independent degradation of yeast Pah1 phosphatidate phosphatase by the 20S proteasome. J. Biol. Chem. 2015, 290, 11467–11478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos-Rosa, H.; Leung, J.; Grimsey, N.; Peak-Chew, S.; Siniossoglou, S. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 2005, 24, 1931–1941. [Google Scholar] [CrossRef] [Green Version]
- O’Hara, L.; Han, G.S.; Peak-Chew, S.; Grimsey, N.; Carman, G.M.; Siniossoglou, S. Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J. Biol. Chem. 2006, 281, 34537–34548. [Google Scholar] [CrossRef] [Green Version]
- Karanasios, E.; Han, G.S.; Xu, Z.; Carman, G.M.; Siniossoglou, S. A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc. Natl. Acad. Sci. USA 2010, 107, 17539–17544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.S.; Su, W.M.; Morgan, J.M.; Han, G.S.; Xu, Z.; Karanasios, E.; Siniossoglou, S.; Carman, G.M. Phosphorylation of phosphatidate phosphatase regulates its membrane associate and physiological functions in Saccharomyces cerevisiae: Identification of Ser(602), Thr(723), and Ser(744) as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J. Biol. Chem. 2011, 286, 1486–1498. [Google Scholar] [CrossRef] [Green Version]
- Su, W.M.; Han, G.S.; Casciano, J.; Carman, G.M. Protein kinase A-mediated phosphorylation of Pah1p phosphatidate phosphatase functions in conjugation with the Pho85p-Pho80p and Cdc28p-cyclin B kinases to regulate lipid synthesis in yeast. J. Biol. Chem. 2012, 287, 33364–33376. [Google Scholar] [CrossRef] [Green Version]
- Su, W.M.; Han, G.S.; Carman, G.M. Cross-talk phosphorylations by protein kinase C and Pho85p-Pho80p kinase regulate Pah1p phosphatidate phosphatase abundance in Saccharomyces cerevisiae. J. Biol. Chem. 2014, 289, 18818–18830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsieh, L.S.; Su, W.M.; Han, G.S.; Carman, G.M. Phosphorylation of yeast Pah1 phosphatidate phosphatase by casein kinase II regulates its function in lipid metabolism. J. Biol. Chem. 2016, 291, 9974–9990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassaninasab, A.; Hsieh, L.S.; Su, W.M.; Han, G.S.; Carman, G.M. Yck1 casein kinase I regulates the activity and phosphorylation of Pah1 phosphatidate phosphatase from Saccharomyces cerevisiae. J. Biol. Chem. 2019, 294, 18256–18268. [Google Scholar] [CrossRef]
- Su, W.M.; Han, G.S.; Carman, G.M. Yeast Nem1-Spo7 protein phosphatase activity on Pah1 phosphatidate phosphatase is specific for the Pho85-Pho80 protein kinase phosphorylation sites. J. Biol. Chem. 2014, 289, 34699–34708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, W.M.; Han, G.S.; Dey, P.; Carman, G.M. Protein kinas A phosphorylates the Nem1-Spo7 protein phosphatase complex that regulates the phosphorylation state of the phosphatidate phosphatase Pah1 in yeast. J. Biol. Chem. 2018, 293, 15801–15814. [Google Scholar] [CrossRef] [Green Version]
- Pascual, F.; Hsieh, L.S.; Soto-Cardalda, A.; Carman, G.M. Yeast Pah1p phosphatidate phosphatase is regulated by proteasome-mediated degradation. J. Biol. Chem. 2014, 289, 9811–9822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Csaki, L.S.; Reue, K. Lipins: Multifunctional lipid metabolism proteins. Annu. Rev. Nutr. 2010, 30, 257–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, G.-S.; Carman, G.M. Characterization of the Human LPIN1-encoded phosphatidate phosphatase isoforms. J. Biol. Chem. 2010, 285, 14628–14638. [Google Scholar] [CrossRef] [Green Version]
- Jang, C.H.; Kim, K.M.; Yang, J.H.; Cho, S.S.; Kim, S.J.; Shin, S.M.; Cho, I.J.; Ki, S.H. The role of Lipin-1 in the regulation of fibrogenesis and TGF-β signaling in hepatic stellate cells. Toxicol. Sci. 2016, 153, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, K.; Fukushima, H.; Ogura, K.; Lien, E.C.; Nihira, N.T.; Zhang, J.; North, B.J.; Guo, A.; Nagashima, K.; Nakagawa, T.; et al. The SCFβ-TRCP E3 ubiquitin ligase complex targets Lipin1 for ubiquitination and degradation to promote hepatic lipogenesis. Sci. Signal. 2017, 10, eaah4117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishimoto, K.; Hayase, A.; Kumagai, F.; Kawai, M.; Okuno, H.; Hino, N.; Okada, Y.; Kawamura, T.; Tanaka, T.; Hamakubo, T.; et al. Degradation of human Lipin-1 by BTRC E3 ubiquitin ligase. Biochem. Biophys. Res. Commun. 2017, 488, 159–164. [Google Scholar] [CrossRef]
- Watahiki, A.; Shimizu, K.; Hoshikawa, S.; Chiba, M.; Kitamura, H.; Egusa, H.; Fukumoto, S.; Inuzuka, H. Lipin-2 degradation elicits a proinflammatory gene signature in macrophages. Biochem. Biophys. Res. Commun. 2020, 524, 477–483. [Google Scholar] [CrossRef]
- Michot, C.; Hubert, L.; Brivet, M.; De Meirleir, L.; Valayannopoulos, V.; Müller-Felber, W.; Venkateswaran, R.; Ogier, H.; Desguerre, I.; Altuzarra, C.; et al. LPIN1 gene mutations: A major cause of severe rhabdomyolysis in early childhood. Hum. Mutat. 2010, 31, E1564–E1573. [Google Scholar] [CrossRef] [Green Version]
- Michot, C.; Mamoune, A.; Vamecq, J.; Viou, M.T.; Hsieh, L.S.; Testet, E.; Lainé, J.; Hubert, L.; Dessein, A.F.; Fontaine, M.; et al. Combination of lipid metabolism alterations and their sensitivity to inflammatory cytokines in human lipin-1-deficient myoblasts. Biochim. Biophys. Acta 2013, 1832, 2103–2114. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Verity, M.A.; Reue, K. Lipin-1 regulates autophagy clearance and intersects with statin drug effects in skeletal muscle. Cell Metab. 2014, 20, 267–279. [Google Scholar] [CrossRef] [Green Version]
- Schweitzer, G.G.; Collier, S.L.; Chen, Z.; Eaton, J.M.; Connolly, A.M.; Bucelli, R.C.; Pestronk, A.; Harris, T.E.; Finck, B.N. Rhabdomyolysis-associated mutations in human LPIN1 lead to loss of phosphatidic acid phosphohydrolase activity. JIMD Rep. 2015, 23, 113–122. [Google Scholar] [CrossRef] [Green Version]
- Tong, K.; Yu, G.S. Acute recurrent rhabdomyolysis in a Chinese boy associated with a novel compound heterozygous LPIN1 variant: A case report. BMC Neurol. 2021, 21, 42. [Google Scholar] [CrossRef]
- Pillai, A.N.; Shukla, S.; Gautam, S.; Rahaman, A. Small phosphatidate phosphatase (TtPAH2) of Tetrahymena complements respiratory function and not membrane biogenesis function of yeast. PAH1. J. Biosci. 2017, 43, 613–621. [Google Scholar] [CrossRef]
- Khayyo, V.I.; Hoffmann, R.M.; Wang, H.; Bell, J.A.; Burke, J.E.; Reue, K.; Airola, M.V.V. Crystal structure of a lipin/Pah phosphatidic acid phosphatase. Nat. Commun. 2020, 11, 1309. [Google Scholar] [CrossRef] [Green Version]
- John, D.T.; Cozzetto, D. DISPRED3: Precise disordered region predictions with annotated protein-binding activity. Bioinformatics 2015, 31, 857–863. [Google Scholar] [CrossRef]
- Kulkarni, P.; Uversky, V.N. Intrinsically disordered proteins: The dark horse of the dark proteome. Proteomics 2018, 18, e1800061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uversky, V.N. Intrinsic disorder, protein-protein interactions, and disease. Adv. Protein Chem. Struct. Biol. 2018, 110, 85–121. [Google Scholar] [CrossRef] [PubMed]
- LaVallie, E.R.; Lu, Z.; Diblasio-Smith, E.A.; Collins-Racie, L.A.; McCoy, J.M. Thioredoxin as a fusion partner for production of soluble recombinant proteins in Escherichia coli. Methods Enzymol. 2000, 326, 322–340. [Google Scholar] [CrossRef]
- Hsiao, C.J.; Hsieh, C.Y.; Hsieh, L.S. Cloning and characterization of the Bambusa oldhamii BoMDH-encoded malate dehydrogenase. Protein Expr. Purif. 2020, 174, 105665. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye-binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Hsieh, C.Y.; Huang, Y.H.; Lin, Z.Y.; Hsieh, L.S. Insights into the substrate selectivity of Bambusa oldhamii phenylalanine ammonia-lyase 1 and 2 through mutational analysis. Phytochem. Lett. 2020, 38, 140–143. [Google Scholar] [CrossRef]
- Lineweavwer, H.; Burk, D. The determination of enzyme dissociation constants. J. Am. Chem. Soc. 1934, 56, 658–666. [Google Scholar] [CrossRef]


| Plasmid | Relevant Characteristics | Source/Ref. |
|---|---|---|
| pGH313 | Full-length PAH1 (1-862) coding sequence inserted into pET15b | [7] |
| pET32b | E. coli expression vector with thioredoxin (Trx) fusion protein and both N-terminal and C-terminal His6-tag fusions | Novagen |
| pET32b-ScPah1-1-862 | Full-length PAH1 (1-862) coding sequence inserted into pET32b | This study |
| pET32b-ScPah1-1-591 | PAH1 (1-591 truncation) inserted into pET32b | This study |
| pET32b-ScPah1-1-550 | PAH1 (1-550 truncation) inserted into pET32b | This study |
| pET32b-ScPah1-1-525 | PAH1 (1-525 truncation) inserted into pET32b | This study |
| pET32b-ScPah1-1-500 | PAH1 (1-500 truncation) inserted into pET32b | This study |
| pET32b-ScPah1-1-98 | PAH1 (1-98 truncation) inserted into pET32b | This study |
| pET32b-ScPah1-1-98-360-591 | PAH1 (1-98-360-591 truncation) inserted into pET32b | This study |
| pET32b-ScPah1-360-591 | PAH1 (360-591 truncation) inserted into pET32b | This study |
| pGH322 | Full-length human LPIN1-α (1-890) coding sequence inserted into pET28b | [33] |
| pET32b-Lipin1-α-1-890 | Full-length human LPIN1-α (1-890) coding sequence inserted into pET32b | This study |
| pET32b-Lipin1-α-1-114-601-890 | LPIN1-α (1-114-601-890 truncation) inserted into pET32b | This study |
| Protein | Km (Mol %) | kcat (s−1) | kcat/Km (s−1 Mol %−1) |
|---|---|---|---|
| Pah1-1-862 | 1.8 | 7.22 | 4.01 |
| Pah1-1-591 | 2.6 | 3.25 | 1.25 |
| Pah1-1-98-360-591 | 0.18 | 0.74 | 4.11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, W.-H.; Huang, Y.-H.; Chen, P.-R.; Hsieh, L.-S. NLIP and HAD-like Domains of Pah1 and Lipin 1 Phosphatidate Phosphatases Are Essential for Their Catalytic Activities. Molecules 2021, 26, 5470. https://doi.org/10.3390/molecules26185470
Hsu W-H, Huang Y-H, Chen P-R, Hsieh L-S. NLIP and HAD-like Domains of Pah1 and Lipin 1 Phosphatidate Phosphatases Are Essential for Their Catalytic Activities. Molecules. 2021; 26(18):5470. https://doi.org/10.3390/molecules26185470
Chicago/Turabian StyleHsu, Wei-Hsin, Yi-Hao Huang, Pin-Ru Chen, and Lu-Sheng Hsieh. 2021. "NLIP and HAD-like Domains of Pah1 and Lipin 1 Phosphatidate Phosphatases Are Essential for Their Catalytic Activities" Molecules 26, no. 18: 5470. https://doi.org/10.3390/molecules26185470






