Transcriptomics-Based Repositioning of Natural Compound, Eudesmin, as a PRC2 Modulator
Abstract
:1. Introduction
2. Results
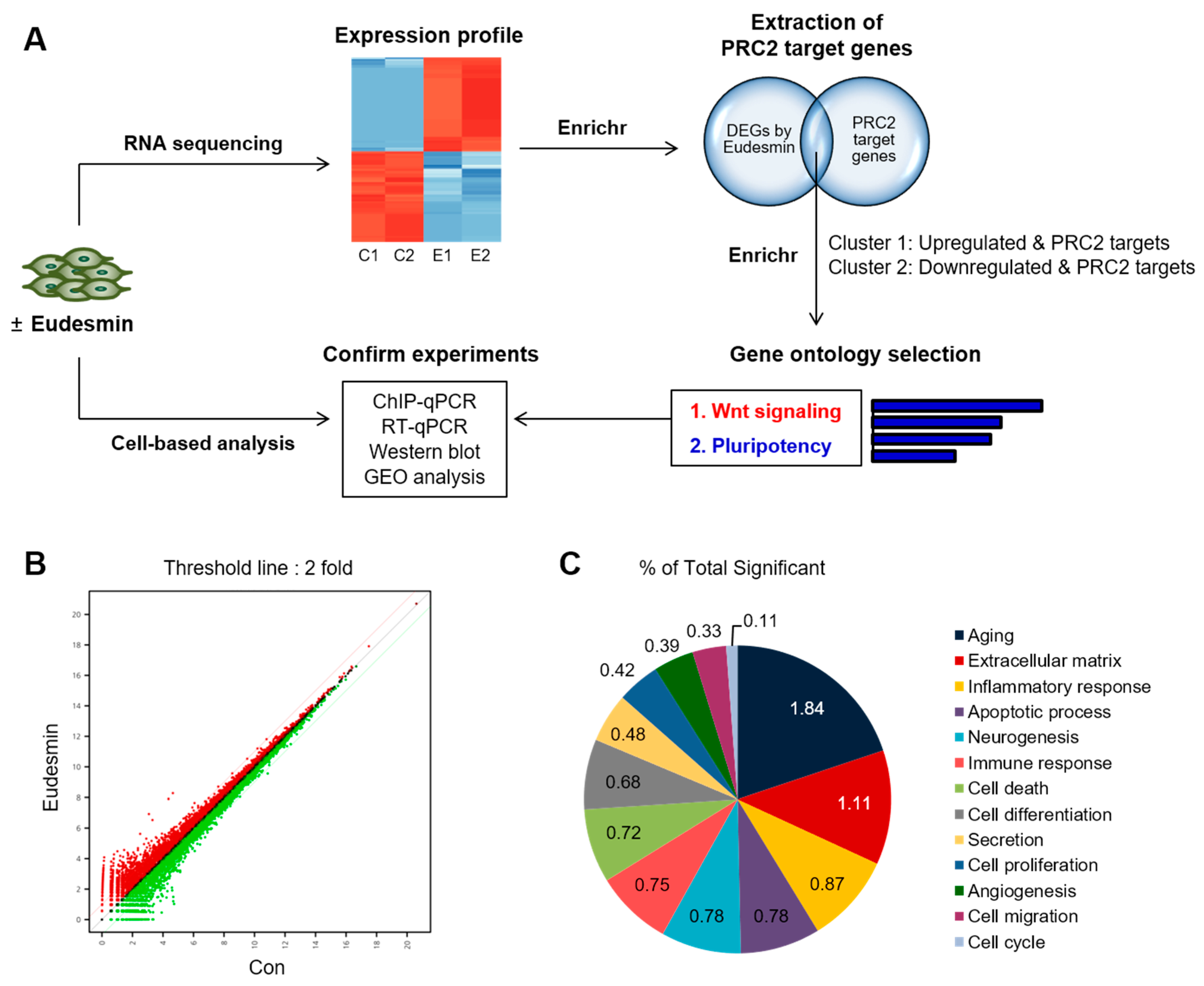
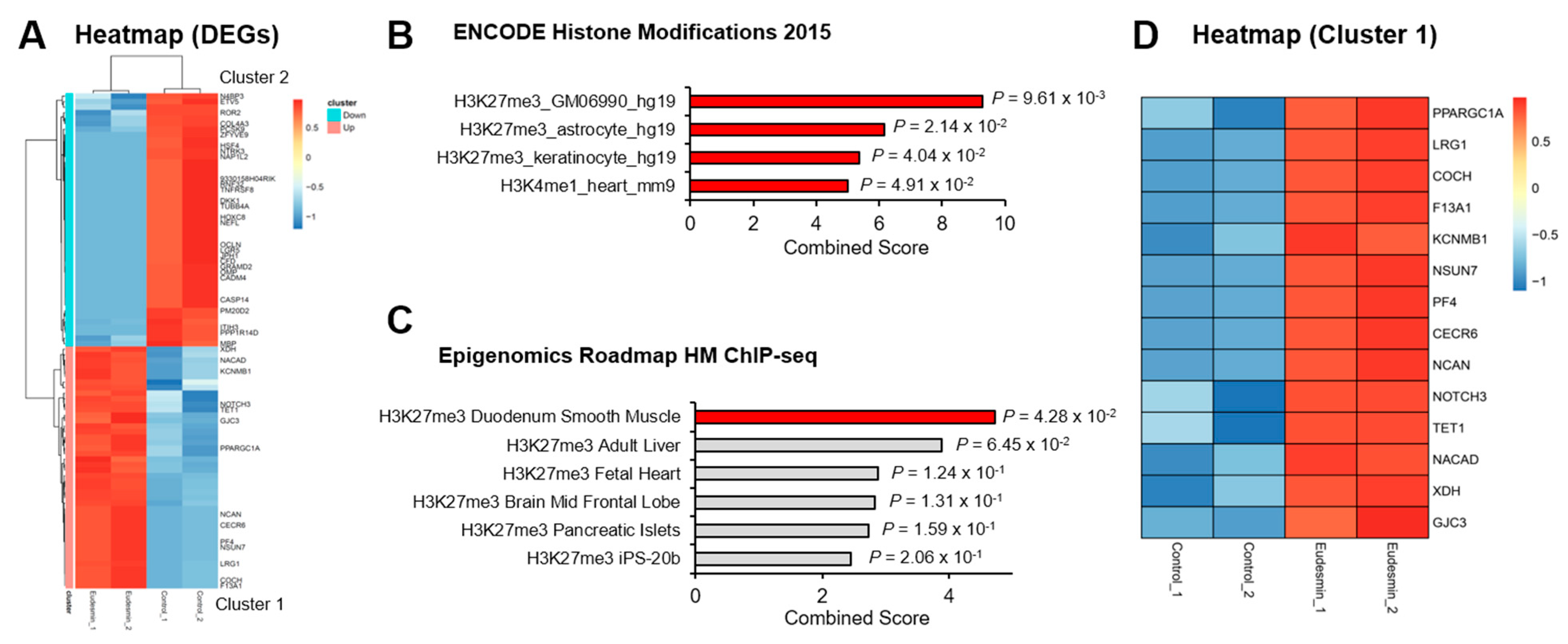
2.1. Enrichment in PRC2 Target Genes by Eudesmin Treatment
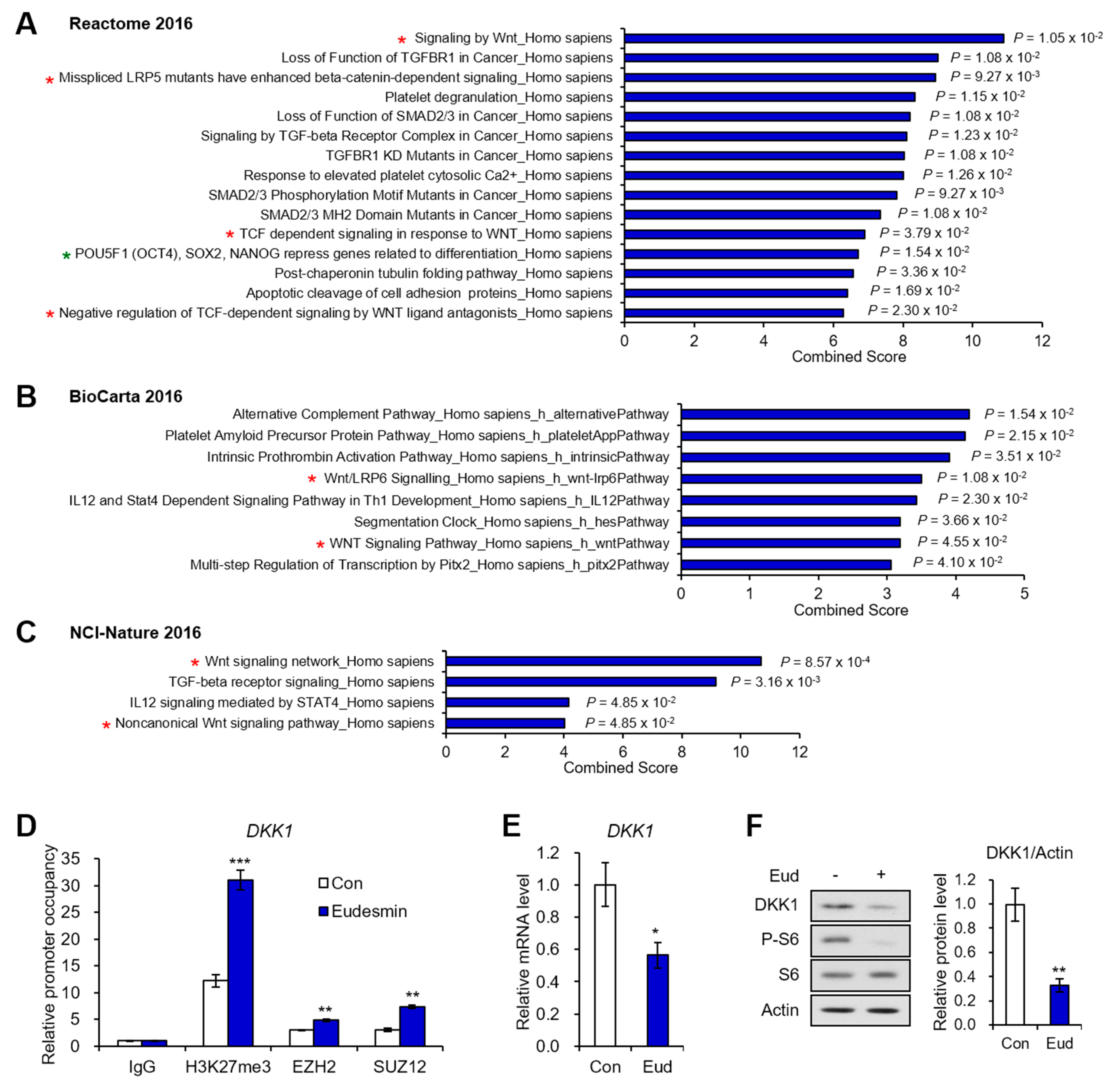
2.2. PRC2 Target Genes Downregulated by Eudesmin Are Involved in Wnt Signaling
2.3. Eudesmin Induces the Expression of Pluripotency Marker Genes
3. Discussion
4. Materials and Methods
4.1. Antibodies and Reagents
4.2. Cell Lines and Culture Condition
4.3. RNA Sequencing
4.4. Enrichr Analyses of Differentially Expressed Genes by Eudesmin
4.5. Reverse Transcription (RT) and Quantitative Real-Time PCR (qPCR)
4.6. Chromatin Immunoprecipitation and Real-Time PCR (ChIP-qPCR)
4.7. Immunoblotting
4.8. Public Data Availability
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Surface, L.C.; Thornton, S.R.; Boyer, L.A. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell 2010, 7, 288–298. [Google Scholar] [CrossRef] [Green Version]
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasini, D.; Bracken, A.P.; Hansen, J.B.; Capillo, M.; Helin, K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell Biol. 2007, 27, 3769–3779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, X.; Liu, Y.; Hsu, Y.J.; Fujiwara, Y.; Kim, J.; Mao, X.; Yuan, G.C.; Orkin, S.H. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 2008, 32, 491–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Carroll, D.; Erhardt, S.; Pagani, M.; Barton, S.C.; Surani, M.A.; Jenuwein, T. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell Biol. 2001, 21, 4330–4336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasini, D.; Bracken, A.P.; Jensen, M.R. Lazzerini Denchi, E.; Helin, K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004, 23, 4061–4071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, K.H.; Yi, S.A.; Lee, J.; Lee, M.G.; Park, J.H.; Oh, H.; Lee, J.; Park, J.W.; Han, J.W. Eudesmin impairs adipogenic differentiation via inhibition of S6K1 signaling pathway. Biochem. Biophys. Res. Commun. 2018, 505, 1148–1153. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. Wnt/beta-catenin signaling in development and disease. Cell 2006, 127, 469–480. [Google Scholar] [CrossRef] [Green Version]
- Aldiri, I.; Moore, K.B.; Hutcheson, D.A.; Zhang, J.; Vetter, M.L. Polycomb repressive complex PRC2 regulates Xenopus retina development downstream of Wnt/β-catenin signaling. Development 2013, 140, 2867–2878. [Google Scholar] [CrossRef] [Green Version]
- Oittinen, M.; Popp, A.; Kurppa, K.; Lindfors, K.; Mäki, M.; Kaikkonen, M.U.; Viiri, K. Polycomb Repressive Complex 2 Enacts Wnt Signaling in Intestinal Homeostasis and Contributes to the Instigation of Stemness in Diseases Entailing Epithelial Hyperplasia or Neoplasia. Stem Cells 2017, 35, 445–457. [Google Scholar] [CrossRef] [Green Version]
- Rothberg, J.L.M.; Maganti, H.B.; Jrade, H.; Porter, C.J.; Palidwor, G.A.; Cafariello, C.; Battaion, H.L.; Khan, S.T.; Perkins, T.J.; Paulson, R.F.; et al. Mtf2-PRC2 control of canonical Wnt signaling is required for definitive erythropoiesis. Cell Discov. 2018, 4, 21. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.L.; Sun, B.R.; Zheng, C.; Yang, G.L. The antitumour effects of eudesmin on lung cancer by inducing apoptosis via mitochondria-mediated pathway in the tumour cells. Pharm. Biol. 2017, 55, 2259–2263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, S.; Grassi, J.; Akhmedjanova, V.; Debiton, E.; Balansard, G.; Beliveau, R.; Barthomeuf, C. Reversal of P-glycoprotein-mediated drug efflux by eudesmin from Haplophyllum perforatum and cytotoxicity pattern versus diphyllin, podophyllotoxin and etoposide. Planta Med. 2007, 73, 1563–1567. [Google Scholar] [CrossRef] [PubMed]
- Cantrell, C.L.; Schrader, K.K.; Mamonov, L.K.; Sitpaeva, G.T.; Kustova, T.S.; Dunbar, C.; Wdege, D.E. Isolation and identification of antifungal and antialgal alkaloids from Haplophyllum sieversii. J. Agric. Food Chem. 2005, 53, 7741–7748. [Google Scholar] [CrossRef]
- Céspedes, C.L.; Ávila, J.G.; Garcia, A.M.; Becerra, J.; Flores, C.; Aqueveque, P.; Bittner, M.; Hoeneisen, M.; Martinez, M.; Silva, M. Antifungal and antibacterial activities of Araucaria araucana (Mol.) K. Koch heartwood lignans. Z. Naturforsch. C J. Biosci. 2006, 61, 35–43. [Google Scholar] [CrossRef]
- Yang, J.S.; Wang, C.M.; Su, C.H.; Ho, H.C.; Chang, C.H.; Chou, C.H.; Hsu, Y.M. Eudesmin attenuates Helicobacter pylori-induced epithelial autophagy and apoptosis and leads to eradication of H. pylori infection. Exp. Ther. Med. 2018, 15, 2388–2396. [Google Scholar]
- Wassef, M.; Michaud, A.; Margueron, R. Association between EZH2 expression, silencing of tumor suppressors and disease outcome in solid tumors. Cell Cycle 2016, 15, 2256–2262. [Google Scholar] [CrossRef] [Green Version]
- Simon, J.A.; Lange, C.A. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008, 647, 21–29. [Google Scholar] [CrossRef]
- Velichutina, I.; Shaknovich, R.; Geng, H.; Johnson, N.A.; Gascoyne, R.D.; Melnick, A.M.; Elemento, O. EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood 2010, 116, 5247–5255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, J.Z.; Yan, Y.; Wang, X.X.; Jiang, Y.; Xu, H.E. EZH2: Biology, disease, and structure-based drug discovery. Acta Pharmacol. Sin. 2014, 35, 161–174. [Google Scholar] [CrossRef] [Green Version]
- Hoy, S.M. Tazemetostat: First Approval. Drugs 2020, 80, 513–521. [Google Scholar] [CrossRef]
- Shahabipour, F.; Caraglia, M.; Majeed, M.; Derosa, G.; Maffioli, P.; Sahebkar, A. Naturally occurring anti-cancer agents targeting EZH2. Cancer Lett. 2017, 400, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Li, Y.; Li, M.; Lu, D. Eudesmin exerts antitumor effects by down-regulating EZH2 expression in nasopharyngeal carcinoma cells. Chem. Biol. Interact. 2019, 307, 51–57. [Google Scholar] [CrossRef]
- Yi, S.A.; Um, S.H.; Lee, J.; Yoo, J.H.; Bang, S.Y.; Park, E.K.; Lee, M.G.; Nam, K.H.; Jeon, Y.J.; Park, J.W.; et al. S6K1 Phosphorylation of H2B Mediates EZH2 Trimethylation of H3: A Determinant of Early Adipogenesis. Mol. Cell 2016, 62, 443–452. [Google Scholar] [CrossRef] [Green Version]
- Marucci, L.; Pedone, E.; Di Vicino, U.; Sanuy-Escribano, B.; Isalan, M.; Cosma, M.P. β-catenin fluctuates in mouse ESCs and is essential for Nanog-mediated reprogramming of somatic cells to pluripotency. Cell Rep. 2014, 8, 1686–1696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyer, L.A.; Lee, T.I.; Cole, M.F.; Johnstone, S.E.; Levine, S.S.; Zucker, J.P.; Guenther, M.G.; Kumar, R.M.; Murray, H.L.; Jenner, R.G.; et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 2005, 122, 947–956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enrichr. Available online: http://amp.pharm.mssm.edu/Enrichr/ (accessed on 14 September 2021).
- Yi, S.A.; Lee, D.H.; Kim, G.W.; Ryu, H.W.; Park, J.W.; Lee, J.; Han, J.; Park, J.H.; Oh, H.; Lee, J.; et al. HPV-mediated nuclear export of HP1γ drives cervical tumorigenesis by downregulation of p53. Cell Death. Differ. 2020, 27, 2537–2551. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| No. | Gene Symbol | Description | Eudesmin/Con | |
|---|---|---|---|---|
| Fold Change | p-Value | |||
| 1 | PPARGC1A | Peroxisome proliferative activated receptor, gamma, coactivator 1 alpha | 9.221 | 0.009 |
| 2 | LRG1 | Leucine-rich alpha-2-glycoprotein 1 | 4.717 | 0.002 |
| 3 | NPAS1 | Neuronal PAS domain protein 1 | 4.251 | 0.010 |
| 4 | COCH | Cochlin | 3.807 | 0.002 |
| 5 | F13A1 | Coagulation factor XIII, A1 subunit | 3.807 | 0.002 |
| 6 | KCNMB1 | Potassium large conductance calcium-activated channel, subfamily M, beta member 1 | 3.018 | 0.005 |
| 7 | NSUN7 | NOL1/NOP2/Sun domain family, member 7 | 2.884 | 0.002 |
| 8 | PF4 | Platelet factor 4 | 2.884 | 0.002 |
| 9 | CECR6 | Cat eye syndrome chromosome region, candidate 6 | 2.884 | 0.002 |
| 10 | NCAN | Neurocan | 2.884 | 0.002 |
| 11 | NOTCH3 | Notch3 | 2.695 | 0.007 |
| 12 | TET1 | Tet methylcytosine dioxygenase 1 | 2.691 | 0.008 |
| 13 | NACAD | NAC alpha domain containing | 2.595 | 0.003 |
| 14 | GLIS1 | GLIS family zinc finger 1 | 2.482 | 0.010 |
| 15 | XDH | Xanthine dehydrogenase | 2.206 | 0.005 |
| 16 | GJC3 | Gap junction protein, gamma 3 | 2.198 | 0.009 |
| No. | Gene Symbol | Description | Eudesmin/Con | |
|---|---|---|---|---|
| Fold Change | p-Value | |||
| 1 | ZFYVE9 | Zinc finger, FYVE domain containing 9 | 0.098 | 0.005 |
| 2 | PCSK9 | Proprotein convertase subtilisin/kexin type 9 | 0.155 | 0.005 |
| 3 | NAP1L2 | Nucleosome assembly protein 1-like 2 | 0.163 | 0.005 |
| 4 | PM20D2 | Peptidase M20 domain containing 2 | 0.178 | 0.001 |
| 5 | NTRK3 | Neurotrophic tyrosine kinase, receptor, type 3 | 0.195 | 0.005 |
| 6 | N4BP3 | NEDD4 binding protein 3 | 0.217 | 0.007 |
| 7 | PPP1R14D | Protein phosphatase 1, regulatory (inhibitor) subunit 14D | 0.218 | 0.005 |
| 8 | ITIH3 | Inter-alpha trypsin inhibitor, heavy chain 3 | 0.218 | 0.005 |
| 9 | HSF4 | Heat shock transcription factor 4 | 0.244 | 0.005 |
| 10 | GRAMD2 | GRAM domain containing 2 | 0.326 | 0.006 |
| 11 | OMP | Olfactory marker protein | 0.326 | 0.006 |
| 12 | CADM4 | Cell adhesion molecule 4 | 0.326 | 0.006 |
| 13 | CASP14 | Caspase 14 | 0.326 | 0.006 |
| 14 | MBP | Myelin basic protein | 0.419 | 0.008 |
| 15 | ROR2 | Receptor tyrosine kinase-like orphan receptor 2 | 0.425 | 0.006 |
| 16 | 9330158H04RIK | RIKEN cDNA 9330158H04 gene | 0.491 | 0.006 |
| 17 | RNF32 | Ring finger protein 32 | 0.491 | 0.006 |
| 18 | TNFRSF8 | Tumor necrosis factor receptor superfamily, member 8 | 0.491 | 0.006 |
| 19 | DKK1 | Dickkopf WNT signaling pathway inhibitor 1 | 0.491 | 0.006 |
| 20 | TUBB4A | Tubulin, beta 4A class IVA | 0.491 | 0.006 |
| 21 | HOXC8 | Homeobox C8 | 0.491 | 0.006 |
| 22 | NEFL | Neurofilament, light polypeptide | 0.491 | 0.006 |
| 23 | OCLN | Occludin | 0.491 | 0.006 |
| 24 | LGR5 | Leucine rich repeat containing G protein coupled receptor 5 | 0.491 | 0.006 |
| 25 | CFD | Complement factor D (adipsin) | 0.491 | 0.006 |
| 26 | JPH1 | Junctophilin 1 | 0.491 | 0.006 |
| 27 | COL4A3 | Collagen, type IV, alpha 3 | 0.495 | 0.004 |
| 28 | ETV5 | Ets variant 5 | 0.499 | 0.007 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yi, S.A.; Nam, K.H.; Lee, M.G.; Oh, H.; Noh, J.S.; Jeong, J.K.; Kwak, S.; Jeon, Y.J.; Kwon, S.H.; Lee, J.; et al. Transcriptomics-Based Repositioning of Natural Compound, Eudesmin, as a PRC2 Modulator. Molecules 2021, 26, 5665. https://doi.org/10.3390/molecules26185665
Yi SA, Nam KH, Lee MG, Oh H, Noh JS, Jeong JK, Kwak S, Jeon YJ, Kwon SH, Lee J, et al. Transcriptomics-Based Repositioning of Natural Compound, Eudesmin, as a PRC2 Modulator. Molecules. 2021; 26(18):5665. https://doi.org/10.3390/molecules26185665
Chicago/Turabian StyleYi, Sang Ah, Ki Hong Nam, Min Gyu Lee, Hwamok Oh, Jae Sung Noh, Jae Kyun Jeong, Sangwoo Kwak, Ye Ji Jeon, So Hee Kwon, Jaecheol Lee, and et al. 2021. "Transcriptomics-Based Repositioning of Natural Compound, Eudesmin, as a PRC2 Modulator" Molecules 26, no. 18: 5665. https://doi.org/10.3390/molecules26185665
APA StyleYi, S. A., Nam, K. H., Lee, M. G., Oh, H., Noh, J. S., Jeong, J. K., Kwak, S., Jeon, Y. J., Kwon, S. H., Lee, J., & Han, J.-W. (2021). Transcriptomics-Based Repositioning of Natural Compound, Eudesmin, as a PRC2 Modulator. Molecules, 26(18), 5665. https://doi.org/10.3390/molecules26185665







