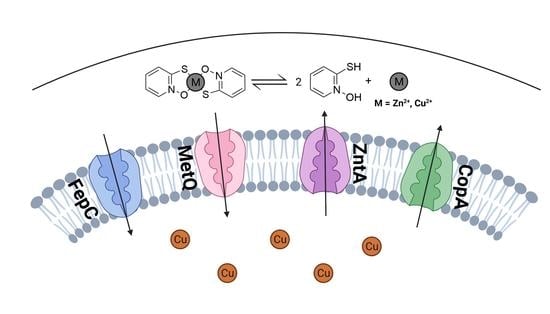
Membrane Transporters Involved in the Antimicrobial Activities of Pyrithione in Escherichia coli
Abstract
:1. Introduction
2. Results
2.1. Screening Membrane Protein Genes of the E. coli Keio Collection: Identification of Membrane Proteins Potentially Involved in the Pharmacological Actions of Pyrithione
2.2. Growth Kinetics of Knockout and Overexpression Strains for metQ, fepC, copA, and zntA Genes at Variable Concentrations of Pyrithione Expanded on the Observations from the Initial Knockout Library Screening
2.3. Effects of the Gene Knockouts on the Minimum Inhibitory Concentrations of Pyrithione in the Presence and Absence of Relevant Metals
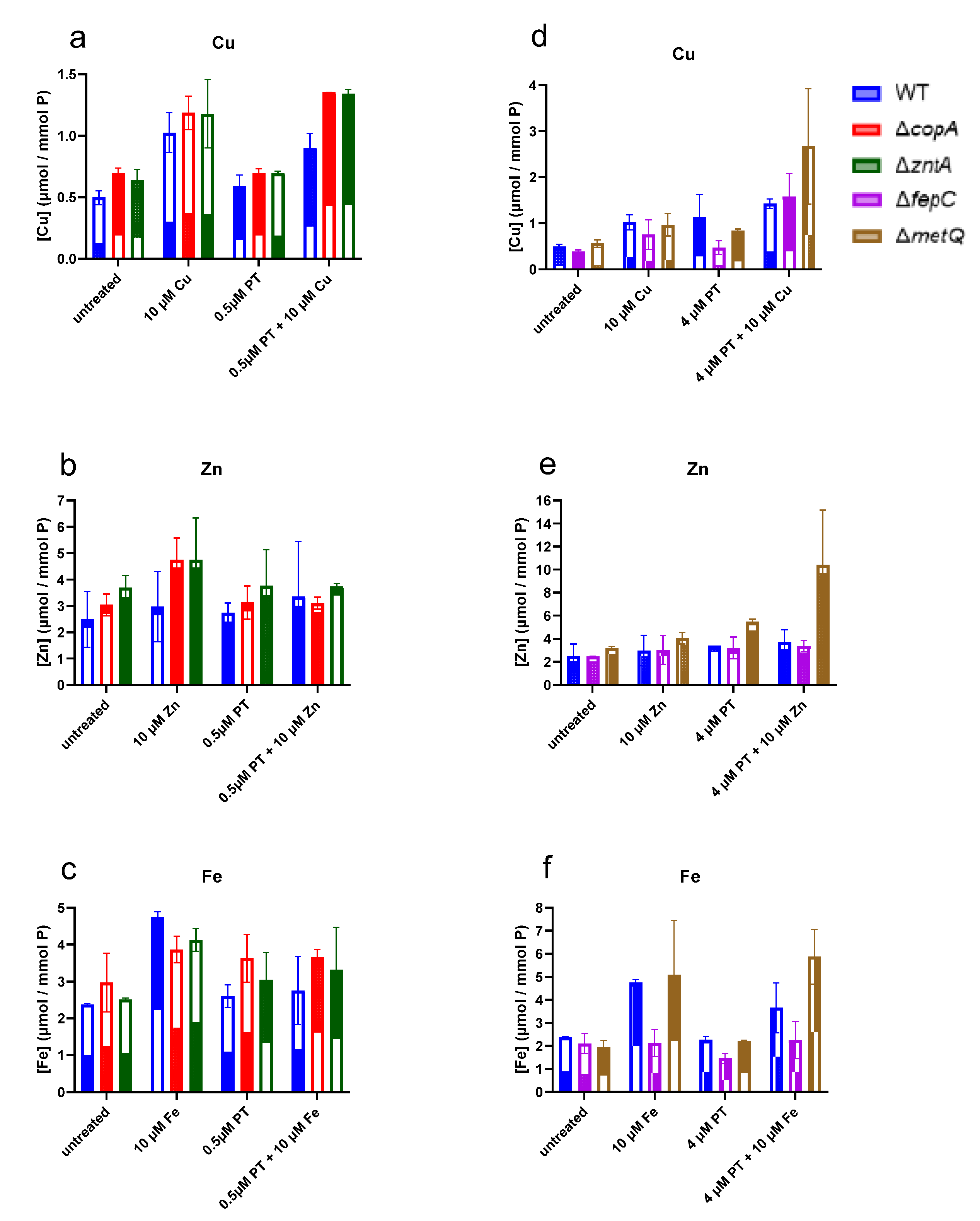
2.4. Levels of Cell-Associated Metals in the Absence of Membrane Transporters CopA, ZntA, FepC, and MetQ
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Strains and Culture
5.2. Growth Profiling
5.3. Measurement of Cell Associated Copper
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Holmes, A.H.; Moore, L.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.; Piddock, L. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef]
- Mushtaq, A. UN commits to tackling antimicrobial resistance. Lancet Infect. Dis. 2016, 16, 1229–1230. [Google Scholar] [CrossRef]
- Wells, V.; Piddock, L.J.V. Addressing antimicrobial resistance in the UK and Europe. Lancet Infect. Dis. 2017, 17, 1230–1231. [Google Scholar] [CrossRef] [Green Version]
- Roope, L.S.J.; Smith, R.D.; Pouwels, K.B.; Buchanan, J.; Abel, L.; Eibich, P.; Butler, C.; Tan, P.S.; Walker, A.S.; Robotham, J.; et al. The challenge of antimicrobial resistance: What economics can contribute. Science 2019, 364, eaau4679. [Google Scholar] [CrossRef]
- Baker, S. Infectious disease. A return to the pre-antimicrobial era? Science 2015, 347, 1064–1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyers, M.; Wright, G.D. Drug combinations: A strategy to extend the life of antibiotics in the 21st century. Nat. Rev. Genet. 2019, 17, 141–155. [Google Scholar] [CrossRef]
- Imai, Y.; Meyer, K.; Iinishi, A.; Favre-Godal, Q.; Green, R.; Manuse, S.; Caboni, M.; Mori, M.; Niles, S.; Ghiglieri, M.; et al. A new antibiotic selectively kills Gram-negative pathogens. Nature 2019, 576, 459–464. [Google Scholar] [CrossRef]
- Baker, S.; Thomson, N.; Weill, F.-X.; Holt, K.E. Genomic insights into the emergence and spread of antimicrobial-resistant bacterial pathogens. Science 2018, 360, 733–738. [Google Scholar] [CrossRef] [Green Version]
- Piddock, L.; Garneau-Tsodikova, S.; Garner, C. Ask the experts: How to curb antibiotic resistance and plug the antibiotics gap? Future Med. Chem. 2016, 8, 1027–1032. [Google Scholar] [CrossRef] [Green Version]
- Annunziato, G. Strategies to overcome antimicrobial resistance (AMR) making use of non-essential target inhibitors: A review. Int. J. Mol. Sci. 2019, 20, 5844. [Google Scholar] [CrossRef] [Green Version]
- Salcedo-Sora, J.E.; Kell, D.B. A quantitative survey of bacterial persistence in the presence of antibiotics: Towards antipersister antimicrobial discovery. Antibiotics 2020, 9, 508. [Google Scholar] [CrossRef] [PubMed]
- Mendes, P.; Girardi, E.; Superti-Furga, G.; Kell, D.B. Why most transporter mutations that cause antibiotic resistance are to efflux pumps rather than to import transporters. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Kell, D.B.; Swainston, N.; Pir, P.; Oliver, S.G. Membrane transporter engineering in industrial biotechnology and whole cell biocatalysis. Trends Biotechnol. 2015, 33, 237–246. [Google Scholar] [CrossRef]
- Kell, D.B. Control of Metabolite Efflux in Microbial Cell Factories. Current Advances and Future Prospects, 1st ed.; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Piddock, L.J.V. Clinically Relevant Chromosomally Encoded Multidrug Resistance Efflux Pumps in Bacteria. Clin. Microbiol. Rev. 2006, 19, 382–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piddock, L.J.V. The 2019 Garrod lecture: MDR efflux in gram-negative bacteria—How understanding resistance led to a new tool for drug discovery. J. Antimicrob. Chemother. 2019, 74, 3128–3134. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Nawaz, N.; Dermani, F.K.; Kohlan, A.K.; Saidijam, M.; Patching, S.G. Bacterial multidrug efflux proteins: A major mechanism of antimicrobial resistance. Curr. Drug Targets 2018, 20, 16–28. [Google Scholar] [CrossRef]
- Phillips-Jones, M.K.; Harding, S.E. Antimicrobial resistance (AMR) nanomachines—Mechanisms for fluoroquinolone and glycopeptide recognition, efflux and/or deactivation. Biophys. Rev. 2018, 10, 347–362. [Google Scholar] [CrossRef] [Green Version]
- Poole, K. Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 2007, 39, 162–176. [Google Scholar] [CrossRef]
- Rahman, T.; Yarnall, B.; Doyle, D.A. Efflux drug transporters at the forefront of antimicrobial resistance. Eur. Biophys. J. 2017, 46, 647–653. [Google Scholar] [CrossRef]
- Willers, C.; Wentzel, J.; du Plessis, L.; Gouws, C.; Hamman, J.H. Efflux as a mechanism of antimicrobial drug resistance in clinical relevant microorganisms: The role of efflux inhibitors. Expert Opin. Ther. Targets 2017, 21, 23–36. [Google Scholar] [CrossRef]
- Vrancianu, C.O.; Gheorghe, I.; Dobre, E.-G.; Barbu, I.C.; Cristian, R.E.; Popa, M.; Lee, S.H.; Limban, C.; Vlad, I.M.; Chifiriuc, M.C. Emerging strategies to combat β-lactamase producing ESKAPE pathogens. Int. J. Mol. Sci. 2020, 21, 8527. [Google Scholar] [CrossRef]
- Gharavi, M.J.; Zarei, J.; Roshani-Asl, P.; Yazdanyar, Z.; Sharif, M.; Rashidi, N. Comprehensive study of antimicrobial susceptibility pattern and extended spectrum beta-lactamase (ESBL) prevalence in bacteria isolated from urine samples. Sci. Rep. 2021, 11, 578. [Google Scholar] [CrossRef]
- Eiamphungporn, W.; Schaduangrat, N.; Malik, A.A.; Nantasenamat, C. Tackling the antibiotic resistance caused by class A β-lactamases through the use of β-lactamase inhibitory protein. Int. J. Mol. Sci. 2018, 19, 2222. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Ye, L.; Kromann, S.; Meng, H. Occurrence of extended-spectrum β-lactamases, plasmid-mediated quinolone resistance, and disinfectant resistance genes in Escherichia coli isolated from ready-to-eat meat products. Foodborne Pathog. Dis. 2017, 14, 109–115. [Google Scholar] [CrossRef]
- Wright, G.D. Antibiotic adjuvants: Rescuing antibiotics from resistance. Trends Microbiol. 2016, 24, 862–871. [Google Scholar] [CrossRef]
- Lemire, J.A.; Harrison, J.; Turner, R.J. Antimicrobial activity of metals: Mechanisms, molecular targets and applications. Nat. Rev. Microbiol. 2013, 11, 371–384. [Google Scholar] [CrossRef]
- Vincent, M.; Duval, R.; Hartemann, P.; Engels-Deutsch, M. Contact killing and antimicrobial properties of copper. J. Appl. Microbiol. 2018, 124, 1032–1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arendsen, L.P.; Thakar, R.; Sultan, A.H. The use of copper as an antimicrobial agent in health care, including obstetrics and gynecology. Clin. Microbiol. Rev. 2019, 32, e00125-18. [Google Scholar] [CrossRef] [PubMed]
- Portelinha, J.; Duay, S.S.; Yu, S.I.; Heilemann, K.; Libardo, M.D.J.; Juliano, S.A.; Klassen, J.L.; Angeles-Boza, A.M. Antimicrobial peptides and copper(II) ions: Novel therapeutic opportunities. Chem. Rev. 2021, 121, 2648–2712. [Google Scholar] [CrossRef] [PubMed]
- Gant, V.A.; Wren, M.W.D.; Rollins, M.S.M.; Jeanes, A.; Hickok, S.S.; Hall, T.J. Three novel highly charged copper-based biocides: Safety and efficacy against healthcare-associated organisms. J. Antimicrob. Chemother. 2007, 60, 294–299. [Google Scholar] [CrossRef] [Green Version]
- Grass, G.; Rensing, C.; Solioz, M. Metallic copper as an antimicrobial surface. Appl. Environ. Microbiol. 2011, 77, 1541–1547. [Google Scholar] [CrossRef] [Green Version]
- Wiebelhaus, N.; Zaengle-Barone, J.M.; Hwang, K.K.; Franz, K.J.; Fitzgerald, M.C. Protein folding stability changes across the proteome reveal targets of cu toxicity in E. coli. ACS Chem. Biol. 2021, 16, 214–224. [Google Scholar] [CrossRef]
- Zaengle-Barone, J.M.; Jackson, A.C.; Besse, D.M.; Becken, B.; Arshad, M.; Seed, P.C.; Franz, K.J. Copper influences the antibacterial outcomes of a β-lactamase-activated prochelator against drug-resistant bacteria. ACS Infect. Dis. 2018, 4, 1019–1029. [Google Scholar] [CrossRef]
- Quezada, L.L.; Li, K.; McDonald, S.L.; Nguyen, Q.; Perkowski, A.J.; Pharr, C.W.; Gold, B.; Roberts, J.; McAulay, K.; Saito, K.; et al. Dual-pharmacophore pyrithione-containing cephalosporins kill both replicating and nonreplicating mycobacterium tuberculosis. ACS Infect. Dis. 2019, 5, 1433–1445. [Google Scholar] [CrossRef] [Green Version]
- Helsel, M.E.; White, E.J.; Razvi, S.Z.A.; Alies, B.; Franz, K.J. Chemical and functional properties of metal chelators that mobilize copper to elicit fungal killing of Cryptococcus neoformans. Metallomics 2017, 9, 69–81. [Google Scholar] [CrossRef] [Green Version]
- Oliveri, V. Biomedical applications of copper ionophores. Coord. Chem. Rev. 2020, 422, 213474. [Google Scholar] [CrossRef]
- Djoko, K.Y.; Achard, M.E.S.; Phan, M.-D.; Lo, A.W.; Miraula, M.; Prombhul, S.; Hancock, S.J.; Peters, K.M.; Sidjabat, H.E.; Harris, P.N.; et al. Copper ions and coordination complexes as novel carbapenem adjuvants. Antimicrob. Agents Chemother. 2018, 62, e02280-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandler, C.J.; Segel, I.H. Mechanism of the antimicrobial action of pyrithione: Effects on membrane transport, ATP levels, and protein synthesis. Antimicrob. Agents Chemother. 1978, 14, 60–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reeder, N.L.; Kaplan, J.; Xu, J.; Youngquist, R.S.; Wallace, J.; Hu, P.; Juhlin, K.D.; Schwartz, J.R.; Grant, R.A.; Fieno, A.; et al. Zinc pyrithione inhibits yeast growth through copper influx and inactivation of iron-sulfur proteins. Antimicrob. Agents Chemother. 2011, 55, 5753–5760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albert, A.; Rees, C.W.; Tomlinson, A.J. The influence of chemical constitution on antibacterial activity. VIII. 2-mercaptopyridine-n-oxide, and some general observations on metalbinding agents. Br. J. Exp. Pathol. 1956, 37, 500–511. [Google Scholar]
- Lanthaler, K.; Bilsland, E.; Dobson, P.; Moss, H.J.; Pir, P.; Kell, D.B.; Oliver, S.G. Genome-wide assessment of the carriers in-volved in the cellular uptake of drugs: A model system in yeast. BMC Biol. 2011, 9, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winter, G.E.; Radic, B.; Mayor-Ruiz, C.; Blomen, A.V.; Jessen-Trefzer, C.; Kandasamy, R.K.; Huber, K.V.; Gridling, M.; Chen, D.; Klampfl, T.; et al. The solute carrier SLC35F2 enables YM155-mediated DNA damage toxicity. Nat. Chem. Biol. 2014, 10, 768–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giacomini, K.M.; Huang, S.M.; Tweedie, D.J.; Benet, L.Z.; Brouwer, K.L.; Chu, X.; Dahlin, A.; Evers, R.; Fischer, V.; Hillgren, K.M.; et al. Membrane transporters in drug development. Nat. Rev. Drug Discov. 2010, 9, 215–236. [Google Scholar] [PubMed]
- Giacomini, K.M.; Galetin, A.; Huang, S.M. The international transporter consortium: Summarizing advances in the role of transporters in drug development. Clin. Pharmacol. Ther. 2018, 104, 766–771. [Google Scholar] [CrossRef] [PubMed]
- Dobson, P.D.; Kell, D.B. Carrier-mediated cellular uptake of pharmaceutical drugs: An exception or the rule? Nat. Rev. Drug Discov. 2008, 7, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Dobson, P.; Lanthaler, K.; Oliver, S.G.; Kell, D.B. Implications of the dominant role of cellular transporters in drug uptake. Curr. Top. Med. Chem. 2009, 9, 163–184. [Google Scholar] [CrossRef] [Green Version]
- Kell, D.B.; Dobson, P.D.; Bilsland, E.; Oliver, S.G. The promiscuous binding of pharmaceutical drugs and their transporter-mediated uptake into cells: What we (need to) know and how we can do so. Drug Discov. Today 2013, 18, 218–239. [Google Scholar] [CrossRef]
- Kell, D.B.; Oliver, S.G. How drugs get into cells: Tested and testable predictions to help discriminate between transporter-mediated uptake and lipoidal bilayer diffusion. Front. Pharmacol. 2014, 5, 231. [Google Scholar] [CrossRef] [Green Version]
- Girardi, E.; César-Razquin, A.; Lindinger, S.; Papakostas, K.; Konecka, J.; Hemmerich, J.; Kickinger, S.; Kartnig, F.; Gürtl, B.; Klavins, K.; et al. A widespread role for SLC transmembrane transporters in resistance to cytotoxic drugs. Nat. Chem. Biol. 2020, 16, 469–478. [Google Scholar] [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, A.K.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 469–478. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, N.; Nakahigashi, K.; Nakamichi, T.; Yoshino, M.; Takai, Y.; Touda, Y.; Furubayashi, A.; Kinjyo, S.; Dose, H.; Hasegawa, M.; et al. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol. Syst. Biol. 2009, 5, 335. [Google Scholar] [CrossRef] [Green Version]
- Kitagawa, M.; Ara, T.; Arifuzzaman, M.; Ioka-Nakamichi, T.; Inamoto, E.; Toyonaga, H.; Mori, H. Complete set of ORF clones of Escherichia coli ASKA library (A complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005, 12, 291–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, H.; Baba, T.; Yokoyama, K.; Takeuchi, R.; Nomura, W.; Makishi, K.; Otsuka, Y.; Dose, H.; Wanner, B.L. Identification of essential genes and synthetic lethal gene combinations in Escherichia coli K-12. Methods Mol. Biol. 2015, 1279, 45–65. [Google Scholar] [CrossRef]
- Davey, H.M.; Kell, D.B. Flow cytometry and cell sorting of heterogeneous microbial populations: The importance of single-cell analysis. Microbiol. Rev. 1996, 60, 641–696. [Google Scholar] [CrossRef] [PubMed]
- Jindal, S.; Thampy, H.; Day, P.J.R.; Kell, D.B. Very rapid flow cytometric assessment of antimicrobial susceptibility during the apparent lag phase of microbial (re)growth. Microbiology 2019, 165, 439–454. [Google Scholar] [CrossRef] [PubMed]
- Jindal, S.; Yang, L.; Day, P.J.; Kell, D.B. Involvement of multiple influx and efflux transporters in the accumulation of cationic fluorescent dyes by Escherichia coli. BMC Microbiol. 2019, 19, 195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salcedo-Sora, J.E.; Jindal, S.; O’Hagan, S.; Kell, D.B. A palette of fluorophores that are differentially accumulated by wild-type and mutant strains of Escherichia coli: Surrogate ligands for profiling bacterial membrane transporters. Microbiology 2021, 167, 001016. [Google Scholar] [CrossRef]
- Khattar, M.; Salt, W.; Stretton, R. The influence of pyrithione on the growth of micro-organisms. J. Appl. Bacteriol. 1988, 64, 265–272. [Google Scholar] [CrossRef]
- Ermolayeva, E.; Sanders, D. Mechanism of pyrithione-induced membrane depolarization in Neurospora crassa. Appl. Environ. Microbiol. 1995, 61, 3385–3390. [Google Scholar] [CrossRef] [Green Version]
- Tarrant, E.; Riboldi, G.P.; McIlvin, M.R.; Stevenson, J.; Barwinska-Sendra, A.; Stewart, L.J.; Saito, M.A.; Waldron, K.J. Copper stress in Staphylococcus aureus leads to adaptive changes in central carbon metabolism. Metallomics 2019, 11, 183–200. [Google Scholar] [CrossRef] [Green Version]
- Reeder, N.; Xu, J.; Youngquist, R.; Schwartz, J.; Rust, R.; Saunders, C. The antifungal mechanism of action of zinc pyrithione. Br. J. Dermatol. 2011, 165 (Suppl. 2), 9–12. [Google Scholar] [CrossRef]
- Wijekoon, C.J.K.; Udagedara, S.R.; Knorr, R.L.; Dimova, R.; Wedd, A.G.; Xiao, Z. Copper ATPase CopA from Escherichia coli: Quantitative correlation between ATPase activity and vectorial copper transport. J. Am. Chem. Soc. 2017, 139, 4266–4269. [Google Scholar] [CrossRef]
- Giachino, A.; Waldron, K.J. Copper tolerance in bacteria requires the activation of multiple accessory pathways. Mol. Microbiol. 2020, 114, 377–390. [Google Scholar] [CrossRef]
- Rosenzweig, A.C.; Argüello, J.M. Toward a molecular understanding of metal transport by P1B-Type ATPases. Curr. Top Membr. 2012, 69, 113–136. [Google Scholar] [CrossRef] [Green Version]
- Hou, Z.; Mitra, B. The metal specificity and selectivity of ZntA from Escherichia coli using the acylphosphate intermediate. J. Biol. Chem. 2003, 278, 28455–28461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rensing, C.; Fan, B.; Sharma, R.; Mitra, B.; Rosen, B.P. CopA: An Escherichia coli Cu(I)-translocating P-type ATPase. Proc. Natl. Acad. Sci. USA 2000, 97, 652–656. [Google Scholar] [CrossRef] [Green Version]
- Fan, B.; Rosen, B.P. Biochemical characterization of CopA, the Escherichia coli Cu(I)-translocating P-type ATPase. J. Biol. Chem. 2002, 277, 46987–46992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banci, L.; Bertini, I.; Baffoni, S.C.; Del Conte, R.; Gonnelli, L. Understanding copper trafficking in bacteria: Interaction between the copper transport protein CopZ and the N-terminal domain of the copper ATPase CopA from Bacillus subtilis. Biochemistry 2003, 42, 1939–1949. [Google Scholar] [CrossRef]
- Padilla-Benavides, T.; Thompson, A.M.G.; McEvoy, M.M.; Argüello, J.M. Mechanism of ATPase-mediated Cu+ export and delivery to periplasmic chaperones: The interaction of Escherichia coli CopA and CusF. J. Biol. Chem. 2014, 289, 20492–20501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radford, D.S.; Kihlken, M.A.; Borrelly, G.P.; Harwood, C.R.; Le Brun, N.; Cavet, J.S. CopZ from Bacillus subtilisinteracts in vivo with a copper exporting CPx-type ATPase CopA. FEMS Microbiol. Lett. 2003, 220, 105–112. [Google Scholar] [CrossRef] [Green Version]
- Kirakosyan, G.; Trchounian, A. Redox sensing by Escherichia coli: Effects of copper ions as oxidizers on proton-coupled membrane transport. Bioelectrochemistry 2007, 70, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Bader, M.L.; Drew, D.; de Gier, J.-W. Rationalizing membrane protein overexpression. Trends Biotechnol. 2006, 24, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Grisshammer, R.; Tate, C. Overexpression of integral membrane proteins for structural studies. Q. Rev. Biophys. 1995, 28, 315–422. [Google Scholar] [CrossRef] [PubMed]
- Beard, S.J.; Hashim, R.; Membrillo-Hernández, J.; Hughes, M.N.; Poole, R.K. Zinc(II) tolerance in Escherichia coli K-12: Evidence that the ZntA gene (o732) encodes a cation transport ATPase. Mol. Microbiol. 1997, 25, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Noll, M.; Lutsenko, S.; Noll, S.L.M. Expression of ZntA, a zinc-transporting P 1-type ATPase, is specifically regulated by zinc and cadmium. IUBMB Life 2000, 49, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Binet, M.R.; Poole, R.K. Cd(II), Pb(II) and Zn(II) ions regulate expression of the metal-transporting P-type ATPase ZntA in Escherichia coli. FEBS Lett. 2000, 473, 67–70. [Google Scholar] [CrossRef] [Green Version]
- Dutta, S.J.; Liu, J.; Mitra, B. Kinetic analysis of metal binding to the amino-terminal domain of ZntA by monitoring metal−thiolate charge-transfer complexes. Biochemistry 2005, 44, 14268–14274. [Google Scholar] [CrossRef]
- Sharma, R.; Rensing, C.; Rosen, B.P.; Mitra, B. The ATP hydrolytic activity of purified ZntA, a Pb(II)/Cd(II)/Zn(II)-translocating ATPase from Escherichia coli. J. Biol. Chem. 2000, 275, 3873–3878. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, P.T.; Lai, J.Y.; Lee, A.T.; Kaiser, J.T.; Rees, D.C. Noncanonical role for the binding protein in substrate uptake by the MetNI methionine ATP Binding Cassette (ABC) transporter. Proc. Natl. Acad. Sci. USA 2018, 115, E10596–E10604. [Google Scholar] [CrossRef] [Green Version]
- Merlin, C.; Gardiner, G.; Durand, S.; Masters, M. The Escherichia coli metD Locus encodes an ABC transporter which includes Abc (MetN), YaeE (MetI), and YaeC (MetQ). J. Bacteriol. 2002, 184, 5513–5517. [Google Scholar] [CrossRef] [Green Version]
- Carlson, M.L.; Stacey, R.G.; Young, J.W.; Wason, I.S.; Zhao, Z.; Rattray, D.G.; Scott, N.; Kerr, C.H.; Babu, M.; Foster, L.J.; et al. Profiling the Escherichia coli membrane protein interactome captured in peptidisc libraries. Elife 2019, 8, e46615. [Google Scholar] [CrossRef] [PubMed]
- Pierce, J.R.; Earhart, C.F. Escherichia coli K-12 envelope proteins specifically required for ferrienterobactin uptake. J. Bacteriol. 1986, 166, 930–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 16 June 2021).
- Ritz, C.; Baty, F.; Streibig, J.C.; Gerhard, D. Dose-response analysis using R. PLoS ONE 2015, 10, e0146021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sebaugh, J.L. Guidelines for accurate EC50/IC50 estimation. Pharm. Stat. 2011, 10, 128–134. [Google Scholar] [CrossRef]
- Hall, B.G.; Acar, H.; Nandipati, A.; Barlow, M. Growth rates made easy. Mol. Biol. Evol. 2013, 31, 232–238. [Google Scholar] [CrossRef] [Green Version]
- Vaas, L.A.I.; Sikorski, J.; Michael, V.; Göker, M.; Klenk, H.-P. Visualization and curve-parameter estimation strategies for efficient exploration of phenotype microarray kinetics. PLoS ONE 2012, 7, e34846. [Google Scholar] [CrossRef] [Green Version]
- Petzoldt, T. Growthrates: Estimate Growth Rates from Experimental Data. 2020. Available online: https://github.com/tpetzoldt/growthrates (accessed on 16 June 2021).







| Media | CuCl2 (µM) | IC50 (µM) |
|---|---|---|
| Lysogeny Broth (LB) | 0 | 6.05 ± 1.02 |
| 5 | 5.22 ± 0.95 | |
| 10 | 0.78 ± 0.25 | |
| 20 | 0.81 ± 0.28 |
| Strain (KO) | Growth Rate Ratio | Gene Annotation | |
|---|---|---|---|
| Mean | SD | ||
| metQ | 2.41 | 0.92 | L-methionine/D-methionine ABC transporter membrane anchored binding protein |
| fepC | 2.12 | 0.90 | ferric enterobactin ABC transporter ATP binding subunit |
| chbA | 1.75 | 0.70 | N,N’-diacetylchitobiose-specific PTS enzyme IIA component |
| araJ | 1.67 | 0.64 | putative transport protein AraJ |
| phoR | 1.66 | 0.46 | sensor histidine kinase |
| lldP | 1.59 | 0.41 | lactate/glycolate:H+ symporter LldP |
| potB | 1.59 | 0.46 | spermidine preferential ABC transporter membrane subunit PotB |
| glvB | 1.54 | 0.45 | putative PTS enzyme II component GlvB |
| hofC | 1.54 | 0.38 | inner membrane protein HofC |
| mdtC | 1.52 | 0.31 | multidrug efflux pump RND permease subunit MdtC |
| WT | 1.04 | 0.11 | E. coli BW25113 (Keio collection reference strain) |
| tatC | 0.70 | 0.11 | twin arginine protein translocation system-TatC protein |
| yjeP | 0.70 | 0.13 | mini conductance mechanosensitive channel MscM |
| fucP | 0.69 | 0.19 | L-fucose:H+ symporter |
| ybiT | 0.69 | 0.14 | putative ATP-binding protein YbiT |
| yheS | 0.60 | 0.20 | putative ATP-binding protein YheS |
| malF | 0.54 | 0.10 | maltose ABC transporter membrane subunit MalF |
| xylE | 0.52 | 0.16 | D-xylose:H+ symporter |
| atpA | 0.44 | 0.12 | ATP synthase F1 complex subunit α |
| copA | 0.44 | 0.12 | Cu+ exporting P-type ATPase (copA;ybaR;atcU) |
| zntA | 0.09 | 0.03 | Zn2+/Cd2+/Pb2+ exporting P-type ATPase |
| Conditions | BW25113 | metQ | fepC | copA | zntA |
|---|---|---|---|---|---|
| PT alone | 17.5 | 17.5 | 17.5 | 8.8 | 2.2 |
| PT and 10 µM Cu | 17.5 | 17.5 | 17.5 | 2.2 | 8.8 |
| PT and 10 µM Fe | 17.5 | 17.5 | 17.5 | 8.8 | 2.2 |
| PT and 10 µM Zn | 17.5 | 8.8 | 17.5 | 8.8 | 1.1 |
| PT and 1.92 mM BCS | >140 | >140 | >140 | >140 | 140 |
| PT and 1.92 mM BCS and 10 µM Zn | >140 | >140 | 140 | >140 | 17.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salcedo-Sora, J.E.; Robison, A.T.R.; Zaengle-Barone, J.; Franz, K.J.; Kell, D.B. Membrane Transporters Involved in the Antimicrobial Activities of Pyrithione in Escherichia coli. Molecules 2021, 26, 5826. https://doi.org/10.3390/molecules26195826
Salcedo-Sora JE, Robison ATR, Zaengle-Barone J, Franz KJ, Kell DB. Membrane Transporters Involved in the Antimicrobial Activities of Pyrithione in Escherichia coli. Molecules. 2021; 26(19):5826. https://doi.org/10.3390/molecules26195826
Chicago/Turabian StyleSalcedo-Sora, Jesus Enrique, Amy T. R. Robison, Jacqueline Zaengle-Barone, Katherine J. Franz, and Douglas B. Kell. 2021. "Membrane Transporters Involved in the Antimicrobial Activities of Pyrithione in Escherichia coli" Molecules 26, no. 19: 5826. https://doi.org/10.3390/molecules26195826
APA StyleSalcedo-Sora, J. E., Robison, A. T. R., Zaengle-Barone, J., Franz, K. J., & Kell, D. B. (2021). Membrane Transporters Involved in the Antimicrobial Activities of Pyrithione in Escherichia coli. Molecules, 26(19), 5826. https://doi.org/10.3390/molecules26195826