Searching Anti-Zika Virus Activity in 1H-1,2,3-Triazole Based Compounds
Abstract
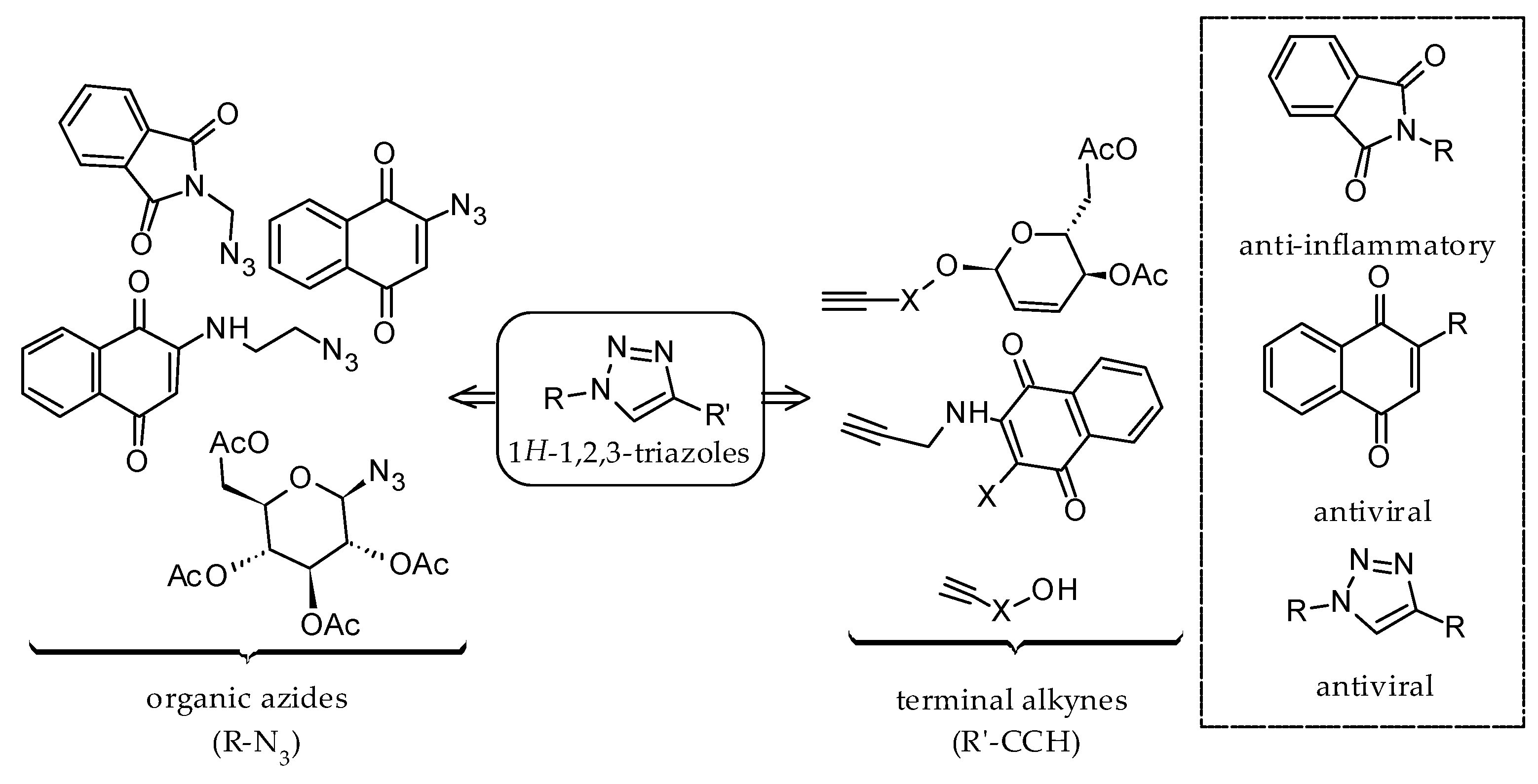
:1. Introduction
2. Results
2.1. Synthesis of the Compounds 1–7
2.2. Cytotoxicity of Triazole Derivatives in Vero Cells
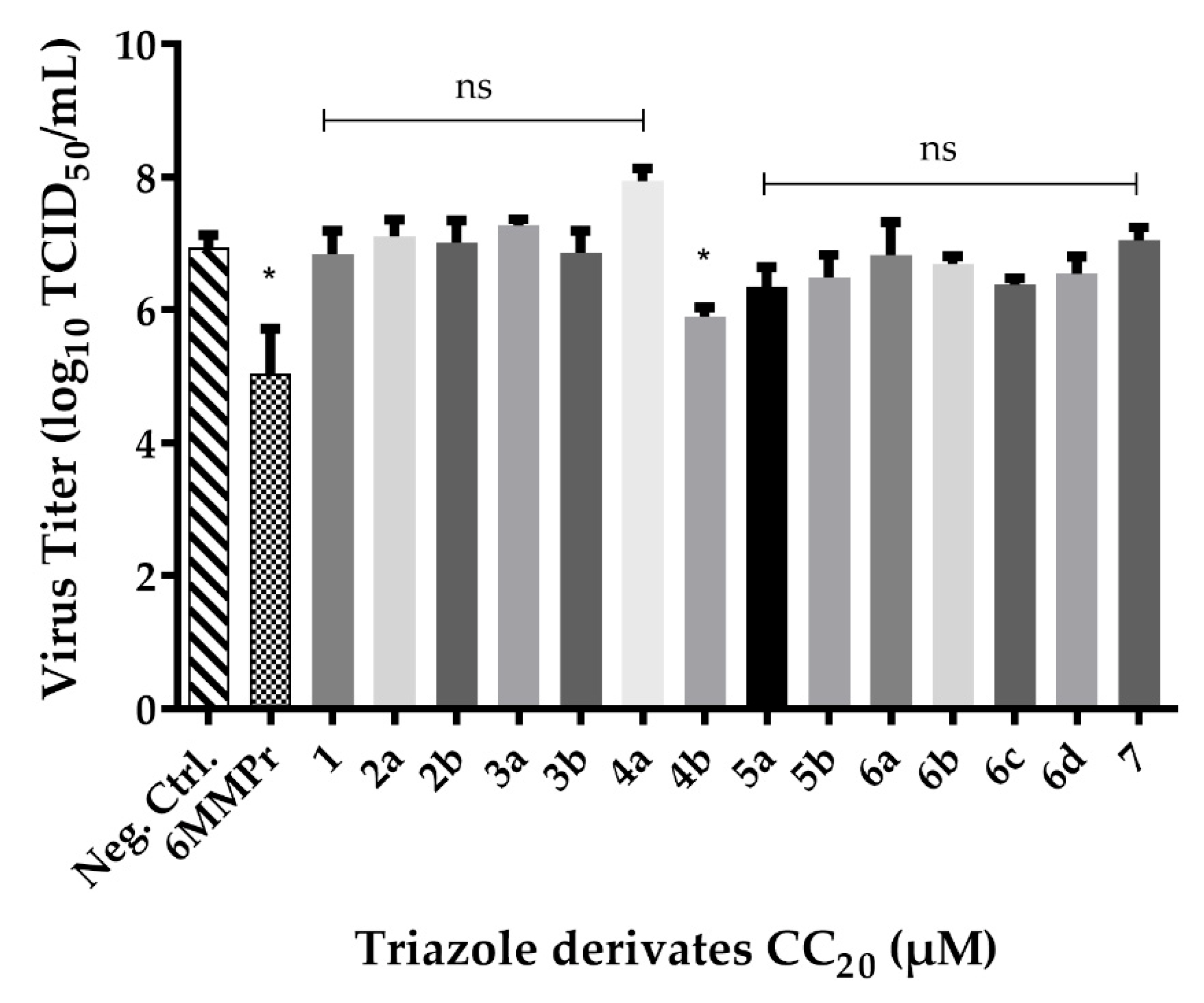
2.3. Screening of Triazole Derivates
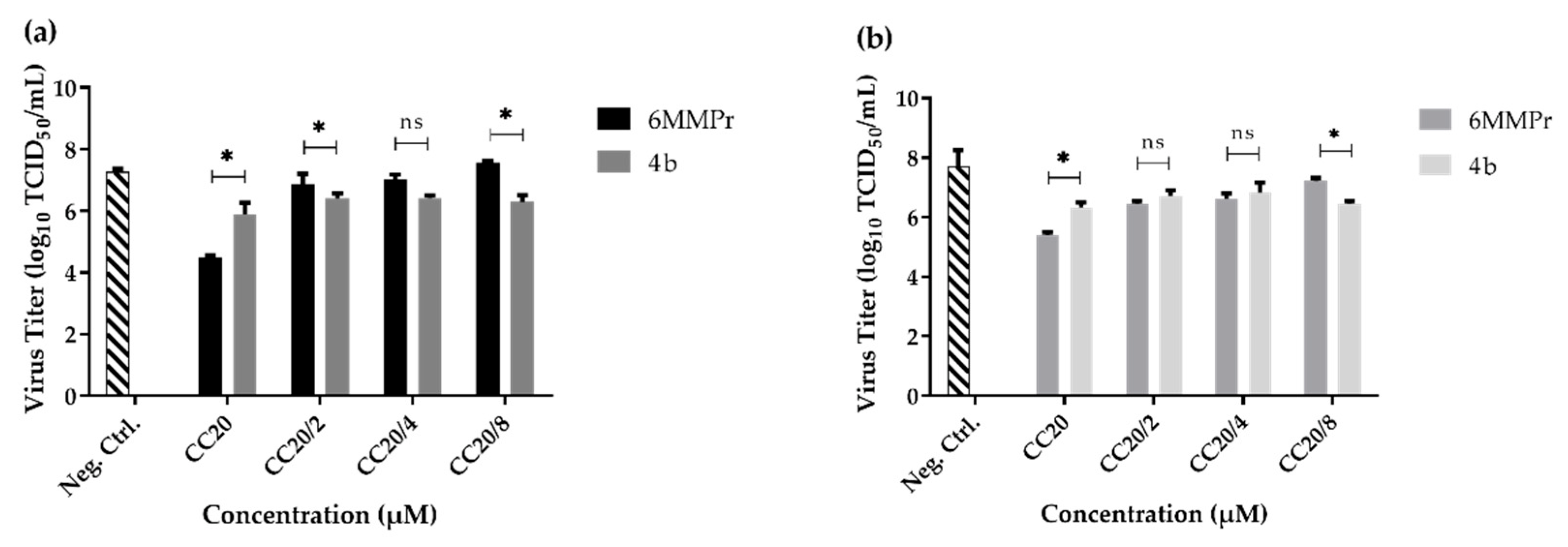
2.4. Pre-Treatment with Compound 4b
2.5. Post-Infection Studies of Compound 4b
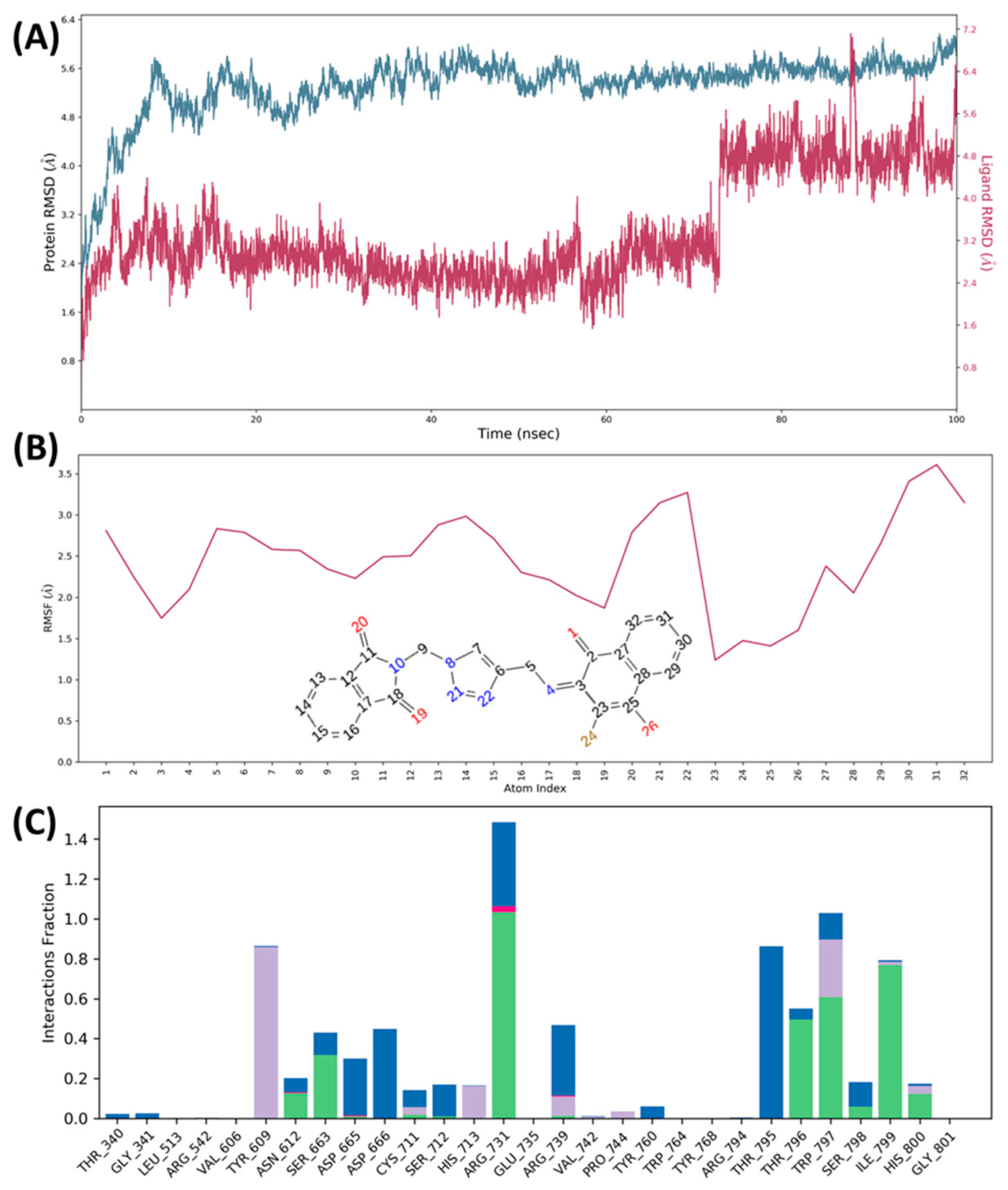
2.6. Molecular Docking
3. Discussion
4. Materials and Methods
4.1. Chemistry
4.2. Cells and Viruses
4.3. Titration of Stock and Samples Virus
4.4. Cytotoxicity Test with MTT Method
4.5. Screening Test
4.6. Pre-Treatment Activity
4.7. Post-Infection Activity
4.8. Molecular Docking
4.8.1. Molecular Docking of Designed Chemical Library
4.8.2. Molecular Dynamics (MD) Simulation
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Guedes, D.R.; Paiva, M.H.; Donato, M.M.; Barbosa, P.P.; Krokovsky, L.; Rocha, S.W.D.S.; LA Saraiva, K.; Crespo, M.M.; Rezende, T.M.; Wallau, G.L.; et al. Zika virus replication in the mosquito Culex quinquefasciatus in Brazil. Emerg. Microbes Infect. 2017, 6, e69. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Sun, L. Systematic Analysis of Structure Similarity between Zika Virus and Other Flaviviruses. ACS Infect. Dis. 2019, 5, 1070–1080. [Google Scholar] [CrossRef]
- Dick, G.W.A.; Kitchen, S.F.; Haddow, A.J. Zika Virus (I). Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952, 46, 509–520. [Google Scholar] [CrossRef]
- Duffy, M.R.; Chen, T.-H.; Hancock, W.T.; Powers, A.M.; Kool, J.L.; Lanciotti, R.S.; Pretrick, M.; Marfel, M.; Holzbauer, S.; Dubray, C.; et al. Zika Virus Outbreak on Yap Island, Federated States of Micronesia. N. Engl. J. Med. 2009, 360, 2536–2543. [Google Scholar] [CrossRef] [PubMed]
- Cao-Lormeau, V.-M.; Roche, C.; Teissier, A.; Robin, E.; Berry, A.-L.; Mallet, H.-P.; Sall, A.A.; Musso, D. Zika virus, French Polynesia, South Pacific, 2013. Emerg. Infect. Dis. 2014, 20, 1085–1086. [Google Scholar] [CrossRef] [PubMed]
- Campos, T.D.L.; Durães-Carvalho, R.; Rezende, A.M.; De Carvalho, O.V.; Kohl, A.; Wallau, G.L.; Pena, L.J. Revisiting key entry routes of human epidemic arboviruses into the mainland americas through large-scale phylogenomics. Int. J. Genom. 2018, 2018, 6941735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowe, R.; Barcellos, C.; Brasil, P.; Cruz, O.G.; Honório, N.A.; Kuper, H.; Carvalho, M.S. The zika virus epidemic in brazil: From discovery to future implications. Int. J. Environ. Res. Public Health 2018, 15, 96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira, M.L.B.; Albuquerque, M.D.F.P.M.D.; de Brito, C.A.A.; França, R.F.D.O.; Moreira, Á.J.P.; Machado, M.; Íris, D.M.; Melo, R.D.P.; Medialdea-Carrera, R.; Mesquita, S.D.; et al. Neurological disease in adults with Zika and chikungunya virus infection in Northeast Brazil: A prospective observational study. Lancet Neurol. 2020, 19, 826–839. [Google Scholar] [CrossRef]
- Cardona-Ospina, J.A.; Henao-SanMartin, V.; Acevedo-Mendoza, W.F.; Nasner-Posso, K.M.; Martínez-Pulgarin, D.F.; Restrepo-López, A.; Valencia-Gallego, V.; Collins, M.H.; Rodriguez-Morales, A.J. Fatal Zika virus infection in the Americas: A systematic review. Int. J. Infect. Dis. 2019, 88, 49–59. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, S.A.; Jamieson, D.J.; Honein, M.A.; Petersen, L.R. Zika Virus and Birth Defects-Reviewing the Evidence for Causality. N. Engl. J. Med. 2016, 374, 1981–1987. [Google Scholar] [CrossRef]
- Akhtar, J.; Khan, A.A.; Ali, Z.; Haider, R.; Yar, M.S. Structure-activity relationship (SAR) study and design strategies of nitrogen-containing heterocyclic moieties for their anticancer activities. Eur. J. Med. Chem. 2017, 125, 143–189. [Google Scholar] [CrossRef]
- Li, C.; Zhu, X.; Ji, X.; Quanquin, N.; Deng, Y.Q.; Tian, M.; Aliyari, R.; Zuo, X.; Yuan, L.; Afridi, S.K.; et al. Chloroquine, a FDA-approved Drug, Prevents Zika Virus Infection and its Associated Congenital Microcephaly in Mice. EBioMedicine 2017, 24, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Bullard-Feibelman, K.M.; Govero, J.; Zhu, Z.; Salazar, V.; Veselinovic, M.; Diamond, M.S.; Geiss, B.J. The FDA-approved drug sofosbuvir inhibits Zika virus infection. Antivir. Res. 2017, 137, 134–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.-A.; Seong, R.-K.; Kumar, M.; Shin, O.S. Favipiravir and ribavirin inhibit replication of Asian and African strains of zika virus in different cell models. Viruses 2018, 10, 72. [Google Scholar] [CrossRef] [Green Version]
- Soto-Acosta, R.; Jung, E.; Qiu, L.; Wilson, D.; Geraghty, R.; Chen, L. 4,7-Disubstituted 7H-Pyrrolo[2,3-D]Pyrimidines and Their Analogs As Antiviral Agents Against Zika Virus. Molecules 2021, 26, 3799. [Google Scholar] [CrossRef]
- Baz, M.; Boivin, G. Antiviral agents in development for zika virus infections. Pharmaceuticals 2019, 12, 101. [Google Scholar] [CrossRef] [Green Version]
- De Carvalho, O.V.; Félix, D.M.; de Mendonça, L.R.; de Araújo, C.M.C.S.; Franca, R.F.D.O.; Cordeiro, M.T.; Júnior, A.S.; Pena, L.J. The thiopurine nucleoside analogue 6-methylmercaptopurine riboside (6MMPr) effectively blocks Zika virus replication. Int. J. Antimicrob. Agents 2017, 50, 718–725. [Google Scholar] [CrossRef] [PubMed]
- Brzuska, G.; Pastuch-Gawolek, G.; Krawczyk, M.; Szewczyk, B.; Krol, E. Anti-tick-borne encephalitis virus activity of novel uridine glycoconjugates containing amide or/and 1,2,3-triazole moiety in the linker structure. Pharmaceuticals 2020, 13, 460. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, V.N.M.; Moura, C.F.D.A.; Peixoto, A.D.S.; Ferreira, V.P.G.; Araújo, H.M.; Pimentel, L.M.L.M.; Pessoa, C.D.Ó.; Nicolete, R.; dos Anjos, J.V.; Sharma, P.P.; et al. Synthesis of alkynylated 1,2,4-oxadiazole/1,2,3-1H-triazole glycoconjugates: Discovering new compounds for use in chemotherapy against lung carcinoma and Mycobacterium tuberculosis. Eur. J. Med. Chem. 2021, 220, 113472. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, G.B.; Guimarães, B.M.; Assis, S.P.O.; Lima, V.L.M.; De Oliveira, R.N. Ultrasound-Assisted Synthesis of 1-N-β-D-Glucopyranosyl-1H-1,2,3-triazole Benzoheterocycles and their Anti-Inflammatory Activities. J. Braz. Chem. Soc. 2013, 24, 914–921. [Google Scholar] [CrossRef]
- Gonzaga, D.; Gomes, R.; Marra, R.; Da Silva, F.; Gomes, M.; Ferreira, D.; Santos, R.; Pinto, A.; Ratcliffe, N.A.; Cirne-Santos, C.; et al. Inhibition of Zika virus replication by synthetic bis-naphthoquinones. J. Braz. Chem. Soc. 2019, 30, 1697–1706. [Google Scholar] [CrossRef]
- Lima, Â.; Teixeira, R.; Silva, B.; Siqueira, R.P.; Silva, Í.; Santos, E.; Fernandes, M.C.; Gonçalves, V.; Bressan, G.; Mendes, T.; et al. Síntese e avaliação das atividades fotoprotetora, citotóxica e antiviral contra o Zika virus de derivados triazólicos da benzofenona. Quim. Nova 2019, 42, 473–484. [Google Scholar] [CrossRef]
- Aguiar, D.F.; Dutra, L.L.A.; Dantas, W.M.; Carvalho, G.; Lemes, R.P.G.; Pessoa, C.D.Ó.; Paier, C.R.K.; Araujo, P.; Araujo, E.; Pena, L.J.; et al. Synthesis, Antitumor and Cytotoxic Activity of New Adamantyl O-Acylamidoximes and 3-Aryl-5-Adamantane-1,2,4-Oxadiazole Derivatives. ChemistrySelect 2019, 4, 9112–9118. [Google Scholar] [CrossRef]
- De Oliveira, V.N.M.; Dos Santos, F.G.; Ferreira, V.P.G.; Araújo, H.M.; Pessoa, C.D.Ó.; Nicolete, R.; Oliveira, R. Focused microwave irradiation-assisted synthesis of N-cyclohexyl-1,2,4-oxadiazole derivatives with antitumor activity. Synth. Commun. 2018, 48, 2522–2532. [Google Scholar] [CrossRef]
- De Oliveira, R.N.; Melo, V.N.; Dantas, W.M.; Camara, C.A. Synthesis of 2,3-unsaturated alkynyl O-glucosides from tri-O-acetyl-d-glucal by using montmorillonite K-10/iron(III) chloride hexahydrate with subsequent copper(I)-catalyzed 1,3-dipolar cycloaddition. Synthesis 2015, 47, 3529–3541. [Google Scholar] [CrossRef]
- De Oliveira, R.; Da Silva, M.; Da Silva, M.; Melo, V.; Valença, W.; Da Paz, J.; Camara, C. New strategies for molecular diversification of 2-[Aminoalkyl-(1H-1,2,3-Triazol-1- yl)]-1,4-naphthoquinones using click chemistry. J. Braz. Chem. Soc. 2017, 28, 681–688. [Google Scholar] [CrossRef]
- Guimarães, T.T.; Pinto, M.D.C.F.; Lanza, J.S.; Melo, M.N.; Neto, R.M.; de Melo, I.M.; Diogo, E.B.; Ferreira, V.; Camara, C.; Valença, W.O.; et al. Potent naphthoquinones against antimony-sensitive and -resistant Leishmania parasites: Synthesis of novel α- And nor-α-lapachone-based 1,2,3-triazoles by copper-catalyzed azide-alkyne cycloaddition. Eur. J. Med. Chem. 2013, 63, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Holanda, V.N.; da Silva, W.V.; Nascimento, P.H.D.; Silva, S.R.B.; Filho, P.E.C.; Assis, S.P.D.O.; da Silva, C.A.; de Oliveira, R.N.; de Figueiredo, R.C.B.Q.; Lima, V.L.D.M. Antileishmanial activity of 4-phenyl-1-[2-(phthalimido-2-yl)ethyl]-1H-1,2,3-triazole (PT4) derivative on Leishmania amazonensis and Leishmania braziliensis: In silico ADMET, in vitro activity, docking and molecular dynamic simulations. Bioorg. Chem. 2020, 105, 104437. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinforma. 2016, 54, 5.6.1–5.6.37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [Green Version]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [Green Version]
- Lüthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of protein models with three-dimensional profiles. Nature 1992, 359, 83–85. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Madhavi Sastry, G.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided. Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Sharma, P.P.; Shankar, U.; Kumar, D.; Joshi, S.K.; Pena, L.; Durvasula, R.; Kumar, A.; Kempaiah, P.; Poonam; et al. Discovery of New Hydroxyethylamine Analogs against 3CLproProtein Target of SARS-CoV-2: Molecular Docking, Molecular Dynamics Simulation, and Structure-Activity Relationship Studies. J. Chem. Inf. Model. 2020, 60, 5754–5770. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger Release 2020-1: Protein Preparation Wizard; Epik; Prime, S., LLC, New York, NY, 2016; Impact, Schrödinger, LLC, New York, NY, 2016; Prime, Schrödinger, LLC, New York, NY. 2020.
- Schrödinger Release 2020-1: LigPrep, S., LLC, New York, NY. 2020.
- Schrödinger Release 2020-1: Epik, S., LLC, New York, NY. 2020.
- Schrödinger Release 2020-1: SiteMap, S., LLC, New York, NY. 2021.
- Schrödinger Release 2020-1: Glide, S., LLC, New York, NY. 2020.
- Schrödinger Release 2020-1: Prime, S., LLC, New York, NY. 2020.
- Schrödinger Release 2020-1: Desmond Molecular Dynamics System, D.E.S.R., New York, NY, 2020. Maestro-Desmond Interoperability Tools, Schrödinger, New York, NY. 2020.
- Mark, P.; Nilsson, L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
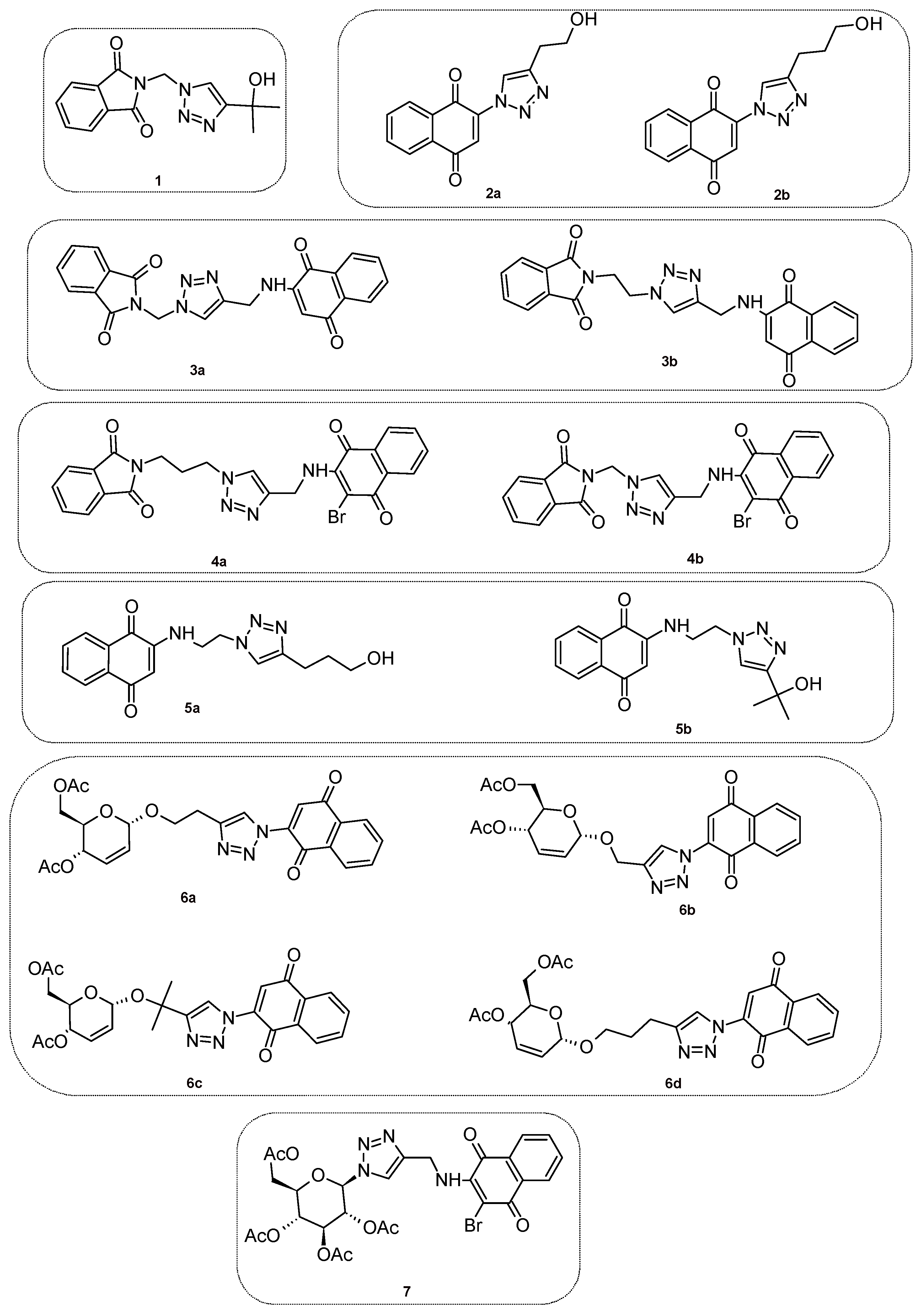



| Entry | Products | Methods | Yields, % | Reference |
|---|---|---|---|---|
| 1 | 1 | A | 92 | [26] |
| 2 | 2a,b | B | 85–95 | [25] |
| 3 | 3a,b | C | 70–95 | [26] |
| 4 | 4a,b | D | 70–88 | [27] |
| 5 | 5a | E | 90 | New compound |
| 5b | E | 86 | [26] | |
| 6 | 6a–d | F | 62–85 | [25] |
| 7 | 7 | D | 85 | [27] |
| Compound | CC20 (µM) 1 | CC50 (µM) 2 | logP 3 |
|---|---|---|---|
| 1 | 177.10 | 680.10 | 1.44 |
| 2a | 8.87 | 38.01 | 0.99 |
| 2b | 9.89 | 38.81 | 1.26 |
| 3a | 55.28 | 469.80 | 2.21 |
| 3b | 527.20 | 1189.00 | 2.22 |
| 4a | 40.69 | 68.78 | 3.23 |
| 4b | 136.90 | 330.20 | 2.94 |
| 5a | 74.85 | 293.90 | 1.05 |
| 5b | 66.93 | 298.20 | 1.38 |
| 6a | 39.92 | 68.05 | 2.07 |
| 6b | 21.18 | 48.69 | 1.86 |
| 6c | 25.73 | 58.14 | 2.67 |
| 6d | 38.61 | 69.50 | 2.34 |
| 7 | 119.10 | 334.50 | 1.80 |
| 6MMPr 4 | 60.5 | 291 | 0.55 |
| Compound | 4b | 6MMPr | |||
|---|---|---|---|---|---|
| Concentration (µM) | 3 d.p.i. 1 % | 5 d.p.i % | Concentration (µM) | 3 d.p.i. % | 5 d.p.i. % |
| CC20 = 136.90 | 94.9 | 97.5 | 60.50 | 99.8 | 99.7 |
| CC20/2 = 68.45 | 85.6 | 93.7 | 30.25 | 54.4 | 96.8 |
| CC20/4 = 34.23 | 85.8 | 90.8 | 15.13 | 42.7 | 95.1 |
| CC20/8 = 17.11 | 88.8 | 96.8 | 7.6 | −93.4 * | 81.1 |
| IC50 1 (µM) | SI 2 | % Viral Inhibition | |
|---|---|---|---|
| 4b | 146.0 | 2.3 | 91.1 |
| 6MMPr | 24.5 | 11.9 | 97.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dantas, W.M.; de Oliveira, V.N.M.; Santos, D.A.L.; Seabra, G.; Sharma, P.P.; Rathi, B.; Pena, L.J.; de Oliveira, R.N. Searching Anti-Zika Virus Activity in 1H-1,2,3-Triazole Based Compounds. Molecules 2021, 26, 5869. https://doi.org/10.3390/molecules26195869
Dantas WM, de Oliveira VNM, Santos DAL, Seabra G, Sharma PP, Rathi B, Pena LJ, de Oliveira RN. Searching Anti-Zika Virus Activity in 1H-1,2,3-Triazole Based Compounds. Molecules. 2021; 26(19):5869. https://doi.org/10.3390/molecules26195869
Chicago/Turabian StyleDantas, Willyenne M., Valentina N. M. de Oliveira, Diogo A. L. Santos, Gustavo Seabra, Prem P. Sharma, Brijesh Rathi, Lindomar J. Pena, and Ronaldo N. de Oliveira. 2021. "Searching Anti-Zika Virus Activity in 1H-1,2,3-Triazole Based Compounds" Molecules 26, no. 19: 5869. https://doi.org/10.3390/molecules26195869
APA StyleDantas, W. M., de Oliveira, V. N. M., Santos, D. A. L., Seabra, G., Sharma, P. P., Rathi, B., Pena, L. J., & de Oliveira, R. N. (2021). Searching Anti-Zika Virus Activity in 1H-1,2,3-Triazole Based Compounds. Molecules, 26(19), 5869. https://doi.org/10.3390/molecules26195869








