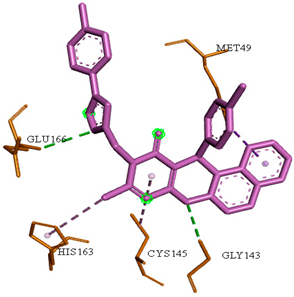
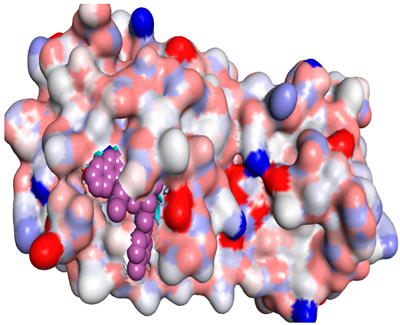
Abstract
A series of novel naphthopyrano[2,3-d]pyrimidin-11(12H)-one containing isoxazole nucleus 4 was synthesized under microwave irradiation and classical conditions in moderate to excellent yields upon 1,3-dipolar cycloaddition reaction using various arylnitrile oxides under copper(I) catalyst. A one-pot, three-component reaction, N-propargylation and Dimroth rearrangement were used as the key steps for the preparation of the dipolarophiles3. The structures of the synthesized compounds were established by 1H NMR, 13C NMR and HRMS-ES means. The present study aims to also predict the theoretical assembly of the COVID-19 protease (SARS-CoV-2 Mpro) and to discover in advance whether this protein can be targeted by the compounds 4a–1 and thus be synthesized. The docking scores of these compounds were compared to those of the co-crystallized native ligand inhibitor (N3) which was used as a reference standard. The results showed that all the synthesized compounds (4a–l) gave interesting binding scores compared to those of N3 inhibitor. It was found that compounds 4a, 4e and 4i achieved greatly similar binding scores and modes of interaction than N3, indicating promising affinity towards SARS-CoV-2 Mpro. On the other hand, the derivatives 4k, 4h and 4j showed binding energy scores (−8.9, −8.5 and −8.4 kcal/mol, respectively) higher than the Mpro N3 inhibitor (−7.0 kcal/mol), revealing, in their turn, a strong interaction with the target protease, although their interactions were not entirely comparable to that of the reference N3.
1. Introduction
Coronaviruses (CoVs) tend to have a high zoonotic potential and according to the World Health Organization (WHO), these viral diseases have emerged as a serious health issue to the world [1]. Consequently, the WHO declared a state of global health emergency to coordinate scientific and medical efforts to rapidly develop a cure for patients [2]. The discovery of an antiviral drug is of immense importance in the current spread of the rapidly modifiable SARS-CoV-2. The aim of the present study was to develop an antiviral drug against the novel COVID-19 virus [3]. One of the ways that several research teams are thinking is to highlight promising molecules and drug compounds by making a virtual screening via molecular docking of drugs approved by the FDA, certain natural substances, and synthetic heterocyclic compounds such as pyranopyrimidinones linked to isoxazoles for probable therapeutic outcome. In this context, antiviral compounds have seriously attracted the attention of organic medicinal chemists, who have thought of chemically modifying them in different ways in order to achieve much more active structural analogues.
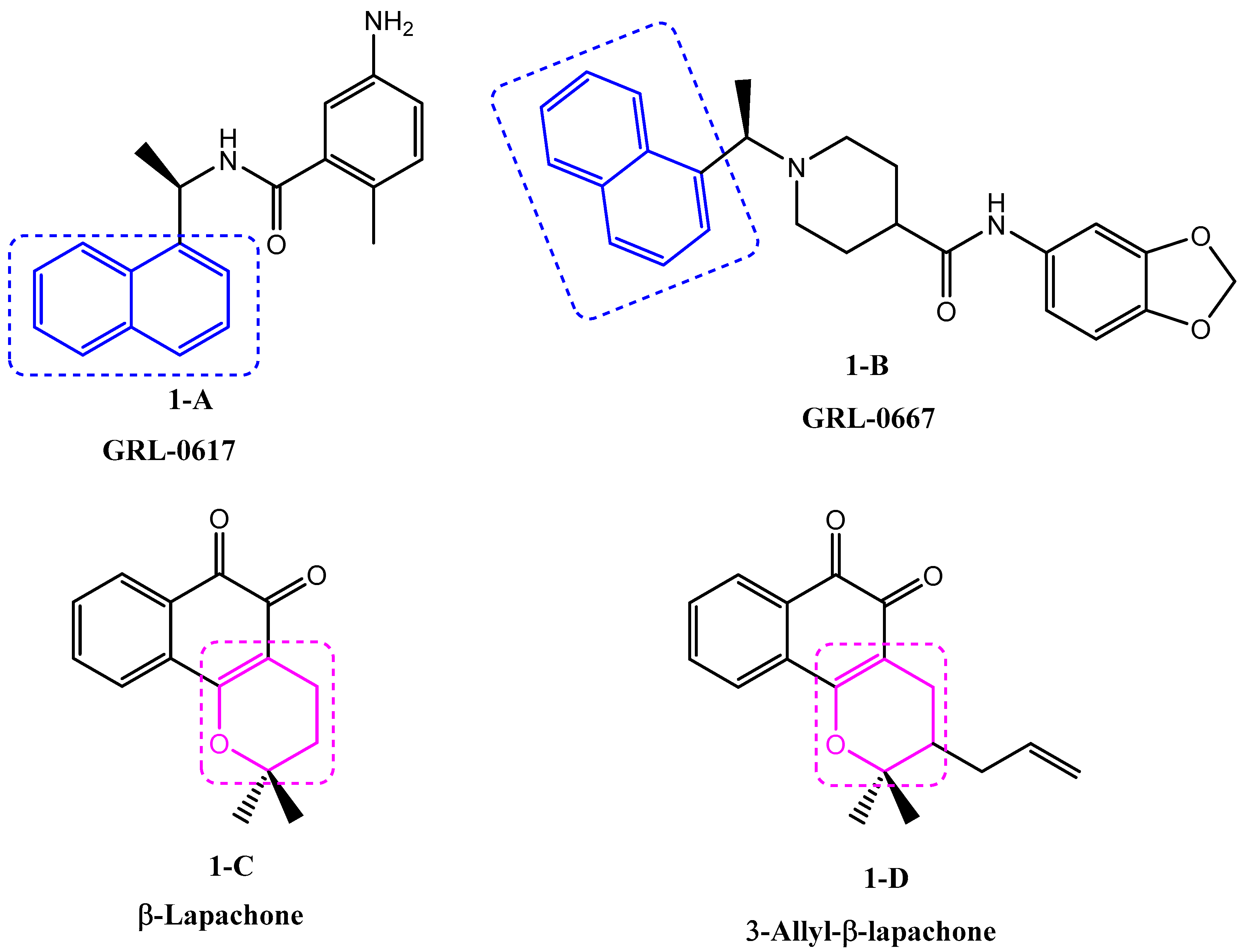
Naphthalenes have, for a long time, attracted the attention of organic chemists and biologists due to their wide spectrum of biological properties, such as antimicrobial, cytotoxic [4], cardiovascular [5], anti-inflammatory [6], and antiviral (Figure 1, 1-A [7], 1-B [8]).
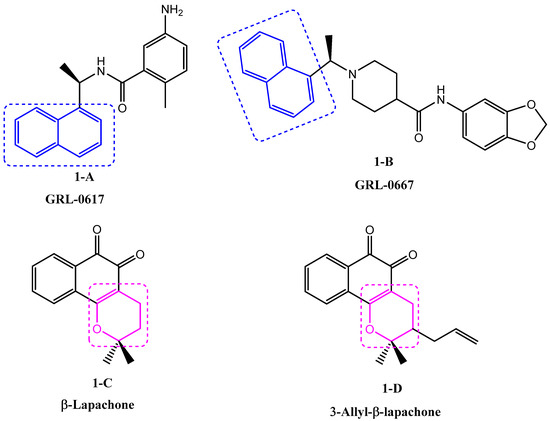
Figure 1.
Previously reported antiviral naphthalene and conjugated pyran derivatives.
On the other hand, heterocyclic scaffolds play a central role in drug discovery and development, thus constituting the key structural component of a majority of biologically active moieties. Conjugated pyranes are heterocyclic compounds arousing great interest due to their multiple biological activities, such as antimicrobial [9], anti-tumor [10], influenza inhibition [11] and antiviral (Figure 1, 1-C, 1-D) [12].
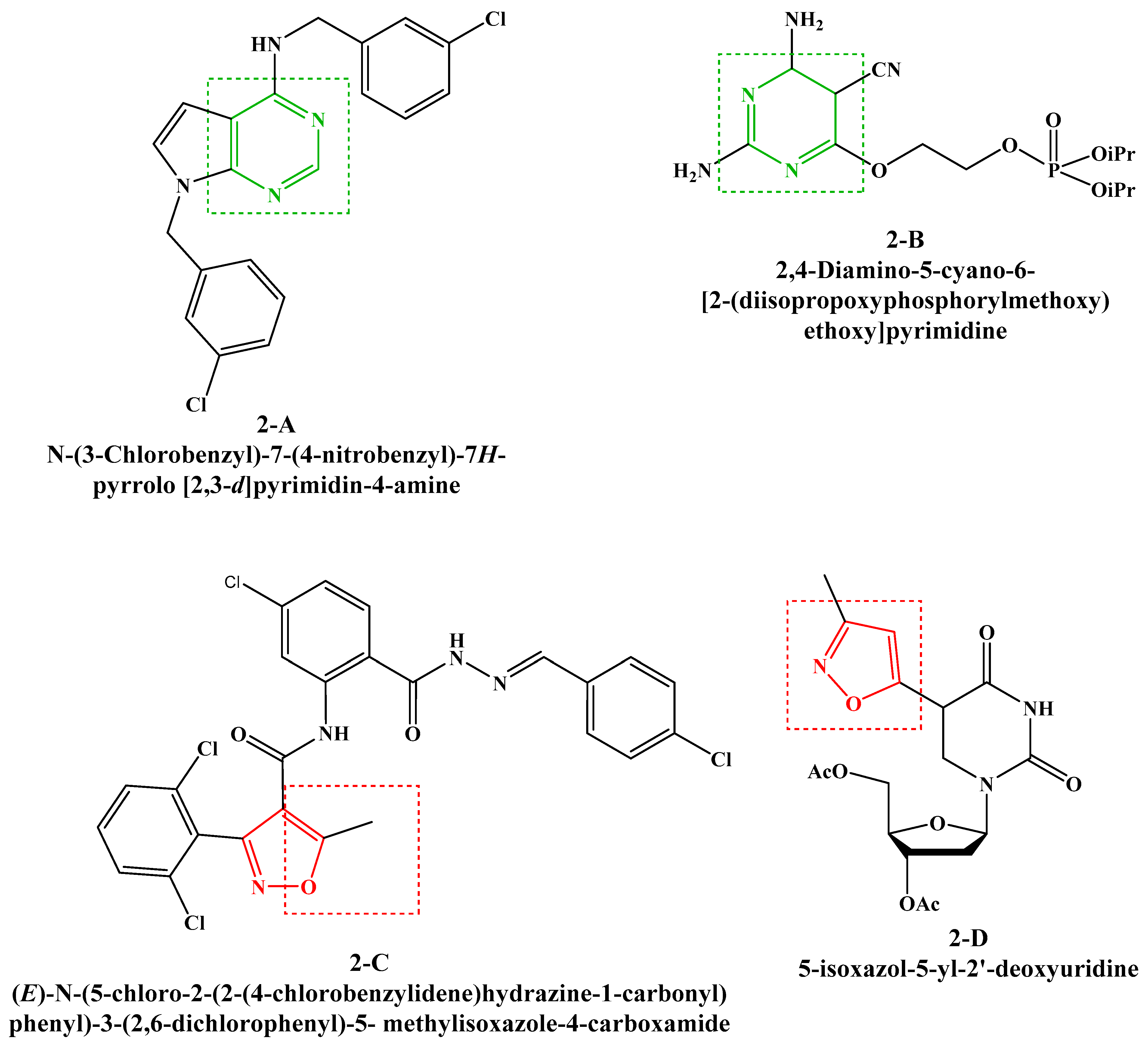
On the other hand, pyrimidines are among the most important nitrogenous heterocyclic classes in medicinal chemistry due to their various extensive biological and therapeutic activities [13]. In this context, the condensed derivatives of pyrimidines are very attractive targets because of their various pharmacological effects, such as anti-tubercular [14], antitumor [15], antityrosinase [16], antiproliferative [17], anti-inflammatory [18] and antiviral (Figure 2, 2-A [19], 2-B [20]).
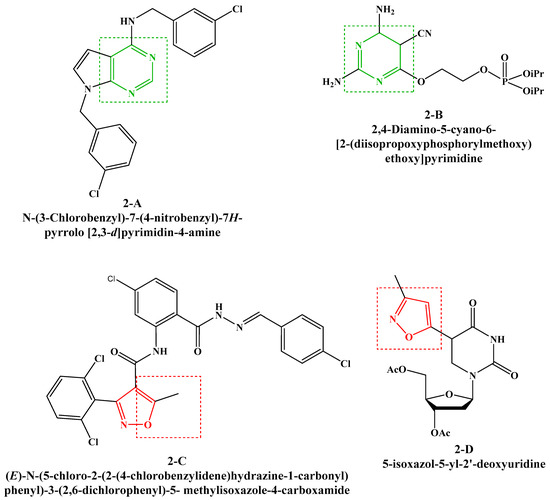
Figure 2.
Previously reported antiviral compounds bearing pyrimidine and isoxazole rings.
Furthermore, isoxazole derivatives possess various biological activities such as anti-inflammatory [21], antileishmanial [22], trypanocidal [23], antimicrobial [24,25] and antiviral (Figure 2, 2-C [26], 2-D [27]).
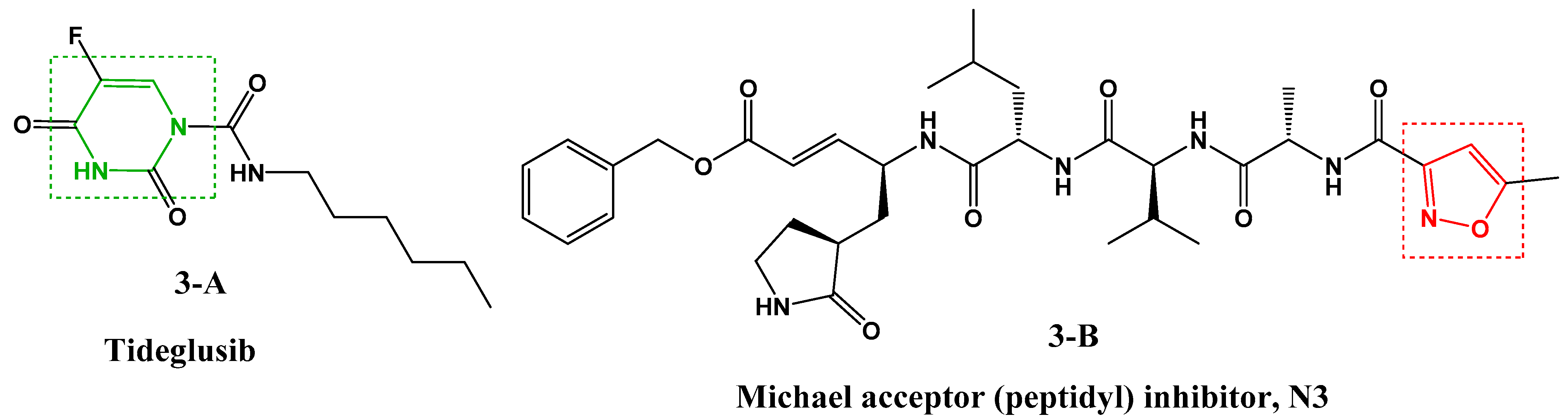
In addition, numerous target-based virtual screenings were performed to discover promising protease inhibitors against SARS-CoV-2. In this context, a recent study showed that high throughput virtual screening of more than 10,000 molecules, based on the SARS-CoV-2 Mpro structure, resulted in the identification of six small inhibitors molecules [28], one of which is a pyrimidine derivative (Figure 3, 3-A [28]) and the other contains the isoxazole nucleus (Figure 3, 3-B [28]).
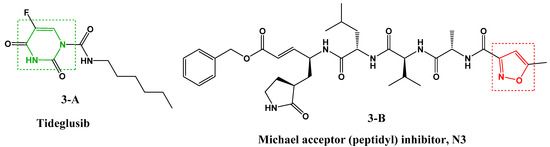
Figure 3.
Structure of SARS-CoV-2 Mpro inhibitors.
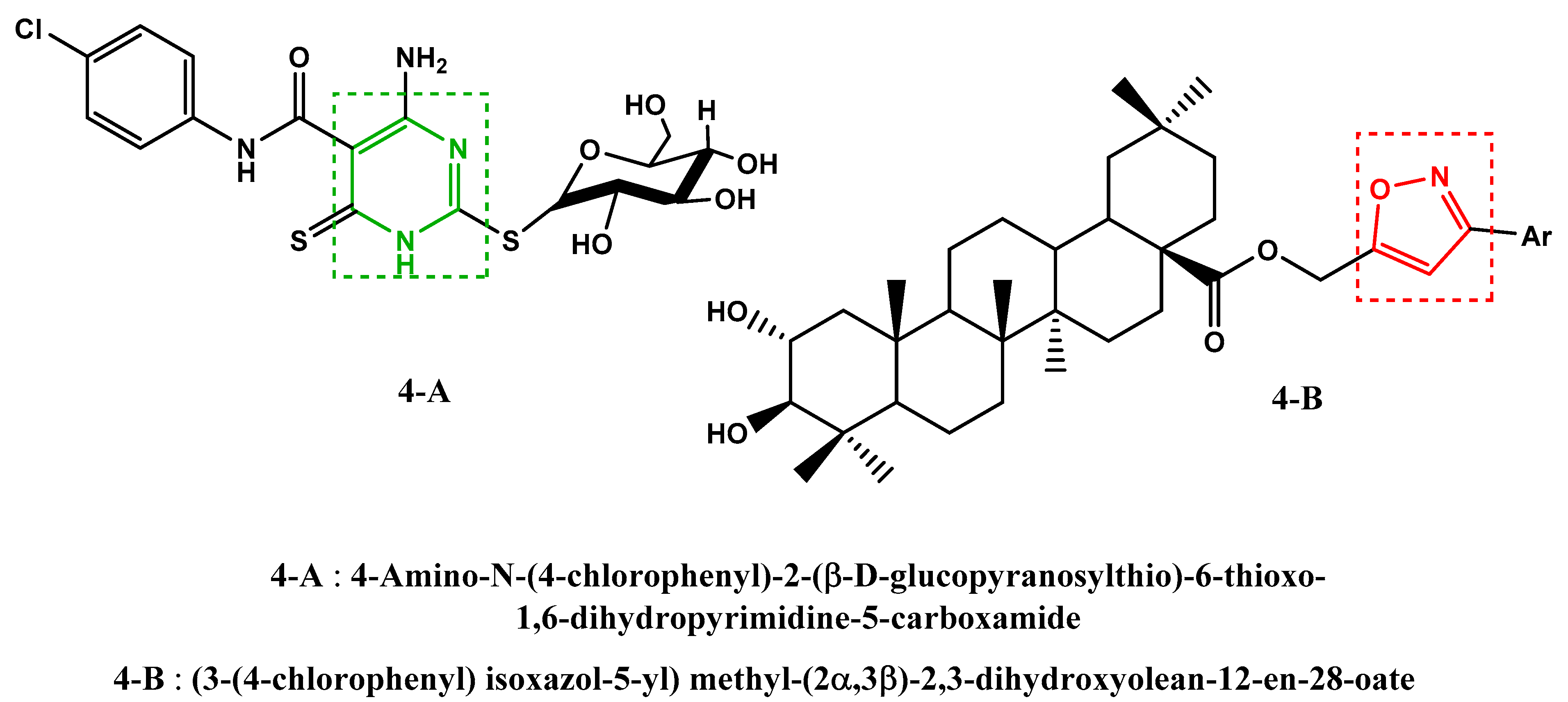
Furthermore, recent work has shown that certain compounds, containing in their structures the pyrimidine (Figure 4, 4-A [29]) and isoxazole (Figure 4, 4-B [30]) units, have shown an anti-SARS-CoV-2 activity.
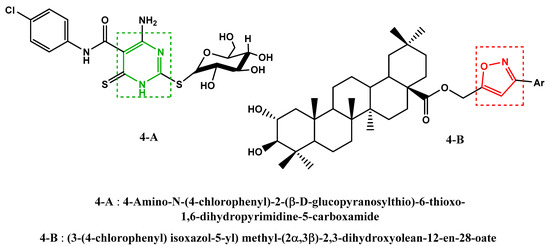
Figure 4.
Previously reported anti-SARS-CoV-2 molecules.
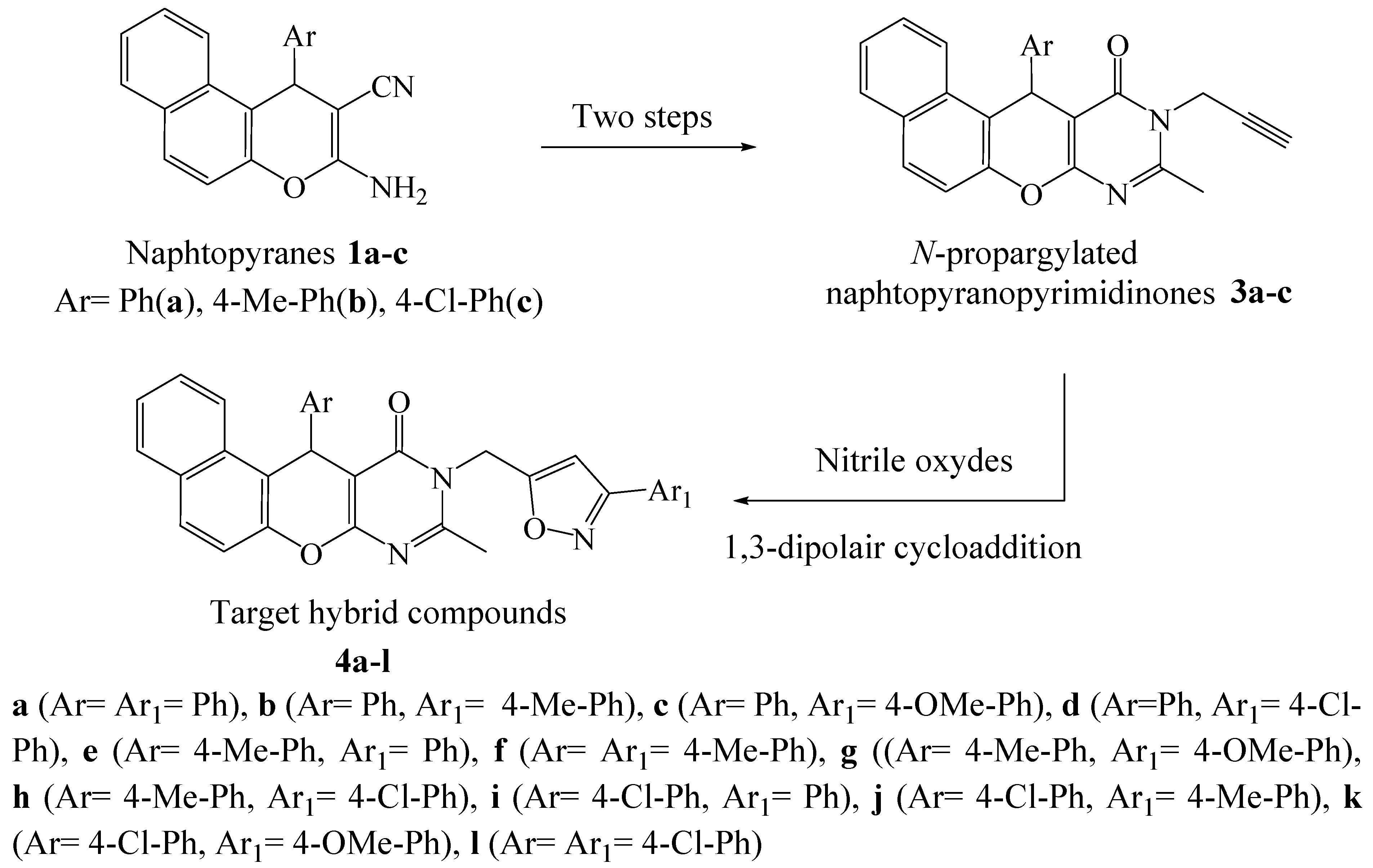
In addition, the junction of the pyrimidine ring with different heterocyclic fragments in the same molecule could lead to a new category of hybrid molecules with pronounced biological power. In this context, and in the continuity of looking for new bioactive pyrimidine compounds [31,32,33], we report herein the synthesis of a new series of hybrid compounds 4, in three steps (Scheme 1), under conventional heating conditions and by microwave irradiation. We have chosen to integrate into the same molecule the naphthalene, pyran, pyrimidine and isoxazole fragments assigned as antiviral and anti-SARS-CoV-2 agents, as indicated above, with the aim to perform their virtual screening, using molecular docking studies, hoping to find out promising protease inhibitors against COVID-19.
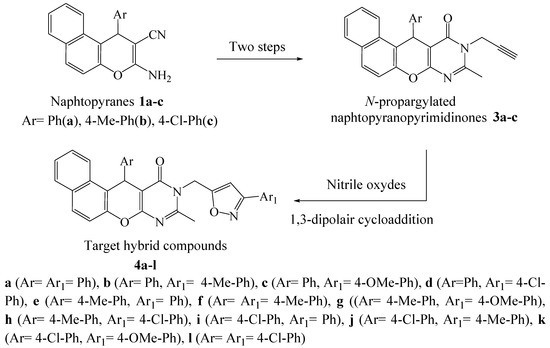
Scheme 1.
The design of target compounds 4a–l.
2. Results and Discussion
2.1. Chemistry
Our key starting materials, 2-amino-3-cyanonaphthopyranes 1a–c, were synthesized by adopting the method of Zayane et al. [34]. It is a multicomponent reaction of equimolar amounts of arylaldehyde, malononitrile and β-naphthol at the reflux of a hydroalcoholic solution in the presence of CuI as a catalyst. Their structures were confirmed on the basis oftheir 1H and 13C NMR spectra and by comparison with literature data [35].
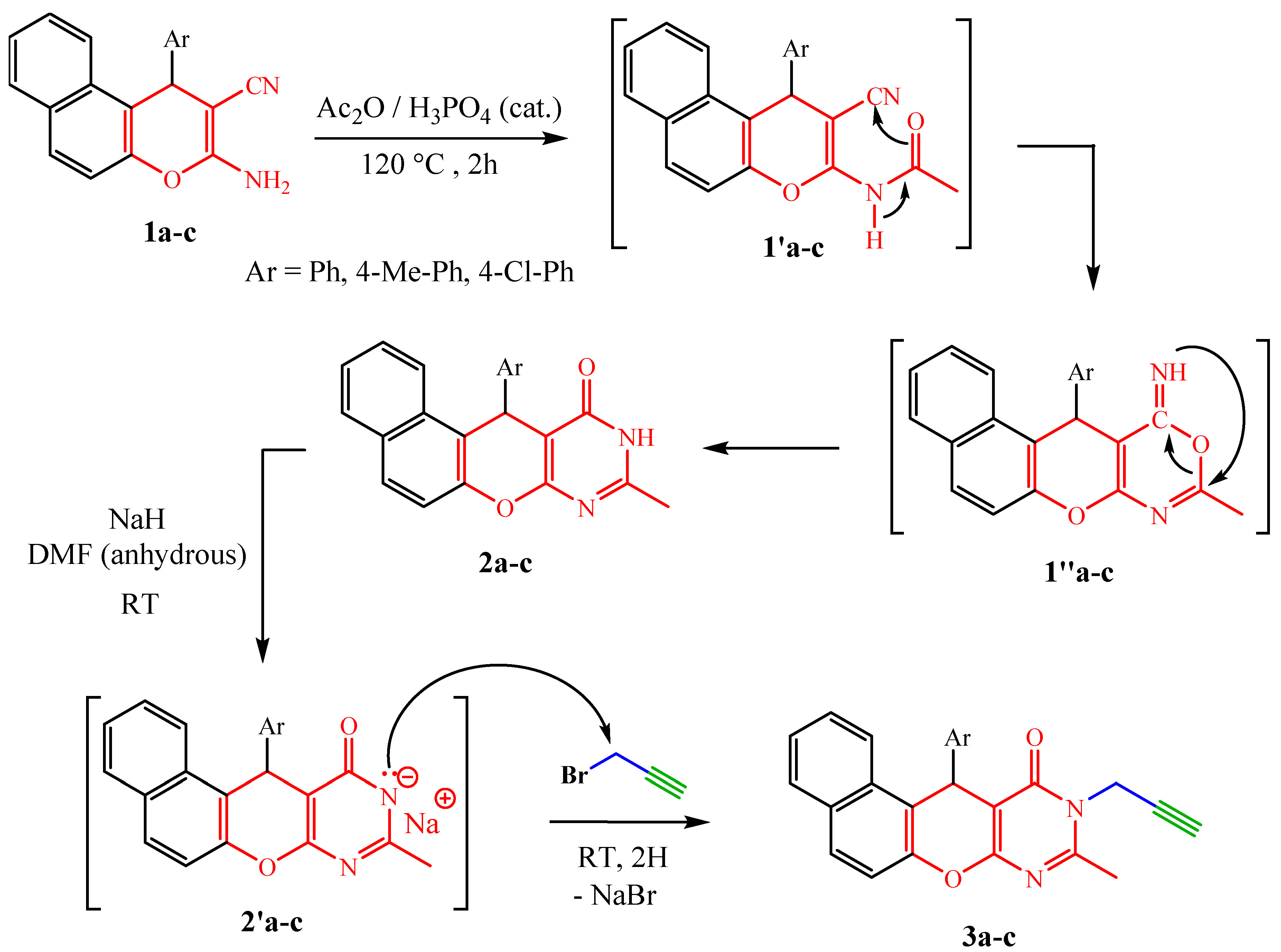
Condensation reaction, of the last precursors, with acetic anhydride in the presence of polyphosphoric acid (PPA) under reflux for 2 h gives pyrimidinones 2a–c in yields ranging from 66% to 80%. From a mechanistic point of view, the reaction is carried out in two steps. The precursors 1a–c react with acetic anhydride to give the non-isolable intermediates 1′a–c which cyclize to 1″a–c after nucleophilic attack at the nitrile group. Dimroth-type intramolecular rearrangement [32,36] of the intermediate thus formed leads to pyranopyrimidinones 2a–c, which reacted with propargyl bromide in the presence of sodium hydride in anhydrous DMF at room temperature to give the corresponding N-propargylated dipolarophiles 3a–c in good yields (80–92%) (Scheme 2) [37].
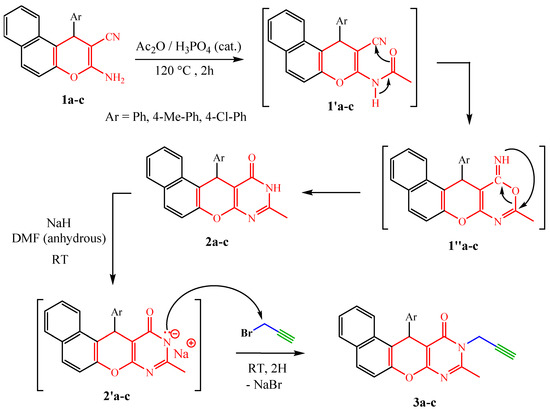
Scheme 2.
Synthetic pathway to compounds 2a–c and 3a–c.
The formed N-propargylatednaphthopyranopyrimidinones 3a–c were identified on the basis of their 1H NMR and 13C NMR spectra. The 1H NMR spectra of these dipolarophiles allows to see, in addition to the signals of the protons introduced by the naphthopyranopyrimidinones 2a–c, the disappearance of a singlet of the NH proton at δH 12.60–12.61 and the appearance of three new signals, two doublets of doublets are observed at δH 4.58–4.61 and 5.08–5.12 (J = 15.9, 2.4 Hz) which were attributed to methylenic protons (CH2) and a characteristic triplet of an acetylenic proton observed at δH 2.29–2.33 (J = 2.4 Hz). The proximity of methylene to the nitrogen atom bearing four different substituents (hybridized sp3) including its free doulet perfectly conjugated with the carbonyl of pyrimidinone is at the origin of the non-isochrony, and thus the non-equivalence of the methylenic protons which logically appear at two different chemical shifts. Moreover, the 13C NMR spectra of these derivatives exhibited the appearance of new signals at δC 31.7–32.7, 71.8–72.4 and 75.3–76.1 relative to new carbons C4′, C5′ and C6′introduced by the propargyl moiety, respectively.
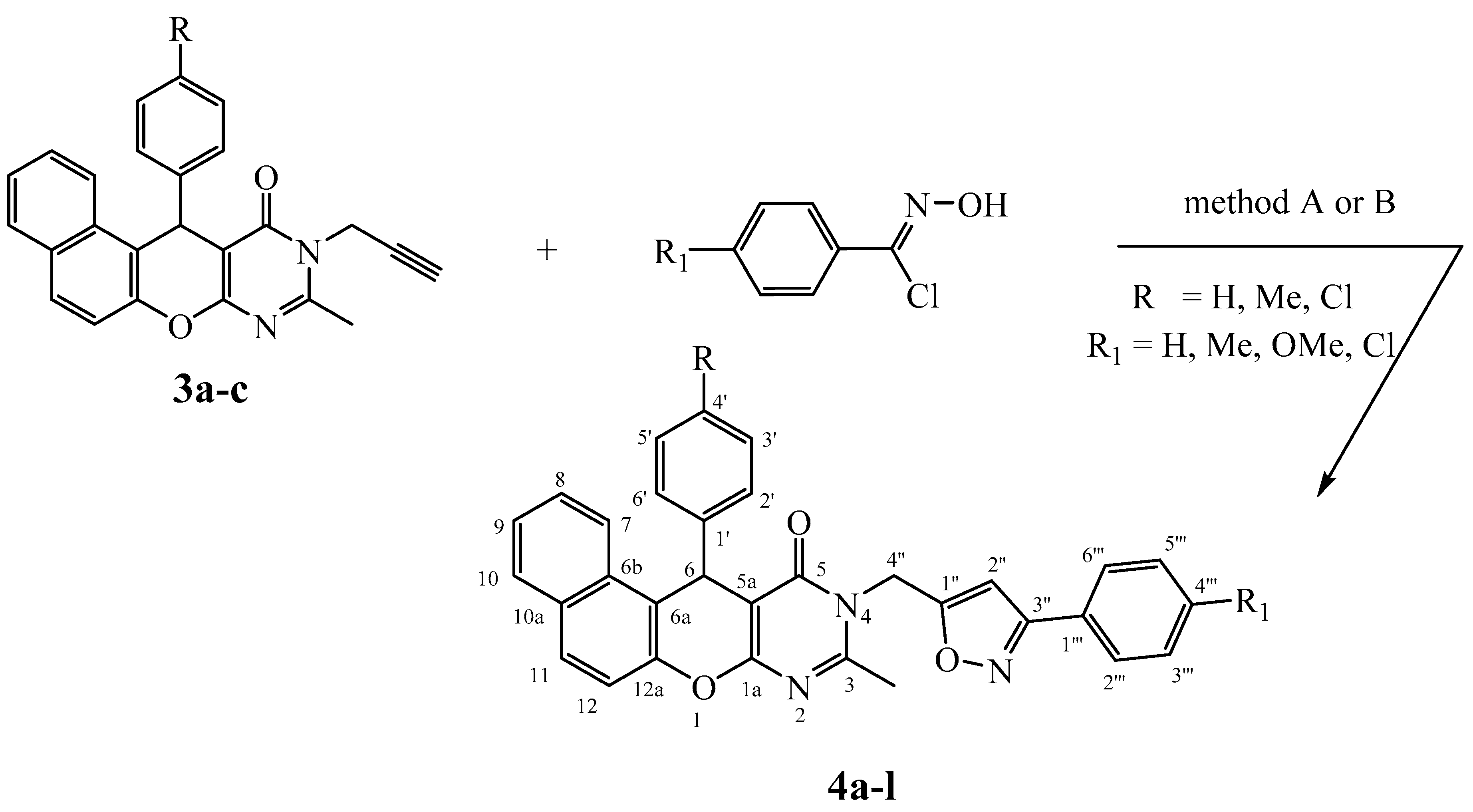
In order to access, via 1,3-dipolar cycloaddition reaction as shown in Scheme 3, to the new expected hybrid molecules 4a–l, the N-propargylated derivatives 3a–c were treated with various arylnitrile oxides, under conventional heating in refluxing DMF for 8–12 h (Method A) or under microwave irradiation in DMF for 3 to 5 min (Method B), both in the presence of copper(I) catalyst and Et3N as a base.
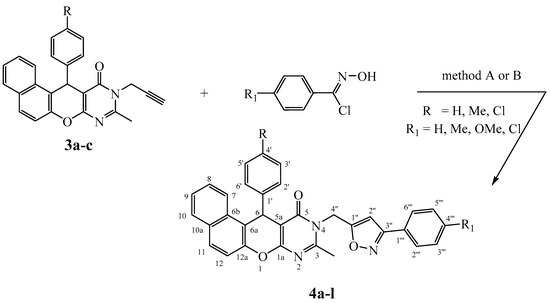
Scheme 3.
Synthesis of 3,5-isoxazole derivatives 4a–l.
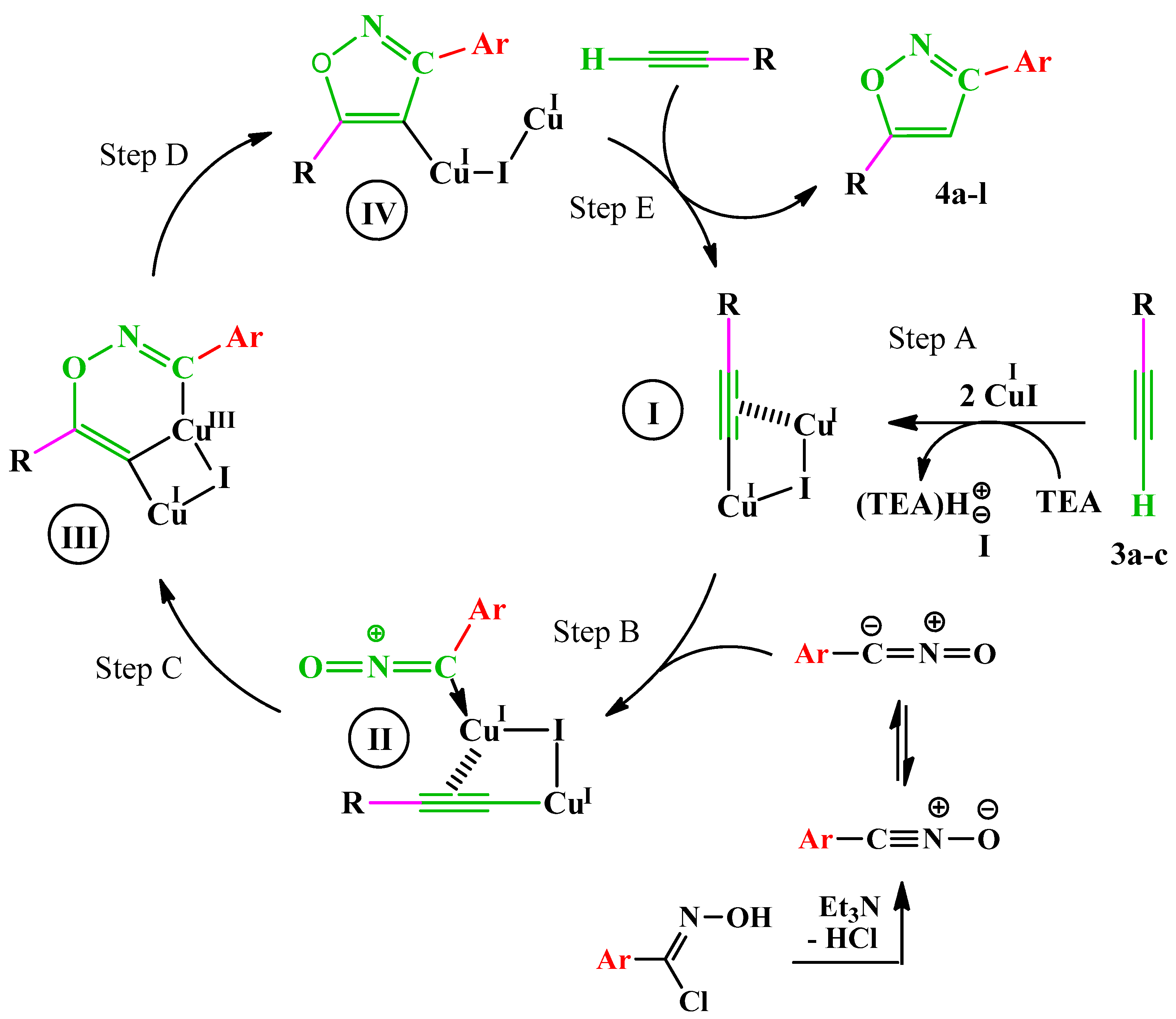
Until now, copper catalyzed nitrile oxide-alkyne click chemistry (CuAAC) is one of the methods of choice for regiospecific access to 3,5-disubstituted isoxazole derivatives [38].
From a mechanistic point of view, the formation of copper(I) acetylide I (step A) occurs through a π-alkyne copper complex intermediate. The π-coordination of alkyne to copper significantly acidifies terminal hydrogen of the alkyne, bringing it into the proper range to be deprotonated in presence of TEA and resulting in the formation of a σ-acetylide with a second copper(I) atom. The added aryl nitrile oxide is then activated by coordination to copper (step B), thus forming the intermediate II. In the next step (C), the formation of the first C–O bond takes place, and a strained copper metallacycle III forms. This intermediate undergoes a rearrangement to lead to copper(I) isoxazole intermediate IV (step D), which in the presence of another molecule of alkyne gives the desired isoxazole while regenerating intermediate I (step E) (Scheme 4).
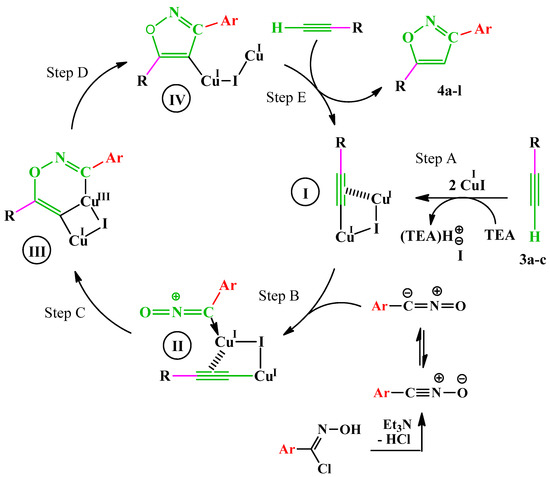
Scheme 4.
Proposed mechanism for the formation of compounds 4a–l.
This reaction sequence generates the new hybrid molecules 4a–l in moderate (method A/58–69%) to excellent yields (method B/91–97%). The absence of any other compound in the reaction mixture implies the strong regiospecificity of this reaction.
These resulting 3,5-regioisomers were confirmed from the NOE H-5isoxazole/Hmethylene and H-5isoxazole/Harom, and the non-observation of any NOE Hmethylene/Harom. Thus, this 3,5-regiospecificity was explained by the uses of copper(I) catalyst.
The comparison of the obtained results shown in Table 1 from the conventional heating method (method A) and those from the MW-assisted synthesis way (method B) showed in all cases much shorter reaction times and higher yields were achieved under MW irradiation compared to classical heating.

Table 1.
Comparison between the conventional heating procedure (method A) and microwave irradiation (method B) for the synthesis of compounds 4a–l.
The structures of the newly prepared compounds 4a–l were raised on the basis of their spectroscopic measurements. In fact, compared to compounds 3, the 1H NMR spectra of isoxazoles derivatives 4a–l indicate the disappearance of the signal at δH 2.30–2.32 relative to the terminal acetylenic proton and the appearance of a new singlet at δH 6.50–6.51 (s, 1H) attributable to H2′ of the isoxazole ring, in addition to signals at δH 6.92–7.98 attributable to the protons of the aromatic moiety linked to the isoxazole system. Moreover, the 13C NMR spectra reinforced the structures of 4a–l by showing the signals of the formed isoxazole moiety, as well as those from the dipolarophiles 3a–c.
The ES-HRMS of compounds 3 and 4 showed the correct protonated molecular ion peaks [M+H]+ that complied with the proposed structures.
2.2. Molecular Docking Study
By studying the binding pocket of the SARS-CoV-2 Mpro, it was found that the co-crystallized native inhibitor (N3) is located inside the pocket of the main protease receptor asymmetrically. The N3 ligand is a designed inhibitor for SARS-CoV-2 Mpro that was built from amino acids based on the Mpro pocket amino acids and cannot used medicinally. The main residues in this pocket were HIS-41, MET-49, PHE-140, GLY-143, HIS-164, MET-165, GLU-166, LEU-167, PRO-168, HIS-172, GLN-189, THR-190 and ALA-191.
Molecular docking of the synthesized molecules (4a–l) and N3 inhibitor into the main protease binding site was performed. The binding energies and interaction details (number of interactions, number of interacting amino acids, interacting amino acids and hydrogen bonds) of ligands with the target enzyme are presented in Table 2.

Table 2.
Binding energy (kcal/mol) and interaction detail of compounds 4a–l docked in the active site of Mpro enzyme (PDB: 6LU7).
All compounds were perfectly placed in the active site by variable scores and binding interactions with the amino acids of the receptor pocket. From the docking results (Table 2), All the tested compounds (4a–l) achieved promising binding scores ranging from −7.6 to −8.9 kcal/mol, compared to the docked co-crystallized N3 inhibitor with a binding score of −7.0 kcal/mol. The synthesized compounds (4a–l) showed a much higher binding affinity towards Mpro enzyme (PDB: 6LU7) than N3.
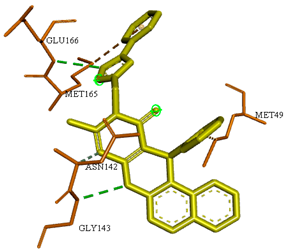
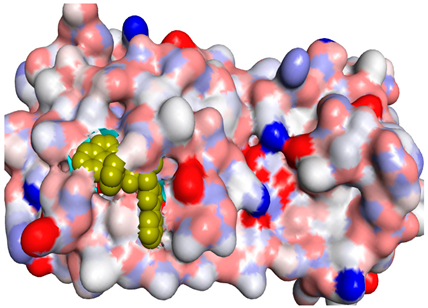
It is worth mentioning that, especially for compounds 4a, 4e and 4i, the obtained binding modes were quite similar to that of the docked N3 inhibitor, as depicted in Table 3. Moreover, these three selected compounds exhibited binding scores of −8.4, −8.5 and −8.5 kcal/mol, respectively, higher than of the docked co-crystallized N3 inhibitor (−7.0 kcal/mol) (Table 2).

Table 3.
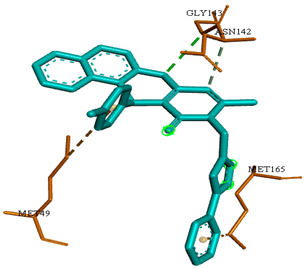
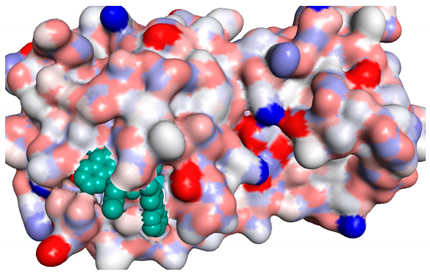
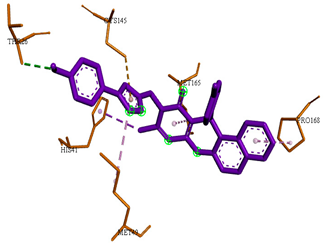
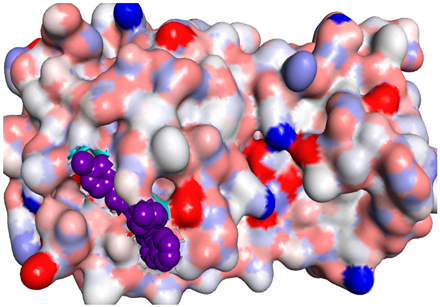
3D representations showing the binding interactions and positioning between the three promising tested compounds (4a, 4e and 4i) and the N3-binding pocket in comparison to the docked N3 inhibitor.
The docked N3 inhibitor was stabilized inside the SARS-CoV-2 Mpro pocket through a three hydrogen bonds formation with GLY-143, HIS-164 and GLN-198 amino acids with the bond lengths of 2.96, 2.67 and 3.25 Å, respectively. Additionally, it formed a Pi-alkyl bond with MET-165 (bond length: 5.05 Å) and two alkyl interactions with HIS-41 (bond length: 4.28 Å) and MET-49 (bond length: 4.45 Å) residues. On the other side, the compound 4a fitted inside the binding site of the SARS-CoV-2 Mpro by two hydrogen bonds formation, with GLY-143 and GLU-166 at 3.37 and 3.05 Å, respectively, with an additional carbon hydrogen bond with ASN-142 residue (bond length: 3.38 Å) and two Pi-sulfur interactions with MET-49 (bond length: 5.38 Å) andMET-165(bond length: 5.23 Å). Moreover, the compound 4e showed a hydrogen bond with the GLY-143 amino acid at 3.19 Å. Additionally, it showed a carbon hydrogen bond with ASN-142 residue (bond length: 3.39 Å) and two Pi-sulfur interactions with MET-49 (bond length: 5.11 Å) and MET-165 (bond length: 5.75 Å). The compound 4i formed a hydrogen bond, a carbon hydrogen bond and two Pi-sulfur interactions with the previously mentioned four amino acids as well. It formed a hydrogen bond with GLY-143 at 3.21 Å, a carbon hydrogen bond with ASN-142 at 3.46 Å and two Pi-sulfur interactions with MET-49 and MET-165 at 5.06 and 5.81 Å, respectively.
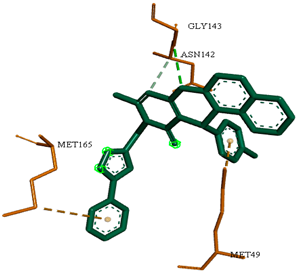
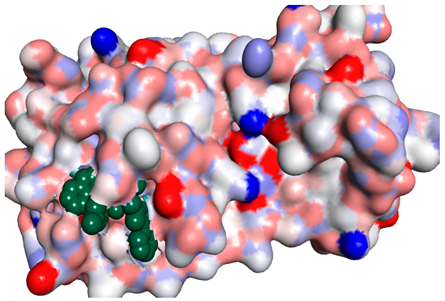
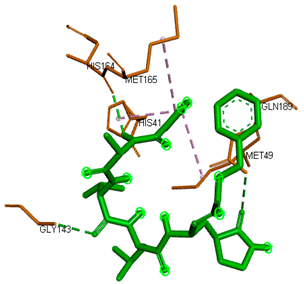
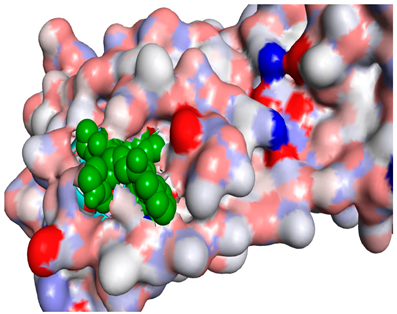
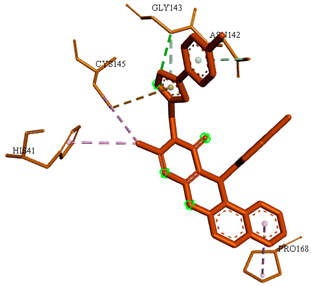
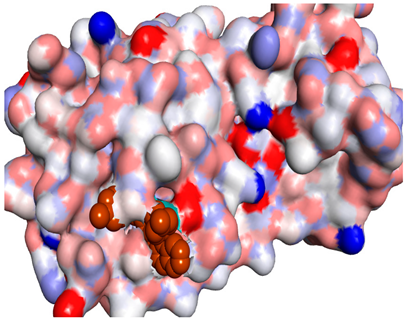
Although compounds 4k, 4h and 4j did not exhibit very comparable interactions to those of N3 towards SARS-CoV-2 Mpro, they, on the other hand, exhibited lower binding energy (−8.9, −8.5 and −8.4 kcal/mol, respectively) (Table 2). This finding can be explained, in part, by the nature of interactions between these compounds and the amino acids that constitute SARS-CoV-2 Mpro (Table 4). Indeed, the compound 4k formed a hydrogen bond with THR-26 (bond length: 3.19 Å), three Pi-alkyl bonds with MET-49 (bond length: 5.39 Å), MET-165 (bond length: 4.82 Å) and PRO-168 (bond length: 4.53 Å), a Pi-sulfur interaction with CYS-145 (bond length: 5.10 Å) and a Pi-sigma interaction with HIS-41 (bond length: 3.71 Å). Moreover, the derivative 4h, was engaged in a hydrogen bond, two Pi-donor hydrogen bonds, a Pi-sulfur interaction, two alkyl interactions and a Pi-alkyl bond with residues GLY-143 (bond length: 2.96 Å), ASN-142 (bond length: 3.65 Å), GLY-143 (bond length: 3.57 Å), CYS-145 (bond length: 4.79 Å), HIS-41 (bond length: 4.09 Å), CYS-145 (bond length: 4.48 Å) and PRO-168 (bond length: 4.88 Å), respectively. The compound 4j interacts with in many amino acids of the main protease of COVID-19 (Mpro) via two Hydrogen Bond, Pi-sigma interaction and a two Pi-alkyl bond with the side chain residues GLY-143 (bond length: 3.19 Å), GLU-166 (bond length: 3.15 Å), MET- 49 (bond length: 3.95 Å), CYS-145 (bond length: 5.25 Å), HIS-163 (bond length: 4.98 Å), respectively.

Table 4.
3D representations showing the binding interactions and positioning between the three promising tested compounds (4k, 4h and 4j) and the N3-binding pocket.
It is important to mention that the nature of the substituents allows the molecule to be placed into space in a well-defined conformation. Indeed, these substituents with different sizes promote or disadvantage possible free rotations, they also increase or decrease the electron density of certain groups essentially the aromatic ones by their indicative and/or mesomeric electronic effects. The spatial arrangement of the two benzene rings in compounds 4a–1 depends on the nature of their substituents (R and R1). This arrangement can, in certain cases, inhibit the delocalization of the electronic doublets by mainly mesomeric effects towards the outside of the ring (case of the aromatic system carried by the isoxazole nucleus), which influences the interaction of these aromatic systems with certain amino acids. Moreover, the R and R1 substituents do not need to engage in direct interactions with amino acids, but their sizes and electronic effects require special interactions of the corresponding aromatic rings with special amino acids. As mentioned in Table 3, the benzene ring at position C-6 of the pyran moiety and that carried by isoxazole both unsubstituted in 4a exhibit Pi-sulfur interactions with MET-49 (bond length: 5.38 Å) andMET-165 (bond length: 5.23 Å), respectively. The same observation was noted with compounds 4e and 4i where the aromatic ring carried by the pyran is para-methylated and para-chlorinated, respectively and the benzene ring carried by the isoxazole in both dervivatives is unsubstituted. This result can be explained by the effect of the size of the methyl group and the chlorine atom on the spatial arrangement of the aromatic ring throughout the molecule. In addition, the inductive donor effect (+ I) exerted by the methyl group and the two opposite electronic effects (-I) and (+ M) exerted by the chlorine atom appear to lead to practically the same result. It was observed that in the case of compound 4h, the benzenic nucleus attached to an isoxazole carrying in para position a chlorine atom (R1) exerts two Pi-donor Hydrogen bonds interactions with ASN-142 (bond length: 3.65 Å). These types of interactions were not observed in the rest of the products with other R and R1 which proves that the nature of these two substituents give the entire molecule a well-defined conformation which also guarantees well-defined interactions. With R = Cl and R1 = OMe, the compound 4k, takes a new conformation which allows, among other things, the OMe group to engage in a hydrogen bond with THR-26 (bond length: 3.19 Å).
According to our findings, the tested compounds, in particular 4a, 4e, 4h, 4i, 4j and 4k, could effectively act as novel antiviral agents against SARS-CoV-2.
3. Methods and Materials
3.1. Chemistry
3.1.1. General Experimental Procedures
All synthesis reactions were checked by TLC using aluminium sheets of silica gel 60 F254, 0.2 mm. Melting points were measured on an electrothermal 9002 apparatus and were reported uncorrected. NMR spectra were run on a Bruker AC-300 spectrometer at 300 MHz (1H) and 75 MHz (13C). All chemical shifts were reported as δ values (ppm) relative to residual non-deuterated solvent. Mass spectra were recorded with HRMS-ES using the reflectron mode in the positive ion mode. Microwave-assisted synthesis was peformed in a Start Synth multimode microwave instrument producing controlled irradiation at 2.45 GHz (Milestone S.r.l., Sorisole, Italy). The device is equipped with an industrial magnetron and a microwave diffuser providing a continuous microwave output power from 0 to 1400 W. An open reaction vessel was employed in all reactions.
The starting materials 1 and 2 were prepared according to the literature [32].
3.1.2. General Procedure for the Preparation of Naphthopyranopyrimidinone N-Propargylated Derivatives 3a–c
1 mmol of naphthopyranopyrimdinone 2a–c was dissolved in 15 mL of anhydrous DMF then 2 equivalents of sodium hydride (NaH) were added and the reaction mixture was stirred at room temperature under an argon atmosphere for 30 min; afterwards, 1.5 eq of propargyl bromide was added slowly, and the mixture was stirred for another 1.5 h under positive atmosphere of argon. Upon completion of the reaction, monitored by TLC, we added distilled water and the formed precipitate was collected by filtration, washed with water and purified by flash column chromatography on silica gel (EtOAc/PE, 1:1, as an eluent).
Compound 3a: 3-methyl-6-phenyl-4-(prop-2-yn-1-yl)-10H-naphtho[2,1-b]pyrano[2,3-d]pyrimidin-11(12H)-one
Brown solid, yield 88%, mp 193–195 °C.1H NMR (300 MHz; CDCl3): δ(ppm) = 2.22 (s, 3H, H9′); 2.32 (t, 1H, H6′’, J = 2.4 Hz); 4.69 (dd, 1H, H4′′a, J = 2.4 Hz, J = 17.4 Hz); 5.12 (dd, 1H, H4′′b, J = 2.4 Hz, J = 17.4 Hz); 5.79 (s, 1H, H6); 7.03 (d, 2H, H2′,6′, J = 8.1 Hz); 7.43 (m, 6H, Harom.); 7.79 (d, 2H, H11,12, J = 7.6 Hz); 7.92 (d, 1H, H7, J = 8.1 Hz). 13C NMR (75 MHz; CDCl3):δ(ppm) = 23.2, 32.7, 35.3, 71.9, 75.3, 102.2, 116.1, 117.0, 123.6, 124.4, 126.6, 127.7, 127.9, 128.6, 128.9, 130.5, 131.2, 135.8, 140.6, 147.5, 157.3, 158.9, 161.0. ESI-HRMS [M+H]+calcd. For (C25H18N2O2)+: 379.1446, found: 379.1456.
Compound 3b: 3-methyl-4-(prop-2-yn-1-yl)-6-(p-tolyl)-10H-naphtho[2,1-b]pyrano[2,3-d]pyrimidin-11(12H)-one
Brown solid, yield 92%, mp 192–194 °C.1H NMR (300 MHz; CDCl3): δ(ppm) = 2.22 (s, 3H, H9′); 2.29 (t, 1H, H6′’, J = 2.4 Hz); 2.67 (s, 3H, H4′); 4.58 (dd, 1H, H4′′a, J = 2.4 Hz, J = 17.4 Hz); 5.08 (dd, 1H, H4′′b, J = 2.4 Hz, J = 17.4 Hz); 5.87 (s, 1H, H6); 7.03 (d, 2H, H3′,5′, J = 8.1 Hz); 7.38 (m, 5H, Haromatic); 7.82 (d, 2H, H11,12, J = 7.4 Hz); 7.92 (d, 1H, H7, J = 8.1 Hz). 13C NMR (75 MHz; CDCl3):δ(ppm) = 20.4, 22.0, 32.7, 35.9, 72.4, 76.1, 101.1, 116.1, 116.9, 123.2, 124.4, 126.6, 127.8, 127.9, 128.6, 128.8, 130.5, 131.0, 135.8, 140.2, 147.5, 157.3, 158.9, 160.9. ESI-HRMS [M+H]+calcd. For (C26H19N2O2)+: 392.1524, found: 392.1531.
Compound 3c: 6-(4′′-chlorophenyl)-3-methyl-4-(prop-2-yn-1-yl)-10H-naphtho[2,1-b]pyrano[2,3-d]pyrimidin-11(12H)-one
Brown solid, yield 80%, mp 194–196 °C.1H NMR (300 MHz; CDCl3): δ(ppm) = 2.24 (s, 3H, CH3); 2.33 (t, 1H, H6′’, J = 2.4 Hz); 4.62 (dd, 1H, H4′′, J = 2.4 Hz, J = 17.4 Hz); 5.18 (dd, 1H, H4′′, J = 2.4 Hz, J = 17.4 Hz); 5.85 (s, 1H, H6); 7.05 (d, 2H, H2′,6′, J = 8.1 Hz); 7.40 (m, 5H, Haromatic); 7.82 (d, 2H, H11,12, J = 7.4 Hz); 7.90 (d, 1H, H7, J = 8.1 Hz). 13C NMR (75 MHz; CDCl3):δ(ppm) = 22.6, 31.7, 35.4, 71.8, 75.4, 102.4, 116.1, 116.9, 123.2, 124.4, 126.6, 127.8, 127.9, 128.7, 128.9, 130.4, 131.0, 136.8, 140.4, 146.5, 156.3, 157.9, 161.3. ESI-HRMS [M+H]+calcd. For (C25H16ClN2O2)+: 412.0978, found: 412.0989.
3.1.3. General Procedure for the Preparation of Isoxazoles Derivatives 4a–l
Method A (conventional method): Alkynes 3a–c (1 mmol) and appropriate hydroximyl chloride (2 mmol) were dissolved in 3 mL DMF. Cuprous iodide (I) (0.1 equiv) was added with stirring followed by Et3N (0.4 mmol) at room temperature. The mixture was refluxed for 8–12 h. After cooling, the mixture was diluted with water and then extracted with ethyl acetate (3 × 30 mL). The organic layer was dried over Na2SO4. After the removal of solvent in vacuo, the resulting residue was purified by silica gel column chromatography eluted with dichloromethane to obtain the desired new products (4a–l).
Method B (microwave-assisted): Alkynes 3a–c (1 mmol) and appropriate hydroximyl chloride (2 mmol) were dissolved in 3 mL DMF. Cuprous iodide (I) (0.1 equiv) was added with stirring followed by Et3N(0.4 mmol) at room temperature. The reaction mixture was then subjected to microwave irradiations at 250 Watts for 3–5 min, after which it was diluted with water and then extracted with ethyl acetate (3 × 30 mL). The organic layer was dried over Na2SO4. After removal of solvent under reduced pressure, the resulting residue was purified by silica gel column chromatography eluted with dichloromethane to abtain the desired new products (4a–l).
Compound 4a: 3-methyl-6-phenyl-4-((3′′-phenylisoxazol-5”-yl)methyl)-10H-naphtho[2,1-b]pyrano[2,3-d]pyrimidin-11(12H)-one
White solid, yield 97%, mp 148–150 °C.1H NMR (300 MHz; CDCl3): δ (ppm) = 2.72 (s, 3H, H9′); 5.13 (d, 1H, H4′′, J = 15.9 Hz); 5.57 (d, 1H, H4′′, J = 15.9 Hz); 5.87 (s, 1H, H6); 6.54 (s, 1H, H2′′);7.19 (d, 2H, H2′′’,6′′’, J = 8.4 Hz); 7.26–7.49 (m, 9H, Harom); 7.70–7.72 (m, 2H, Harom);7.79–7.83 (m, 3H, Harom). 13C NMR (75 MHz; CDCl3): δ (ppm) = 22.4, 35.7, 39.2, 100.7, 101.7, 115.2, 116.9, 122.9, 124.6, 126.3, 126.7, 127.8, 128.0, 128.4, 129.2, 129.3, 129.7, 130.3, 131.1, 132.1, 141.5, 147.5, 157.6, 161.3, 162.4, 165.6. ESI-HRMS [M+H]+ calcd. For (C32H23N3O3)+: 498.1817, found: 498.1822.
Compound 4b: 3-methyl-6-phenyl-4-((3′′-(p-tolyl)isoxazol-5”-yl)methyl)-10H-naphtho[2,1-b]pyrano[2,3-d]pyrimidin-11(12H)-one
White solid, yield 93%, mp 138–140 °C. 1H NMR (300 MHz; CDCl3): δ (ppm) = 2.36 (s, 3H, H9′); 2.73 (s, 3H, H7′′’); 5.11 (d, 1H, H4′′, J = 15.9 Hz); 5.61(d, 1H, H4′′, J = 15.6 Hz); 5.92 (s, 1H, H6); 6.54 (s, 1H, H2′′);7.15 (d, 2H, H2′,6′, J = 7.2 Hz); 7.21–7.28 (m,5H, Harom); 7.35–7.53 (m, 3H, Harom); 7.61 (d, 2H, Harom, J = 8.1 Hz); 7.83 (m, 2H, Harom); 7.97 (d, 1H, H11, J = 7.8 Hz). 13C NMR (75 MHz; CDCl3): δ (ppm) = 21.3, 22.9, 36.8, 39.7, 101.7, 102.1, 116.3, 117.4, 123.6, 125.0, 125.5, 126.7, 126.8, 127.1, 128.4, 128.5, 128.5, 129.5, 131.0, 140.4, 143.6, 148.0, 157.9, 159.5, 161.8, 162.8, 166.0. ESI-HRMS [M+H]+ calcd. For (C33H25N3O3)+: 512.1973, found: 512.1988.
Compound 4c: 4-((3′′-(p-methoxyphenyl)isoxazol-5”-yl)methyl)-3-methyl-6-phenyl-10H-naphtho[2,1-b]pyrano[2,3-d]pyrimidin-11(12H)-one
White solid, yield 92%, mp 166–168 °C. 1H NMR (300 MHz; CDCl3): δ (ppm) = 2.71 (s, 3H, H9′); 3.84 (s, 3H, H7′′’); 5.07 (d, 1H, H4′′, J = 15.6 Hz); 5.58 (d, 1H, H4′′, J = 15.9Hz); 5.91 (s,1H, H6);6.50 (s,1H, H2′′); 6.92 (d, 2H, H3′′’,5′′’, J = 9.6 Hz); 6.96 (d, 1H, Harom, J = 7.6 Hz); 7.12 (m, 2H, Harom); 7.41–7.48 (m, 5H, Harom); 7.64 (d, 2H, H2′′’,6′′’, J = 9.6 Hz); 7.68 (m,2H, Harom); 7.69 (d, 1H, H1, J = 7.4 Hz). 13C NMR (75 MHz; CDCl3): δ (ppm) = 22.4, 36.3, 39.2, 54.8, 101.1, 101.5, 113.8, 115.8, 116.9, 120.3, 123.1, 124.5, 126.2, 126.6, 127.7, 127.9, 128.0, 129.0, 130.5, 131.1, 143.1, 147.5, 157.4, 158.9, 160.6, 161.3, 161.9, 165.4. ESI-HRMS [M+H]+ calcd. For (C33H25N3O4)+: 528.1923, found: 528.1935.
Compound 4d: 4-((3′′-(p-chlorophenyl)isoxazol-5”-yl)methyl)-3-methyl-6-phenyl-10H-naphtho[2,1-b]pyrano[2,3-d]pyrimidin-11(12H)-one
White solid, yield 92%, mp 150–152 °C. 1H NMR (300 MHz; CDCl3): δ (ppm) = 2.73 (s, 3H, H9′); 5.12 (d, 1H, H4′′, J = 15.6 Hz); 5.60 (d, 1H, H4′′, J = 15.9 Hz); 5.91 (s,1H, H6); 6.54 (s,1H, H2′′); 7.10 (m, 1H, Harom); 7.28 (m, 2H, Harom); 7.37–7.49 (m, 7H, Harom); 7.66 (d, 2H, H2′′’,6′′’, J = 8.7 Hz); 7.67 (m,2H, Harom); 7.96 (d, 1H, H11, J = 7.8 Hz).13C NMR (75 MHz; CDCl3): δ (ppm) = 22.9, 36.8, 39.7, 101.7, 102.1, 116.3, 117.4, 123.6, 125.0, 126.7, 126.8, 127.1, 128.0, 128.4, 128.5, 128.6, 129.2, 129.5, 131.0, 131.6, 136.3, 143.6, 148.0, 157.8, 159.5, 161.8, 161.9, 166.6. ESI-HRMS [M+H]+ calcd. For (C32H22ClN3O3)+: 532.1427, found: 532.1437.
Compound 4e: 3-methyl-4-((3′′-phenylisoxazol-5”-yl)methyl)-6-(p-tolyl)-10H-naphtho[2,1-b]pyrano[2,3-d]pyrimidin-11(12H)-one
White solid, yield 95%, mp 157–159 °C. 1H NMR (300 MHz; CDCl3): δ (ppm) = 2.19 (s, 3H, H9′); 2.73 (s, 3H, H7′); 5.08 (d, 1H, H4′′, J = 15.6 Hz); 5.63 (d, 1H, H4′′, J = 15.6 Hz); 5.88 (s,1H, H6);6.55 (s,1H, H2′′); 7.05 (d, 2H, H2′′’,6′′’, J = 7.8 Hz); 7.14–7.34 (m, 3H, Harom);7.37–7.57 (m, 5H, Harom); 7.66 (d, 2H, H3′′’,5′′’, J = 7.8 Hz); 7.83 (d, 2H, H5,6, J = 9.0 Hz); 7.98 (d, 1H, H1, J = 8.1 Hz).13C NMR (75 MHz; CDCl3): δ (ppm) = 21.4, 22.9, 36.3, 39.7, 101.2, 101.6, 115.9, 116.8, 123.1, 124.5,126.3, 126.5, 127.5, 127.8, 128.0, 128.6, 128.7, 128.9, 130.5, 135.8, 135.9, 140.2, 147.4, 157.2, 158.9, 161.3, 161.4, 166.1. ESI-HRMS [M+H]+ calcd. For (C33H25N3O3)+: 512.1974, found: 512.1981.
Compound 4f: 3-methyl-6-(p-tolyl)-4-((3′′-(p-tolyl)isoxazol-5”-yl)methyl)-10H-naphtho[2,1-b]pyrano[2,3-d]pyrimidin-11(12H)-one
White solid, yield 96%, mp 154–156 °C. 1H NMR (300 MHz; CDCl3): δ (ppm) = 1.72 (s, 3H, H9′); 2.23 (s, 3H, H7′); 2.72 (s, 3H, H7′′’); 5.10 (d, 1H, H4′′, J = 15.9 Hz); 5.63 (d, 1H, H4′′, J = 15,9 Hz); 5.89 (s, 1H, H6); 6.58 (s, 1H, H2′′);7.05 (d, 2H, H3′′’,5′′’, J = 7.8 Hz); 7.26–7.49 (m, 6H, Harom); 7.61–7.83 (m, 5H, Harom); 7.98 (d, 1H, H1, J = 8.1 Hz). 13C NMR (75 MHz; CDCl3): δ (ppm) = 20.9,22.9, 36.3, 39.2, 101.8, 102.2, 116.5, 117.4, 123.6, 125.0, 126.7, 126.8, 127.1, 128.3, 128.4, 128.5, 128.8, 128.9, 129.1, 129.4, 130.2, 131.0, 131.6, 136.3, 140.7, 148.0, 157.7, 159.4, 161.8, 162.8, 166.3. ESI-HRMS [M+H]+ calcd. For (C34H27N3O3)+: 526.2130, found: 536.2137.
Compound 4g: 4-((3′′-(p-methoxyphenyl)isoxazol-5”-yl)methyl)-3-methyl-6-(p-tolyl)-10H-naphtho[2,1-b]pyrano[2,3-d]pyrimidin-11(12H)-one
White solid, yield 94%, mp 144–146 °C. 1H NMR (300 MHz; CDCl3): δ (ppm) = 2.23 (s, 3H, H9′); 2.72 (s, 3H, H7′); 3.85 (s, 3H, H7′′’); 5.06 (d, 1H, H4′′, J = 15.6 Hz); 5.61 (d, 1H, H4′′, J = 15.9 Hz); 5.88 (s,1H, H6);6.52 (s, 1H, H2′′); 6.92 (d, 2H, H3′′’,5′′’, J = 8.7 Hz); 6.96 (d, 2H, Harom, J = 7.4 Hz); 7.02–7.48 (m, 5H, Harom); 7.65 (d, 2H, H2′′’,6′′’, J = 8.7 Hz); 7.82 (m, 2H, Harom); 7.98 (d, 1H, H11, J = 8.4 Hz). 13C NMR (75 MHz; CDCl3): δ (ppm) = 20.9, 22.9, 36.3, 39.7, 55.3, 101.8, 102.0, 114.3, 116.5, 117.4, 120.8, 123.6, 124.9, 127.1,128.2, 128.3, 128.4, 129.1, 129.3, 131.0, 131.5, 136.3, 140.7, 148.0, 157.8, 159.4, 161.1, 161.8, 162.4, 166.0. ESI-HRMS [M+H]+ calcd. For (C34H27N3O4)+: 542.2079, found: 542.2088.
Compound 4h: 4-((3-(4-chlorophenyl)isoxazol-5-yl)methyl)-3-methyl-6-(p-tolyl)-10H-naphtho[2,1-b]pyrano[2,3-d]pyrimidin-11(12H)-one
White solid, yield 97%, mp 140–142 °C. 1H NMR (300 MHz; CDCl3): δ (ppm) = 2.23 (s, 3H, H9′); 2.72 (s, 3H, H7′); 5.09 (d, 1H, H4′′, J = 15.9 Hz); 5.62 (d, 1H, H4′′, J = 15.9 Hz); 5.88 (s,1H, H6);6.56 (s,1H, H2′′); 7.03 (d, 2H, H2′′’,6′′’, J = 8.4 Hz); 7.37 (m, 2H, Harom);7.39–7.48 (m, 5H, Harom); 7.65 (d, 2H, H3′′’,5′′’,J = 8.4 Hz); 7.67 (m,2H, Harom); 7.69 (d, 1H, H11, J = 7.8 Hz). 13C NMR (75 MHz; CDCl3): δ (ppm) = 21.3, 22.9, 36.3, 39.7, 101.8, 102.1, 116.5, 117.4, 123.6, 124.9, 125.5, 126.6, 126.7, 127.1, 128.3, 128.4, 129.1, 129.3, 129.4, 129.6, 131.0, 131.5, 136.3, 140.4, 140.7, 148.0, 157.8, 166.1. ESI-HRMS [M+H]+ calcd. For (C33H24ClN3O3)+: 546.1584, found: 546.1594.
Compound 4i: 6-(4-chlorophenyl)-3-methyl-4-((3′′-phenylisoxazol-5”-yl)methyl)-10H-naphtho[2,1-b]pyrano[2,3-d]pyrimidin-11(12H)-one
White solid, yield 93%, mp 122–124 °C. 1H NMR (300 MHz; CDCl3): δ (ppm) = 2.71 (s, 3H, H9′); 5.09 (d, 1H, H4′′, J = 15.9 Hz); 5.59 (d, 1H, H4′′, J = 15.9 Hz); 5.92 (s, 1H, H6); 6.57 (s, 1H, H2′′);7.14 (d, 2H, H2′′’,6′′’, J = 7.5 Hz); 7.14–7.23 (m, 2H, Harom); 7.26–7.49 (m, 9H, Harom); 7.59–7.72 (m, 2H, Harom); 7.49–7.57 (m, 2H, Harom); 7.74 (d, 1H, H1, J = 7.6 Hz). 13C NMR (75 MHz; CDCl3): δ (ppm) = 22.4,36.3, 39.2, 101.2, 101.7, 115.8, 116.9, 123.1, 124.5, 126.3, 127.8, 127.9, 128.0, 128.4, 129.0, 129.7, 130.5, 131.1, 143.1, 147.5, 157.4, 159.0, 161.3, 162.3, 165.8. ESI-HRMS [M+H]+ calcd. For (C32H23ClN3O3)+: 532.1427, found: 532.1435.
Compound 4j: 6-(p-chlorophenyl)-3-methyl-4-((3′′-(p-tolyl)isoxazol-5”-yl)methyl)-10H-naphtho[2,1-b]pyrano[2,3-d]pyrimidin-11(12H)-one
White solid, yield 91%, mp 147–149 °C. 1H NMR (300 MHz; CDCl3): δ (ppm) = 2.39 (s, 3H, H9′); 2.74 (s, 3H, H7′′’); 5.15 (d, 1H, H4′′, J = 15.9Hz,); 5.59 (d, 1H, H4′′, J = 15.6Hz); 5.89 (s,1H, H6);6.54 (s,1H, H2′′); 7.18–7.28 (m, 4H, Harom); 7.36–7.49 (m, 5H, Harom);7.64 (d, 2H, Harom, J = 8.1 Hz); 7.89 (m, 3H, Harom).13C NMR (75 MHz; CDCl3): δ (ppm) = 21.4, 23.0, 36.2, 39,7, 101.5, 102.2, 114.8, 116.5, 117.4, 123.6, 124.9, 125.5, 126.6, 126.7, 127.1, 128.3, 128.4, 129.1, 129.3, 129.4, 129.6, 131.0, 131.5, 136.3, 140.4, 140.7, 148.0, 157.8, 166.1. ESI-HRMS [M+H]+ calcd. For (C33H24ClN3O3)+: 546.1584, found: 546.1594.
Compound 4k: 6-(p-chlorophenyl)-4-((3′′-(p-methoxyphenyl)isoxazol-5”-yl)methyl)-3-methyl-10H-naphtho[2,1-b]pyrano[2,3-d]pyrimidin-11(12H)-one
White solid, yield 96%, mp 160–162 °C. 1H NMR (300 MHz; CDCl3): δ (ppm) = 2.74 (s, 3H, H9′); 3.80 (s,3H, H7′′’); 5.14 (d, 1H, H4′′, J =15.9 Hz); 5.58 (d, 1H, H4′′, J = 16.5 Hz); 5.89 (s, 1H, H6); 6.50 (s, 1H, H2′′);6.97 (d, 2H, H3′′’,5′′’, J = 8.7 Hz); 7.18 (d, 2H, H2′,6′, J = 8.4 Hz); 7.36 (d, 2H, H2′′’,6′′’, J = 8.7 Hz); 7.26–7.49 (m,4H, Harom); 7.70 (d, 2H, Harom, J = 7.0 Hz); 7.73–7.89 (m, 3H, Harom). 13C NMR (75 MHz; CDCl3): δ (ppm) = 22.7,36.2, 38.7, 55.8, 101.1, 101.5, 113.8, 115.8, 116.9, 120.3, 123.1, 124.5, 126.2, 126.6, 127.7, 127.9, 128.0, 129.0, 130.5, 131.1, 143.1, 147.5, 157.4, 158.9, 160.6, 161.3, 161.9, 165.4. ESI-HRMS [M+H]+ calcd. For (C33H25ClN3O4)+: 562.1533, found: 562.1548.
Compound 4l: 6-(p-chlorophenyl)-4-((3′′-(p-chlorophenyl)isoxazol-5”-yl)methyl)-3-methyl-10H-naphtho[2,1-b]pyrano[2,3-d]pyrimidin-11(12H)-one
White solid, yield 92%, mp 152–154 °C. 1H NMR (300 MHz; CDCl3): δ (ppm) = 2.74 (s, 3H, H9′); 5.16 (d, 1H, H4′′, J = 15.9 Hz); 5.58 (d, 1H, H4′′, J = 15.9 Hz); 5.89 (s, 1H, H6); 6.53 (s, 1H, H2′′); 7.18 (m, 2H, Harom); 7.20–7.69 (m, 8H, Harom); 7.81–7.83 (m, 2H, Harom); 7.85–7.89 (m, 2H, Harom). 13C NMR (75 MHz; CDCl3): δ (ppm) = 22.9, 36.8, 39.7, 101.2, 102.1, 115.6, 117.4, 123.4, 125.2, 126.8, 127.3, 127.7, 127.9, 128.0, 128.5, 128.6, 129.0, 129.1, 129.2, 129.3, 129.8, 130.8, 131.6, 132.6, 136.3, 142.0, 148.0, 158.0, 159.5, 161.7, 161.9, 166.5. ESI-HRMS [M+H]+ calcd. For (C33H21Cl2N3O3)+: 566.1038, found: 566.1050.
The NMR spectra of all synthesized compounds are given as supplementary material.
3.2. Molecular Docking Procedure
The chemical compound structures of the co-crystallized inhibitor (N3) and the synthesized compounds (4a–l) were generated and optimized using ACD (3D viewer) software (Version 2017.2.1, http://www.filefacts.com/acd3d-viewer-freeware-info, accessed on 8 July 2021), where their energies were minimized. The structure 6LU7 for RBVS (receptor-based virtual screening) having a catalytic dyad (Cys145 and His41), was selected because of its easy ability to complex with the co-crystallized peptide-like ligand inhibitor N3 used as a reference standard in the present work [28]. The crystal structure of SARS-CoV-2 Mpro (PDB: 6LU7) was obtained from the RSCB data bank (https://www.rcsb.org, accessed on 8 July 2021). The protein was prepared by removing the complexed inhibitor ligand and water molecules. Then, the polar hydrogens were added followed by appending Kollman charges. Hence, the grid box with dimensions of 40 × 40 × 40 points, spacing of 1.0 Å and centered with coordinates x: −11.993, y: 15.425, and z: 65.951, was generated based on N3 binding position in the target protein binding site. The molecular docking analysis of N3 and the synthesized compounds (4a–l) were performed using AutoDock Vina software (The Scripps Research Institute, La Jolla, CA, USA) [39]. Molecule–enzyme interactions were drawn and construed by employing the Biovia Discovery Studio Visualizer, BIOVIA, San Diego, CA, USA (2017).
4. Conclusions
In conclusion, we have successfully synthesized in this work a new series of 3,5-isoxazole linked naphthopyranopyrimidinones 4a–l, in three steps, using the naphtopyranyl moiety as a building blocks support, via 1,3-dipolar cycloaddition reaction using arylnitrile oxides catalyzed by Cu(I) under classical and microwave irradiation conditions. The last condition offers several advantages including good-to-high yields, cleaner products, and a reduction in reaction time, making this method particularly attractive. In addition, to help fight COVID-19, molecular docking based on virtual screening was performed to identify among the newly synthesized compounds 4a–1 which ones has the potential to interact with COVID-19 Mpro. Our results demonstrate that compounds 4a, 4e, 4h, 4i, 4j and 4k, exhibited good binding affinity towards the main protease of COVID-19 (Mpro) better than that made by the native co-ligand N3 inhibitor. These molecules could possibly be promising in terms of therapeutic effect against SARs-CoV-2. However, in vitro and then in vivo investigation are essential to validate our prediction results and pave the way for the discovery of anti-SARs-CoV-2 drugs.
Supplementary Materials
The following are available online, NMR spectra of the synthesized compounds 4a–l.
Author Contributions
Conceptualization, F.K.A., M.C. and S.J.; methodology, A.R. and N.B.H.; formal analysis, M.R.E., M.A.A. and S.J.; investigation, F.K.A., M.C. and S.J.; resources, F.K.A., H.B.J. and A.R.; data curation, H.B.J. and S.J.; writing—original draft preparation, F.K.A., M.C. and S.J.; writing—review and editing, F.K.A. and H.B.J.; supervision, N.B.H. and H.B.J.; funding acquisition, F.K.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received on external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
This research was supported by the Deanship of Scientific Research, Imam Mohammad IbnSaud Islamic University (IMSIU), Saudi Arabia, Grant No. (21-13-18-063).
Conflicts of Interest
The authors declare no conflict of interests.
Sample Availability
Samples of the compounds 4a–l are available from the authors.
References
- Cascella, M.; Rajnik, M.; Cuomo, A.; Dulebohn, S.C.; Di Napoli, R. Features, Evaluation and Treatment Coronavirus (COVID-19); Stat Pearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Sohrabi, C.; Alsafi, Z.; O’Neill, N.; Khan, M.; Kerwan, A.; Al-Jabir, A.; Iosifidis, C.; Agha, R. World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). Int. J. Surg. 2020, 76, 71–76. [Google Scholar] [CrossRef]
- Li, H.; Zhou, Y.; Zhang, M.; Wang, H.; Zhao, Q.; Liu, J. Updated Approaches against SARS-CoV. Antimicrob. Agents Chemother. 2020, 64, e00483-20. [Google Scholar] [CrossRef] [Green Version]
- Shen, A.Y.; Hwang, M.H.; Roffler, S.; Chen, C.F. Cytotoxicity and antimicrobial activity of some naphthol derivatives. Arch. Pharm. 1995, 328, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Shen, A.Y.; Tsai, C.T.; Chen, C.L. Synthesis and cardiovascular evaluation of N-substituted aminonaphthols. Eur. J. Med. Chem. 1999, 34, 877–882. [Google Scholar] [CrossRef]
- Huang, M.H.; Wu, S.N.; Wang, J.P.; Lin, C.H.; Lu, S.I.; Liao, L.F.; Shen, A.Y. Biological Study of Naphthalene Derivatives with Antiinflammatory Activities. Drug Dev. Res. 2003, 60, 261–269. [Google Scholar] [CrossRef]
- Ratia, K.; Pegan, S.; Takayama, J.; Sleeman, K.; Coughlin, M.; Baliji, S.; Chaudhuri, R.; Fu, W.; Prabhakar, B.S.; Johnson, M.E.; et al. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. USA 2008, 105, 16119–16124. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, A.K.; Takayama, J.; Rao, K.V.; Ratia, K.; Chaudhuri, R.; Mulhearn, D.C.; Lee, H.; Nichols, D.B.; Baliji, S.; Baker, S.C.; et al. Severe Acute Respiratory Syndrome Coronavirus Papain-like Novel Protease Inhibitors: Design, Synthesis, Protein−Ligand X-ray Structure and Biological Evaluation. J. Med. Chem. 2010, 53, 4968–4979. [Google Scholar] [CrossRef] [Green Version]
- Eid, F.A.; Abd El-Wahab, A.H.F.; El-Hagali, G.A.M.; Khafagy, M.M. Syhthesis and antimicrobial evaluation of naphtho[2,1-b]pyrano[2,3-d]pyrimidine and pyrano[3,2-e][1,2,4] triazolo[1,5-c]pyrimidine derivatives. Acta Pharm. 2004, 54, 13–26. [Google Scholar]
- Eiden, F.; Denk, F. Synthesis and CNS-activity of pyrane derivatives 6,8-dioxabicyclo(3,2,1)octanes. Arch. Pharm. 1991, 324, 353–354. [Google Scholar] [CrossRef]
- Taylor, N.R.; Cleasby, A.; Singh, O.; Skarzynski, T.; Wonacott, A.J.; Smith, P.W.; Sollis, S.L.; Howes, P.D.; Cherry, P.C.; Bethell, R.; et al. Dihydropyrancarboxamides related to zanamivir: A new series of inhibitors of influenza virus sialidases. Crystallographic and molecular modeling study of complexes of 4-amino-4H-pyran-6-carboxamides and sialidase from influenza virus types A and B. J. Med. Chem. 1998, 41, 798–807. [Google Scholar] [CrossRef]
- Li, C.J.; Zhang, L.J.; Dezube, B.J.; Crumpacker, C.S.; Pardee, A.B. Three inhibitors of type 1 human immunodeficiency virus long terminal repeat-directed gene expression and virus replication. Proc. Natl. Acad. Sci. USA 1993, 90, 1839–1842. [Google Scholar] [CrossRef] [Green Version]
- Rajendran, A.; Raghupathy, D.; Priyadarshini, M. Green synthesis of biologically active pyrazolopyrimidine derivatives using an ionic liquid 2-methyl-3-butyl imidazolium chloride. Int. J. Chem. Tech. Res. 2011, 3, 293–397. [Google Scholar]
- Modia, P.; Patela, S.; Chhabria, M. Structure-based design, synthesis and biological evaluation of a newer series of pyrazolo[1,5-a]pyrimidine analogues as potential anti-tubercular agents. Bioorg. Chem. 2019, 87, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Dong, Y.; Li, M.; Wang, R.; Zhang, X.; Gong, P.; Zhao, Y. Design, synthesis and biological evaluation of novel thieno[3,2-d] pyrimidine and quinazoline derivatives as potent antitumor agents. Bioorg. Chem. 2019, 90, 103086. [Google Scholar] [CrossRef] [PubMed]
- Mirmortazavi, S.S.; Farvandi, M.; Ghafouri, H.; Mohammadi, A.; Shourian, M. Evaluation of novel pyrimidine derivatives as a new class of mushroom tyrosinase inhibitor. Drug Des. Devel. Ther. 2019, 13, 2169–2178. [Google Scholar] [CrossRef] [Green Version]
- Yanga, F.; Yub, L.Z.; Diaoa, P.C.; Jiana, X.E.; Zhoua, M.F.; Jianga, C.S.; Youa, W.W.; Mab, W.F.; Zhao, P.L. Novel [1,2,4]triazolo[1,5-a]pyrimidine derivatives as potent antitubulin agents: Design, multicomponent synthesis and antiproliferative activities. Bioorg. Chem. 2019, 92, 103260. [Google Scholar] [CrossRef]
- Somakala, K.; Tariq, S.; Amir, M. Synthesis, evaluation and docking of novel pyrazolo pyrimidines as potent p38α MAP kinase inhibitors with improved anti-inflammatory, ulcerogenic and TNF-α inhibitory properties. Bioorg. Chem. 2019, 87, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Acosta, R.S.; Jung, E.; Qiu, L.; Wilson, D.J.; Geraghty, R.J.; Chen, L. 4,7-Disubstituted 7H-Pyrrolo[2,3-d]pyrimidines and Their Analogs as Antiviral Agents against Zika Virus. Molecules 2021, 26, 3779. [Google Scholar] [CrossRef]
- Hockova, D.; Holy, A.; Masojıdkov, M.; Andrei, G.; Snoeck, R.; De Clercqb, E.; Balzarini, J. Synthesis and antiviral activity of 2,4-diamino-5-cyano-6-[2-(phosphonomethoxy)ethoxy]pyrimidine and related compounds. Bioorg. Med. Chem. 2004, 12, 3197–3202. [Google Scholar] [CrossRef]
- Abdelall, E.K.A. Synthesis and biological evaluations of novel isoxazoles and furoxan derivative as anti-inflammatory agents. Bioorg. Chem. 2020, 94, 103441. [Google Scholar] [CrossRef]
- Das Neves, A.R.; Trefzger, O.S.; Barbosa, N.V.; Honorato, A.M.; Carvalho, D.B.; Moslaves, I.S.; Kadri, M.C.T.; Yoshida, N.C.; Kato, M.J.; Arruda, C.C.P.; et al. Effect of isoxazole derivatives of tetrahydrofuran neolignans on intracellular amastigotes of Leishmania (Leishmania) amazonensis: A structure-activity relationship comparative study with triazole-neolignan-based compounds. Chem. Biol. Drug Des. 2019, 94, 2004–2012. [Google Scholar] [CrossRef]
- De Souza, A.N.A.; Xavier, V.F.; Coelho, G.S.; Junior, P.A.S.; Romanha, A.J.; Murta, S.M.F.; Carneiroc, C.M.; Taylor, J.G. Synthesis of 3,5-Diarylisoxazole Derivatives and Evaluation of in vitro Trypanocidal Activity. J. Braz. Chem. Soc. 2018, 29, 269–277. [Google Scholar] [CrossRef]
- Reddy, S.P.; Yamini, G.; Sowmya, D.V.; Padmavathi, V.; Padmaja, A. Synthesis and Antimicrobial Activity of Some New 3,5-Disubstituted Pyrazoles and Isoxazoles. Med. Chem. 2017, 7, 371–380. [Google Scholar] [CrossRef]
- Thotla, K.; Noole, V.G.; Kedika, B.; Reddy, K. An Efficient Synthesis and Antimicrobial Activity of 5-{2-[(1-Aryl-1H-1,2,3-triazol-4-yl)methoxy]- 5-bromophenyl}isoxazoles. Russ. J. Gen. Chem. 2019, 89, 789–793. [Google Scholar] [CrossRef]
- Yang, Z.B.; Li, P.; He, Y.J. Design, Synthesis, and Bioactivity Evaluation of Novel Isoxazole-Amide Derivatives Containing an Acylhydrazone Moiety as New Active Antiviral Agents. Molecules 2019, 24, 3766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.S.; Park, S.M.; Kim, B.H. Synthesis of 5-isoxazol-5-yl-20-deoxyuridines exhibiting antiviral activity against HSV and several RNA viruses. Bioorg. Med. Chem. Lett. 2009, 19, 1126–1128. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abu-Zaied, M.A.; Elgemeie, G.H.; Mahmoud, N.M. Anti-Covid-19 Drug Analogues: Synthesis of Novel Pyrimidine Thioglycosides as Antiviral Agents Against SARS-COV 2 and Avian Influenza H5N1 Viruses. ACS Omega 2021, 6, 16890–16904. [Google Scholar] [CrossRef]
- Soltane, R.; Chrouda, A.; Mostafa, A.; Al-Karmalawy, A.A.; Chouaïb, K.; Dhahri, A.; Pashameah, R.A.; Alasiri, A.; Kutkat, O.; Shehata, M.; et al. Strong Inhibitory Activity and Action Modes of Synthetic Maslinic Acid Derivative on Highly Pathogenic Coronaviruses: COVID-19 Drug Candidate. Pathogens 2021, 10, 623. [Google Scholar] [CrossRef]
- Ben Said, A.; Rahmouni, A.; Daami-Ramadi, M.; Romdhane, A.; Ben Jannet, H. Design and Synthesis of new antimicrobial [1,2,4]triazolo[1,5-c]pyrimidines. J. Tun. Chem. Soc. 2017, 19, 94–104. [Google Scholar]
- Cherif, M.; Debbabi, M.; Chortani, S.; Romdhane, A.; Ben Jannet, H. Design and synthesis of new naphtho[2,1-b]pyrano [2,3-d]pyrimidinones under classical and microwave conditions. Turk. J. Chem. 2018, 42, 1623–1639. [Google Scholar] [CrossRef]
- Debbabi, M.; Nimbarte, V.D.; Chekir, S.; Chortani, S.; Romdhane, A.; Ben jannet, H. Design and synthesis of novel potent anticoagulant and anti-tyrosinase pyranopyrimidines and pyranotriazolopyrimidines: Insights from molecular docking and SAR analysis. Bioorg. Chem. 2019, 82, 129–138. [Google Scholar] [CrossRef]
- Zayane, M.; Romdhane, A.; Remadi, M.D.; Ben Jannet, H. Access to new antimicrobial 4-methylumbelliferone derivatives. J. Chem. Sci. 2015, 127, 1619–1626. [Google Scholar] [CrossRef]
- Romdhane, A.; Martin, M.T.; Ben Said, A.; Jabrane, A.; Ben Jannet, H. Synthesis of new acetylcholinesterase inhibitors Naphto[2,1-B]pyrano[3,2-E][1,2,4]triazolo[1,5-C]pyrimidine derivatives. J. Tun. Chem. Soc. 2012, 14, 127–135. [Google Scholar]
- Mohammed, F.K.; Soliman, A.Y.; Sawy, A.S.; Badre, M.G. 2-Amino-5- hydroxy-4-phenyl-7-methyl-4H[1-chromeno-3-carbonitrile] as a key precursor for the synthesis of several chromene based heterocyclic systems. J. Chem. Pharm. Res. 2009, 1, 213–224. [Google Scholar]
- Hesek, D.; Lee, M.; Noll, B.C.; Fisher, J.F.; Mobashery, S. Complications from Dual Roles of Sodium Hydride as a Base and as a Reducing Agent. J. Org. Chem. 2009, 74, 2567–2570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chouaïb, K.; Romdhane, A.; Delemasure, S.; Dutartre, P.; Elie, N.; Touboul, D.; Ben Jannet, H.; Hamza, M.A. Regiospecific synthesis, anti-inflammatory and anticancer evaluation of novel 3,5-disubstituted isoxazoles from the natural maslinic and oleanolic acids. Ind. Crop. Prod. 2016, 85, 287–299. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. Software News and Update AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).