Abstract
The Glycoside Hydrolase Family 65 (GH65) is an enzyme family of inverting α-glucoside phosphorylases and hydrolases that currently contains 10 characterized enzyme specificities. However, its sequence diversity has never been studied in detail. Here, an in-silico analysis of correlated mutations was performed, revealing specificity-determining positions that facilitate annotation of the family’s phylogenetic tree. By searching these positions for amino acid motifs that do not match those found in previously characterized enzymes from GH65, several clades that may harbor new functions could be identified. Three enzymes from across these regions were expressed in E. coli and their substrate profile was mapped. One of those enzymes, originating from the bacterium Mucilaginibacter mallensis, was found to hydrolyze kojibiose and α-1,2-oligoglucans with high specificity. We propose kojibiose glucohydrolase as the systematic name and kojibiose hydrolase or kojibiase as the short name for this new enzyme. This work illustrates a convenient strategy for mapping the natural diversity of enzyme families and smartly mining the ever-growing number of available sequences in the quest for novel specificities.
1. Introduction
In the carbohydrate-active enzymes database (CAZy) [1], 10 enzyme specificities (Table 1) are grouped to form the Glycoside Hydrolase Family 65 (GH65), which is a member of clan GH-L. All members of this enzyme family break α-glucosidic bonds through a single displacement mechanism that inverts the anomeric configuration (Figure 1), but they differ in the substrates they prefer [2]. Based on the nucleophile that is used in the breakdown reaction, two groups can be distinguished: glucoside hydrolases use water to hydrolyze the glucosidic bond, whereas glucoside phosphorylases employ inorganic phosphate, releasing glucose or β-glucose 1-phosphate (βGlc1P) as a reaction product, respectively [3]. GH65 enzymes are typically active on α-glucobioses (trehalose, kojibiose, nigerose or maltose) or derivatives thereof (trehalose 6-phosphate, α-1,2- or α-1,3-oligoglucans), but a few other natural substrates have also been identified (3-O-α-glucosyl-l-rhamnose, 2-O-α-glucosylglycerol or the α-glucosyl-1,2-β-galactosyl decoration found on hydroxylysine residues of collagen) [2,4,5,6].

Table 1.
Overview of all currently identified GH65 enzyme specificities and their annotated representatives. Gal: d-galactose, Glc: d-glucose, Glc6P: d-glucose 6-phosphate, Hyl: 5-hydroxy-l-lysine, l-Rham: l-rhamnose, βGlc1P: β-d-glucose 1-phosphate.
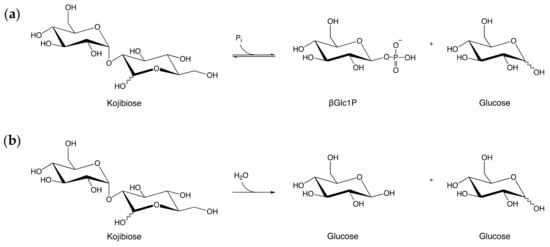
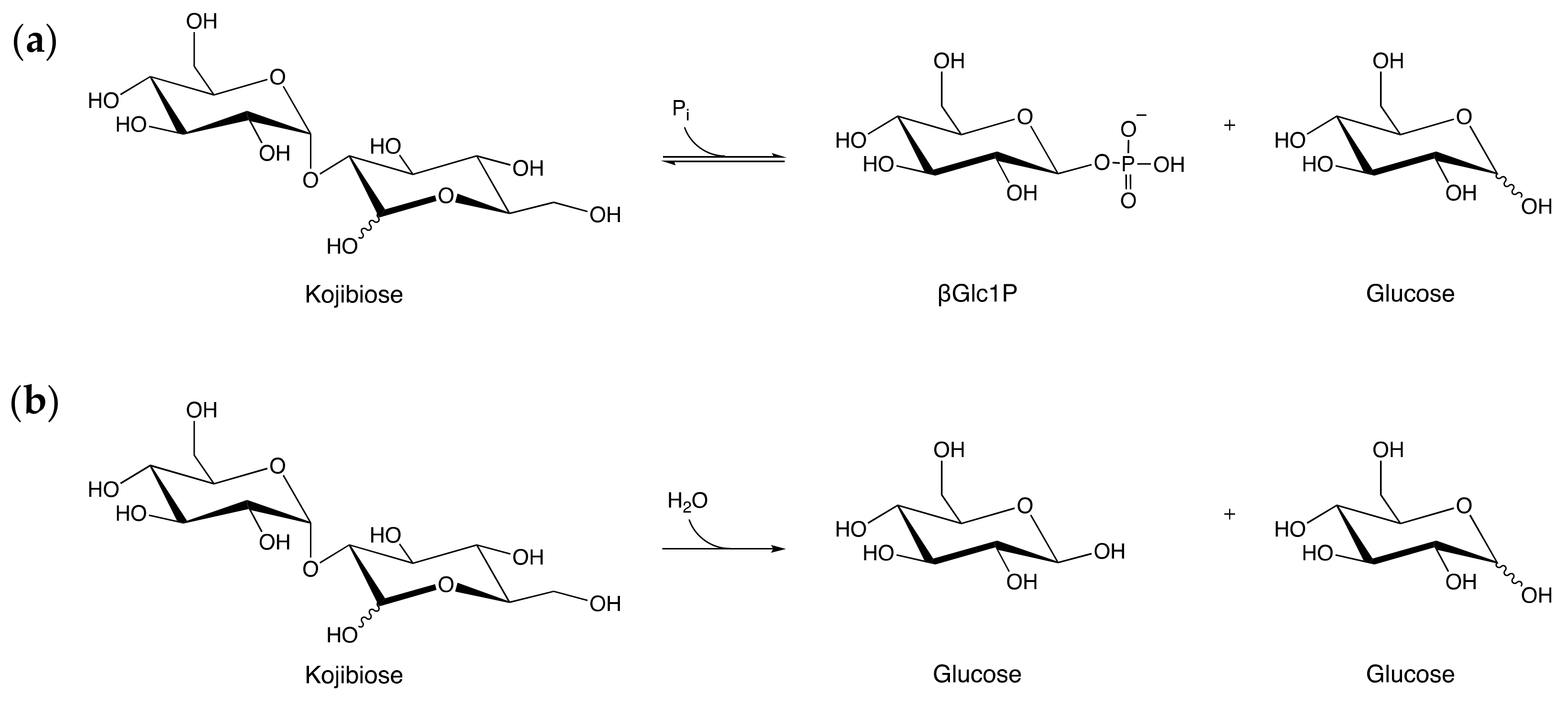
Figure 1.
The two types of reactions catalyzed by enzymes in the Glycoside Hydrolase Family 65 (GH65) illustrated by their respective reactions on kojibiose: (a) Reaction catalyzed by kojibiose phosphorylase; (b) Reaction catalyzed by kojibiose hydrolase. Pi: inorganic phosphate, βGlc1P: β-d-glucose 1-phosphate.
In the past, efforts have been made to elucidate how substrate specificity is controlled in GH65 enzymes. Yamamoto et al. constructed several chimeric enzymes of the kojibiose phosphorylase (KP) and trehalose phosphorylase (TP) of Thermoanaerobacter brockii. Surprisingly, a certain chimera of 785 amino acids contained only one segment of 125 residues originating from the KP (Met384–Thr512) but still exhibited KP activity. This region, ranging from α3 to α6 of the (α,α)6-barrel catalytic domain, was therefore suspected to contain crucial residues for substrate recognition [34]. Within this region, Nakai et al. identified loop 3, a loop that connects α3 and α4 and forms the rim of the active site, as a potential specificity determinant. Loop 3 is typically conserved within one specificity but highly divergent, both in length and amino acid sequence, between different specificities. Through inspection of the crystal structure and mutational analysis, they were able to identify a three-residue motif within this loop (His413, Asn414 and Glu415 in Lactobacillus acidophilus maltose phosphorylase) that is crucial for maltose binding. Replacing this HNE-motif with the corresponding residues in the KP (TPK) or TP (SAY) of Thermoanaerobacter brockii severely impaired the phosphorolytic activity on maltose, while introducing a low activity on kojibiose or trehalose, respectively [35]. Later, Okada et al. determined the crystal structure of the KP of Caldicellulosiruptor saccharolyticus, which resulted in the discovery of another substrate recognition motif. Three residues (Trp391, Glu392 and Thr417) within loop 3 that bind the glucose moiety of kojibiose in the +1 subsite, are highly conserved within all KPs, but show different motifs in other specificities. However, a specificity switch could not be induced by changing the WET-motif into the patterns observed in maltose-, trehalose- or nigerose-active enzymes [4].
Both motifs identified by Nakai et al. and Okada et al. were based on manual inspection of multiple sequence alignments (MSAs) and analysis of crystal structures. Although they were able to identify crucial residues for maltose resp. kojibiose binding, an in-depth search for specificity determinants of the entire GH65 family has never been performed, as it would be practically impossible to investigate the entire sequence diversity with these ad-hoc methods. However, in the context of sequence annotation, enzyme discovery and specificity engineering, it would be extremely beneficial to identify certain positions in the family’s MSA that can be used as an indicator for specificity. Those so-called specificity-determining positions are conserved among all enzymes that share the same specificity but mutate simultaneously across specificity boundaries. If such positions exist, they should be easily picked up by correlated mutations analysis (CMA), a type of statistical analysis that identifies positions in an MSA that underwent interdependent mutations through evolution. As substrate binding is a complex interplay of different residues and interactions, one can expect that a certain function-switching mutation requires one or more compensatory mutations for the protein to remain highly active. These co-evolving positions will thus form a correlation network that can be detected in an MSA through CMA [36].
This technique was already successfully used in the past to uncover specificity fingerprints. Glucokinases could be distinguished from other hexokinases based on six correlated positions that surround the active site [37]. In a similar manner, a network of nine correlated positions was detected in the isocitrate lyase and phosphoenolpyruvate mutase superfamily. One of those nine positions always housed a serine residue in oxaloacetate hydrolases (OAH) and could thus be used as a very specific marker for the OAH subfamily [37]. Bacterial lytic polysaccharide monooxygenases in the Auxiliary Activity Family 10 with activity on cellulose either oxidize exclusively the C1-carbon, or both the C1- and C4-carbon of their substrate. A co-evolving network of 13 positions was shown to be a reliable indicator for this variation in oxidative regioselectivity [38,39]. Recently, we described the so-called heptagonal box model for the NDP-sugar active short-chain dehydrogenase/reductase superfamily based on conservation and correlation patterns. The different subfamilies and specificities can be distinguished based on which amino acids occupy the seven “walls” or fingerprint regions of the model [40].
In this work, we used CMA to identify specificity-determining positions for the GH65 family. Visualization of this specificity fingerprint on the family’s phylogenetic tree did not only facilitate functional annotation of the sequences in this family, but also uncovered a new enzyme specificity. We describe the discovery of a kojibiose glucohydrolase from the bacterium Mucilaginibacter mallensis. Our work demonstrates the potential of CMA for mapping and exploring the natural diversity of an enzyme family and can be beneficial for further explorative endeavors.
2. Results
2.1. Correlated Mutations Analysis
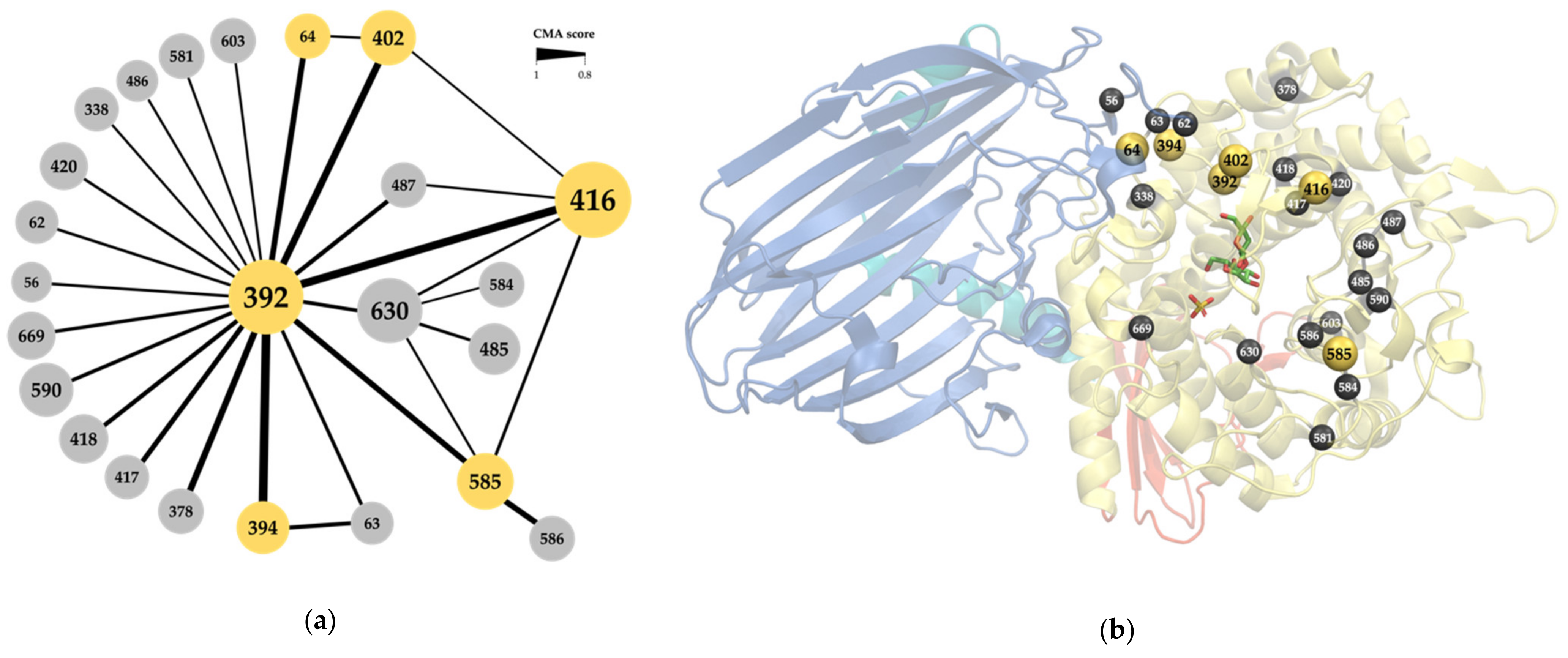
All sequences in family GH65 were extracted from the CAZy database to build a family alignment. In what follows, the sequence numbering of the kojibiose phosphorylase of Caldicellulosiruptor saccharolyticus (CsKP) will be used to refer to MSA positions, unless stated otherwise. CMA was performed with Comulator [37], which revealed 24 positions in the MSA with a correlation score of 0.8 or higher (Figure 2a). Position 392 clearly holds a central position in the correlation network, with 20 co-evolutionary interactions (CMA ≥ 0.8) detected. All correlated positions are part of the (α,α)6-barrel catalytic domain, except for positions 56, 62, 63 and 64, which are located in a loop that emanates from the N-terminal β-sandwich and is known to be involved in the active site architecture of the Levilactobacillus brevis maltose phosphorylase (Figure 2b) [4]. Some of these residues are found within the active site, while others are further away, with the distance between their α-carbon and that of the catalytic acid ranging from 5 to 27 Å. For reasons of clarity and simplicity, this selection of correlated positions was further narrowed down. Since both the correlation strength, measured as the CMA score, and the number of correlated partners are important indicators for overall correlation [36], only the six positions with at least one CMA score higher than 0.9 and at least two correlation partners (CMA ≥ 0.8) will be focused on in the rest of the analysis, namely positions 64, 392, 394, 402, 416 and 585.
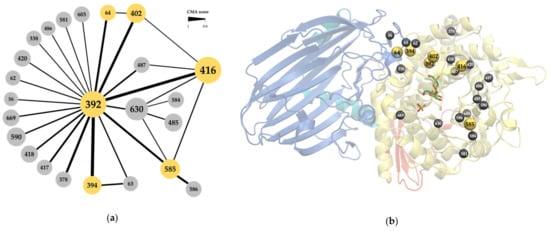
Figure 2.
(a) Correlated position network of the GH65 family alignment, determined with Comulator [37]. Nodes represent the alignment positions, numbering according to the kojibiose phosphorylase of Caldicellulosiruptor saccharolyticus (CsKP). Node size indicates the number of edges. Edge thickness indicates the strength of pair-wise correlation. The six selected positions are highlighted in yellow; (b) Structure of CsKP (PDB ID: 3WIQ) with the location of the correlated positions (yellow and gray spheres). The four structural domains are indicated in different colors: N-terminal domain (blue), linker region (cyan), catalytic domain (yellow) and C-terminal domain (red). Kojibiose (green sticks) and sulphate (yellow sticks) are also shown.
2.2. Correlated Positions as Specificity Determinants
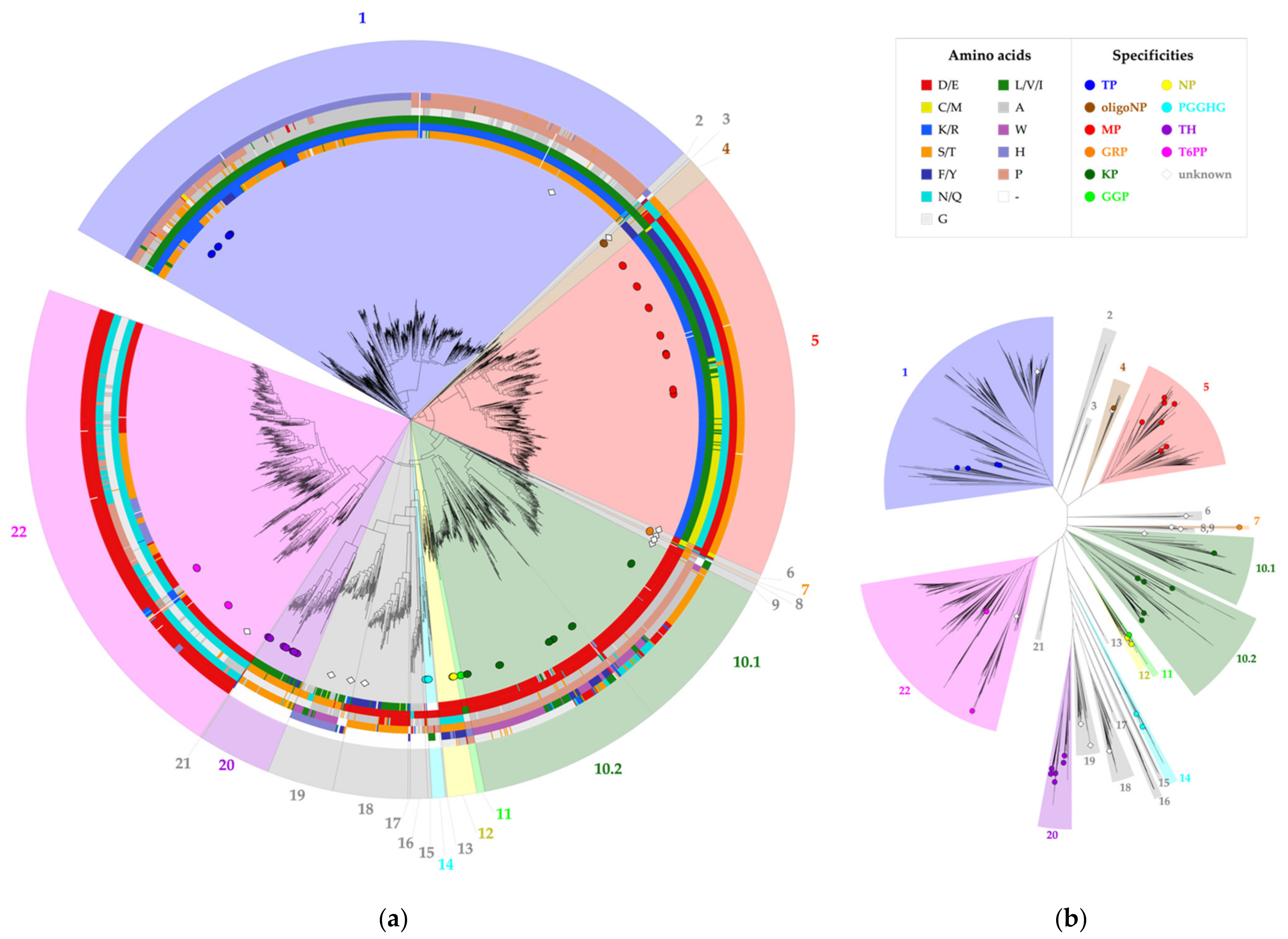
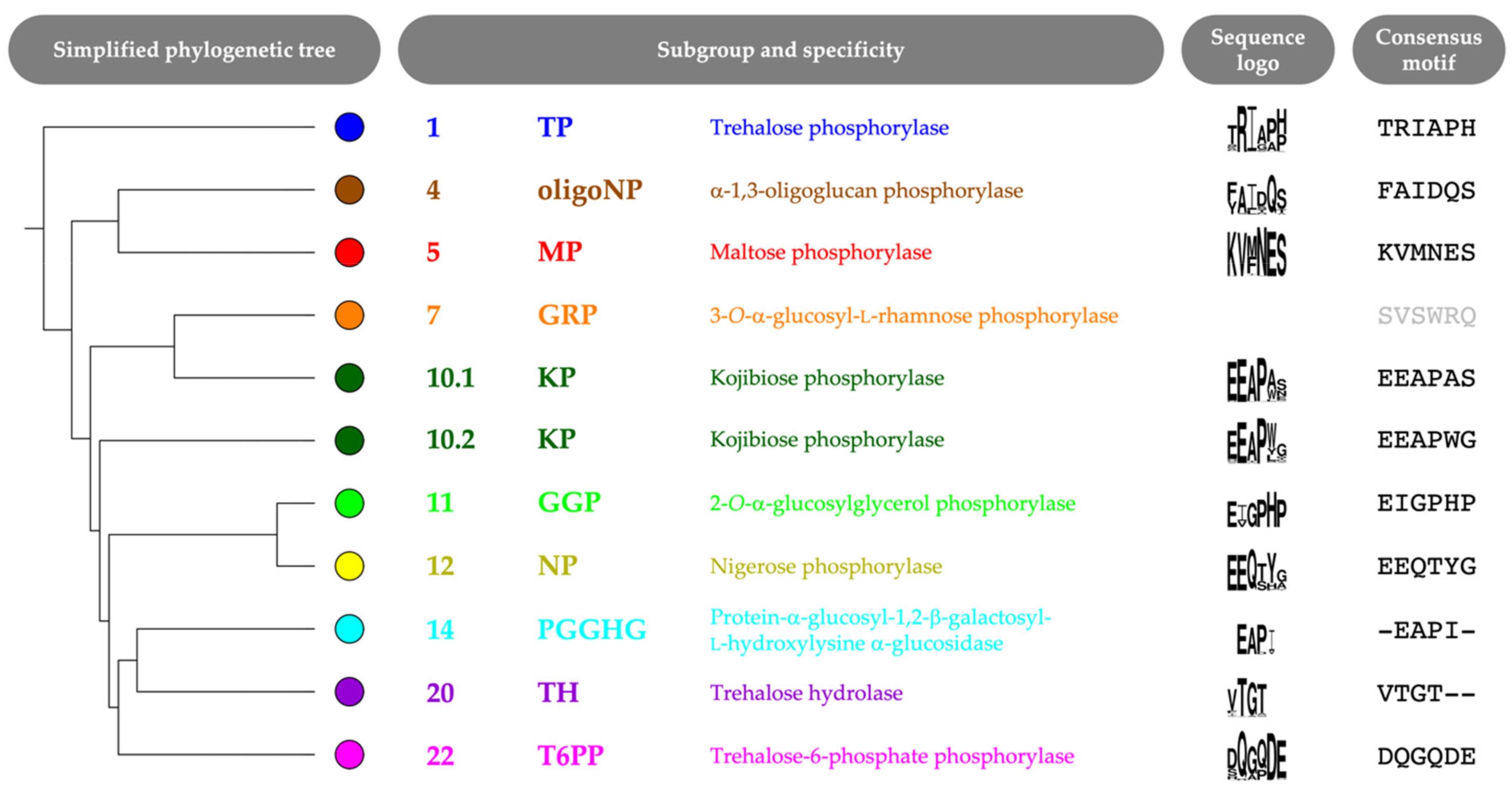
The selected correlated positions were visualized on the family’s phylogenetic tree by means of colored rings (Figure 3, Figure S1). The combination of coevolutionary and phylogenetic information was used to divide the entire GH65 family into 22 subgroups. Subgroups that contain enzymes that have previously been characterized experimentally (Table 1) were annotated with the reported substrate specificity. Taking a closer look at the sequence logos of the correlated positions for these subgroups, it is clear that a comparison of the conserved motifs can easily distinguish all specificities (Figure 4, Figure S2). For example, maltose phosphorylases (MPs) have a highly conserved KV[MF]NES motif, whereas the EEAPxx motif is characteristic of KPs.
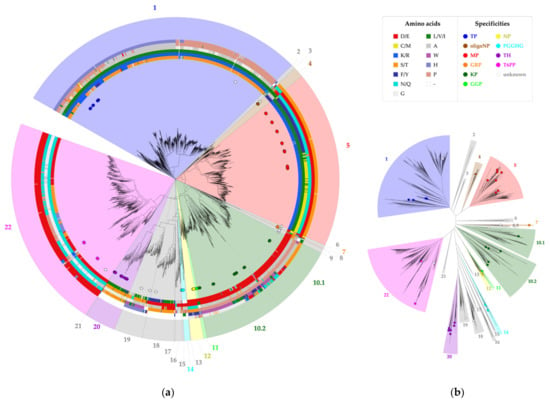
Figure 3.
Phylogenetic tree of family GH65. All annotated representatives and all new enzymes discussed in this study are indicated with circles and diamonds, respectively. The tree is divided into 22 subgroups, which are colored according to their putative specificity. The legend specifies the colors used for each amino acid and each enzyme specificity. (a) Circular tree representation with a visualization of the amino acids present at six correlated positions, shown as colored rings around the phylogenetic tree. From inside to outside, these rings represent positions 64, 392, 394, 402, 416 and 585 (CsKP numbering); (b) Unrooted tree representation.
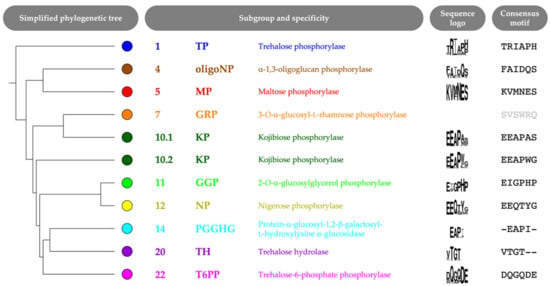
Figure 4.
Simplified phylogenetic tree with sequence logo and consensus motif of six correlated positions (left to right, positions 64, 392, 394, 402, 416 and 585; CsKP numbering) for each subgroup that contains characterized GH65 enzymes. A sequence logo for subgroup 7 is not shown, as this branch contains only one sequence (with motif SVSWRQ).
To verify the predictive power of these motifs to act as specificity indicators, four enzymes were selected from different subgroups that already contain annotated representatives. Sequences originating from thermophilic organisms that reside in functionally uncharacterized branches of the subgroups were preferred. The sequences of choice originated from the organisms Thermobispora bispora (TbGP, GenBank ID: ADG89586.1, in subgroup 1), Caldicellulosiruptor hydrothermalis (ChGP, GenBank ID: ADQ05832.1, in subgroup 4), Halothermothrix orenii (HoGP, GenBank ID: ACL68803.1, in subgroup 10.1) and Caldithrix abyssi (CaGP, GenBank ID: APF18594.1, in subgroup 22). Based on their location in the tree and their specificity motifs (TRIGPP, FAITQA, EEAPWS and DQGQDE), they were predicted to be a trehalose phosphorylase (TP), an α-1,3-oligoglucan phosphorylase (oligoNP), a kojibiose phosphorylase (KP) and a trehalose-6-phosphate phosphorylase (T6PP), respectively. All four enzymes were expressed in E. coli and purified by means of their His6-tag with a yield of 20.2, 41.6, 40.5 and 5.1 mg protein from lysates of a 250 mL-culture, respectively. Their acceptor profile was evaluated by screening them on 46 potential substrates (Table S1). The enzymes were found to show the predicted activity (Figure S3). In a reaction mixture of glucose and βGlc1P, TbGP produced trehalose. ChGP elongated nigerose, and other disaccharides, to form α-1,3-oligoglucans. HoGP showed typical KP behavior, as it was able to elongate both mono- and disaccharides with α-1,2-bound glucose units. In a reaction mixture that contained both kojibiose and βGlc1P, phosphorolytic and synthetic reactions co-occur, both breaking kojibiose down to glucose and βGlc1P and using it as acceptor to produce α-1,2-glucans up to a degree of polymerization (DP) of 6. Finally, our prediction for CaGP was also confirmed, as trehalose 6-phosphate was produced from βGlc1P and glucose 6-phosphate.
2.3. Specificity-Determining Correlated Positions as Tool for Enzyme Discovery
Subgroups 2, 3, 6, 8, 9, 13, 15–19 and 21 could not be annotated as they do not contain any characterized enzymes. Moreover, their sequence motifs at the specificity-determining correlated positions diverge significantly from those of already described enzymes (Figure S2). These subgroups might contain enzymes with new properties or even new enzyme specificities, and are therefore interesting candidates for further exploration. Six enzymes were selected from such subgroups. Based on the residues that occupy positions known to be involved in phosphate binding [4,41,42], three of them were predicted to be phosphorylases, whereas the other three were predicted to be hydrolases (Figure S4). The three putative glycoside phosphorylases originate from the bacteria Mageeibacillus indolicus (MiGP, GenBank ID: ADC90669.1, in subgroup 6), Kiritimatiella glycovorans (KgGP, GenBank ID: AKJ64725.1, in subgroup 8) and Paenibacillus riograndensis (PrGP, GenBank ID: CQR58226.1, in subgroup 9). The three putative hydrolases originate from the bacteria Mucilaginibacter mallensis (MmGH, GenBank ID: SDT07729.1, in subgroup 18), Phyllobacterium zundukense (PzGH, GenBank ID: ATU91641.1, in subgroup 19) and Streptomyces sp. (StreGH, GenBank ID: AEN12156.1, in subgroup 19). Soluble expression of KgGP, PzGH and StreGH in E. coli was not successful, and these enzymes were thus not further investigated. However, MiGP, PrGP and MmGH did express well and were purified by means of their C-terminal His6-tag, with a yield of 6.2, 17.7 and 26.5 mg protein from lysates of a 250 mL-culture, respectively. The acceptor profile of the putative phosphorylases MiGP and PrGP was mapped by screening them in the synthesis direction of the reversible reaction on 46 potential acceptor substrates (Table S1). Both enzymes showed activity towards a diverse set of substrates. Apart from minor activity on glucose and galactose, MiGP prefers α-glucosidic disaccharides, whereas PrGP is mainly active on monosaccharides, even on some l-sugars. The results of this acceptor screening did however not give a clear hint for the natural activity of MiGP and PrGP and their true specificity remains a mystery for now.
The substrate profile of the predicted hydrolase MmGH was evaluated by screening its activity on nine α-glucosides as potential substrates. The enzyme showed very high activity on kojibiose and weak activity on nigerose, but was not capable of breaking down trehalose, maltose, isomaltose, sucrose, isomaltulose, turanose or melezitose (Table 2). MmGH was also able to hydrolyze α-1,2-oligoglucans with a higher DP (Figure S5). Based on these results it was hypothesized that kojibiose might be the natural substrate of MmGH (Figure 1b).

Table 2.
Hydrolytic activity of MmGH on nine potential substrates. Specific activity is reported as mean ± standard deviation of three independent experiments. Fru: d-fructose, Glc: d-glucose, n.d.: not detected (<0.01 U/mg).
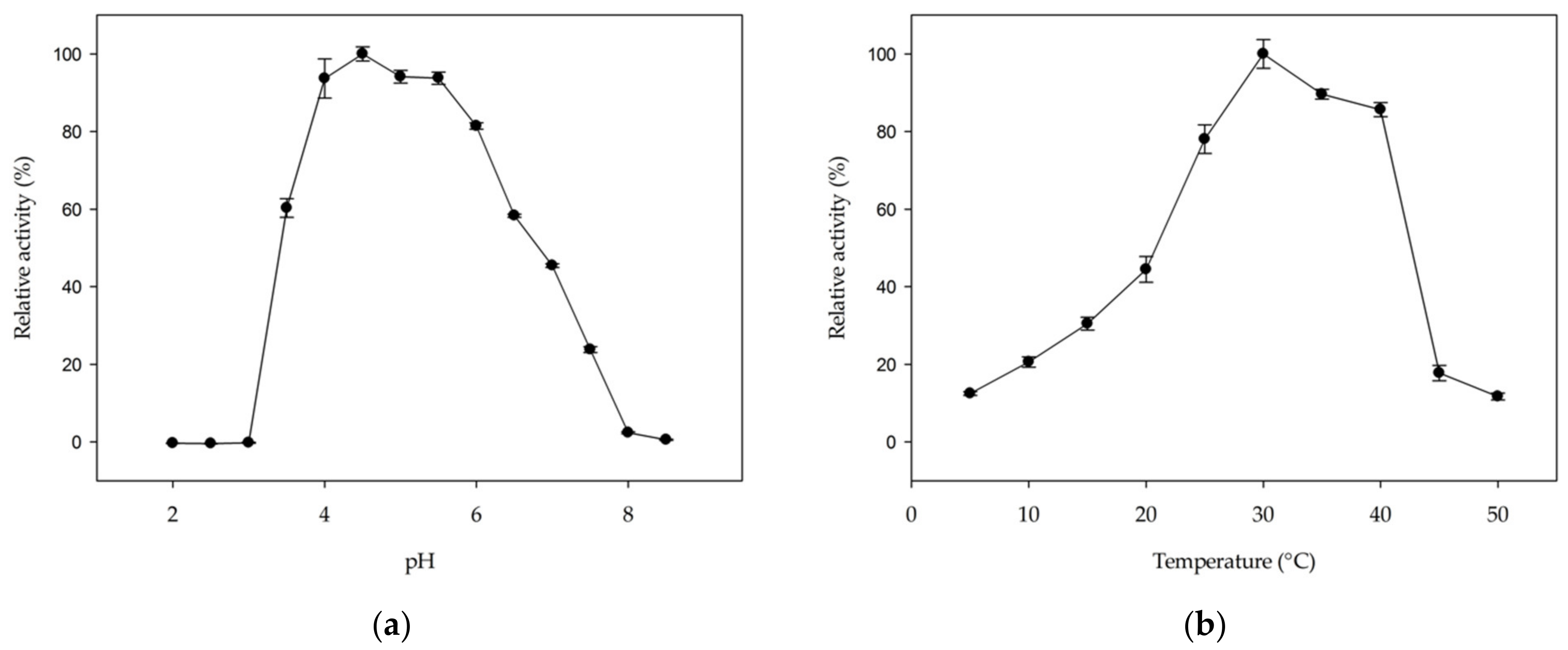
2.4. Optimal pH and Temperature and Kinetic Properties of MmGH
The hydrolytic activity of MmGH on kojibiose is optimal in a pH range from 4 to 5.5 (Figure 5a). The optimal pH range is comparable to that of other hydrolases in family GH65 [27,28,32,33,43,44,45], but differs from GH65 phosphorylases, which typically prefer neutral pH values [5,14,18,19,22,25,26,41]. The optimal temperature was found to be 30 °C (Figure 5b), which is at the higher end of the growth range of Mucilaginibacter mallensis (optimal growth at 25 °C) [46].
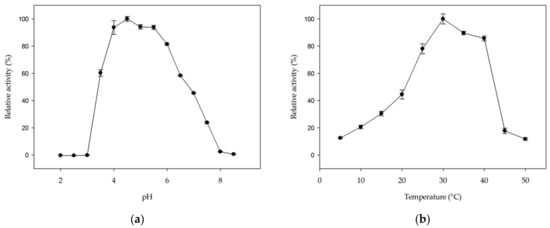
Figure 5.
The effect of (a) pH and (b) temperature on the hydrolytic activity of MmGH on kojibiose. Error bars represent standard error from three independent experiments.
The kinetic parameters of MmGH were determined at the optimal pH (4.5) and temperature (30 °C). The enzyme exhibited Michaelis–Menten kinetics under these conditions, and KM and kcat values of 0.77 ± 0.01 mM and 9.9 ± 0.3 s−1 were deduced. The catalytic efficiency (kcat/KM) equals 13 mM−1 s−1. The affinity of MmGH for kojibiose is higher than that of other GH65 hydrolases for their preferred substrate (KM values between 2.6 and 5.7 mM) [31,33,43,45], which further substantiates that kojibiose is indeed the true substrate of this novel hydrolase.
3. Discussion
In this study, we used CMA to uncover distinct sequence patterns that were applied as specificity fingerprints. Our approach allowed us to analyze the entire GH65 family in a systematic, rather than ad-hoc, manner. This is especially important in light of the continuously growing number of sequences available in databases. For instance, the GH65 family contained 1520 sequences in the CAZy database in 2015 [47], whereas this number has increased to 8189 in 2021 (29 August 2021). In earlier work, we already discovered two new enzyme specificities based on rational comparison of sequence motifs, but information about structure–function relationships was required as an input [48,49]. Here, we report how CMA is a relatively easy method to detect specificity-determining positions in large datasets without any prior knowledge required. The described strategy should be readily applicable to other protein families.
The possible applications of CMA are manifold. Firstly, we showed how analysis of correlated mutations allowed protein annotation and phylogenetic tree analysis. Conservation patterns can be used as an indicator for specificity, which can help to predict the activity of unknown sequences and to identify homologues of a certain protein of interest. Visualizing the correlated positions on the phylogenetic tree resulted in an informative and easy-to-read figure, which facilitated annotation of clades in the tree. Furthermore, the identified specificity fingerprint was also demonstrated to be relevant for enzyme discovery. Guided by the conservation patterns, we discovered a dedicated hydrolase for the breakdown of kojibiose. Future efforts for the discovery of novel enzymes in GH65 could focus on elucidating the natural activity of MiGP and PrGP, and the other unexplored subgroups (2, 3, 13, 15–17 and 21) are of particular interest as well. Finally, CMA might also be valuable for enzyme engineering endeavors, as it provides insight into non-obvious interactions between residues that would not be easily exposed by manual analysis of sequences and crystal structures [36]. The finding that certain positions are entangled in a co-evolving network should sound a cautionary note for mutating these residues in engineering studies. Substituting an amino acid at one position might require compensatory mutations in other positions in the network. Unwittingly disturbing this network can have a dramatic impact on the enzyme’s functionality. This could possibly explain why earlier attempts to mutate positions 392, 402 and 417 of the GH65 correlation network resulted in severely impaired catalytic activity [4,35].
The analysis of correlated positions guided the discovery of a kojibiose hydrolase, for which no EC number is currently available. A few glucosidases have been reported to show some hydrolytic activity on kojibiose, but they typically have a rather relaxed substrate specificity and kojibiose is never their preferred substrate (Table S2). To the best of our knowledge, MmGH is the first glucosidase that is highly specific for kojibiose. We therefore propose kojibiose glucohydrolase as the systematic name and kojibiose hydrolase or kojibiase as the short name for this new enzyme. Kojibiose is now the second sugar, next to trehalose, for which both a dedicated hydrolase and phosphorylase exist in the GH65 family. Therefore, these would make interesting model enzymes for investigating the evolutionary relationship between glycoside hydrolases and phosphorylases [50], even though they only show 15–25% sequence identity. Besides its activity on kojibiose, MmGH is also able to act on α-1,2-oligoglucans, an ability it shares with its phosphorylase counterparts [20,21,22,23]. MmGH’s side-activity on nigerose (0.22% compared to kojibiose) did not come as a surprise either, as KPs have been reported to phosphorolyze nigerose with a similar relative activity (0.23–0.73%) [22,23].
4. Materials and Methods
4.1. Materials
All chemicals were of analytical grade and were obtained from Sigma-Aldrich/Merck (Darmstadt, Germany) or Carbosynth (Berkshire, United Kingdom), except for psicose (Izumori Lab, Kagawa University, Kagawa, Japan), glycerol (Chem-Lab, Zedelgem, Belgium), tagatose (Nutrilab, Giessen, The Netherlands) and trehalose (Cargill, Vilvoorde, Belgium). Kojibiose, nigerose and βGlc1P were produced in-house according to the procedures of Beerens et al. [51], Franceus et al. [52] and Van der Borght et al. [53], respectively.
4.2. Sequence Analysis
All protein sequences classified in family GH65 (January 2020) were extracted from the CAZy database (www.cazy.org) [1]. Redundant sequences with more than 90% sequence identity were removed using CD-HIT with standard parameters [54]. Any annotated GH65 representatives (Table 1) that were removed in this step were added back to the dataset manually. The resulting sequences were aligned with Clustal Omega using default parameters [55]. All 59 sequences lacking the catalytic acid were removed, resulting in a final dataset of 1953 sequences. Those were re-aligned in two steps. First, all characterized GH65 representatives (Table 1) were structurally aligned using MAFFT-DASH [56]. Next, this MSA was used as a skeleton alignment to which the rest of the dataset was aligned with the seed-option of MAFFT [57]. All positions with a gap content of 95% or higher were removed, resulting in a final MSA of 1490 positions.
A phylogenetic tree was generated with FastTree on the BOOSTER server (https://booster.pasteur.fr/) (accessed on 12 January 2021). Branch supports were calculated as transfer bootstrap expectations (TBE) based on 200 bootstrap replicates [58]. The tree was visualized in iTOL [59]. Correlated positions were determined using the Bio-Prodict Comulator tool (https://comulator.bio-prodict.com/) (accessed on 24 January 2021) [37] and visualized as colored rings (RasMol color scheme) around the phylogenetic tree using the built-in function in iTOL. Sequence logos were generated using WebLogo 3 (http://weblogo.threeplusone.com/) (accessed on 19 May 2021) [60]. Figure 2b and Figure S4b were made with PyMOL [61].
4.3. Gene Cloning and Transformation
The genes encoding the enzymes expressed in this paper were codon-optimized for E. coli (Table S3), synthesized and subcloned into a pET21a vector at NheI and XhoI restriction sites, introducing a C-terminal His6-tag, by GeneArt (Thermo Fisher Scientific, Waltham, MA, USA). The plasmid was used for transformation of E. coli BL21(DE3) agp- electrocompetent cells.
4.4. Enzyme Expression and Purification
An overnight culture of the appropriate strain was inoculated (2 v/v%) in 250 mL lysogeny broth (LB) medium (10 g/L tryptone, 5 g/L yeast extract and 5 g/L NaCl) supplemented with 100 µg/mL ampicillin and incubated at 37 °C with continuous shaking at 200 rpm until the OD600 reached ~0.6. Subsequently, enzyme expression from the pET21a vector was induced by adding isopropyl β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mM and continuing incubation at 20 °C overnight. Cells were harvested by centrifugation (30 min at 9000 rpm and 4 °C). Cell pellets were frozen at −20 °C for at least one hour. For enzyme extraction and purification, the pellet of a 250 mL culture was resuspended in 8 mL of lysis buffer (10 mM imidazole, 100 µM phenylmethanesulfonyl fluoride (PMSF), 1 mg/mL lysozyme, 500 mM NaCl, 50 mM sodium phosphate, pH 7.4) and cooled on ice for 30 min. The resulting suspension was sonicated 2 times 3 min (Branson sonifier 450, level 3, 50% duty cycle). Next, the crude cell extract was separated from the cell debris by centrifugation (30 min at 9000 rpm and 4 °C) and subsequently purified by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography. The HisPurTM Ni-NTA resin (1.5 mL, Thermo Fisher Scientific) was washed with 8 mL water and equilibrated twice with 8 mL equilibration buffer (10 mM imidazole, 500 mM NaCl, 50 mM sodium phosphate, pH 7.4) before the crude cell extract was added to allow binding to the resin. Next, the resin was washed three times with 8 mL wash buffer (30 mM imidazole, 500 mM NaCl, 50 mM sodium phosphate, pH 7.4). Protein was eluted with 8 mL elution buffer (250 mM imidazole, 500 mM NaCl, 50 mM sodium phosphate, pH 7.4). The buffer was exchanged to 50 mM 3-(N-morpholino)propanesulfonic acid (MOPS) (pH 7.0) using a 30 kDa cut-off Amicon centrifugal filter unit (Millipore). The protein content was determined by measuring the absorbance at 280 nm with the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). The extinction coefficient and molecular weight of His6-tagged enzymes were calculated using the ProtParam tool on the ExPASy server (https://web.expasy.org/protparam/) (accessed on 24 February 2021). Molecular weight and purity of the protein were verified by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE; 10% gel).
4.5. Detection of Reaction Components
Phosphorylase activity was monitored in the synthetic direction of the reversible reaction by measuring the release of inorganic phosphate using the phosphomolybdate assay described by Gawronski and Benson (2004) [62]. Hydrolytic activity was monitored by the release of glucose, which could be quantified with an enzymatic coupled assay using glucose oxidase and peroxidase (GOD-POD) [63]. The assay solution contained 0.45 mg/mL GOD, 69.2 µg/mL POD and 0.5 mg/mL 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) in 1 M acetate buffer (pH 4.5). As the pH of the assay solution was not sufficient to inactivate MmGH, samples (25 µL) were first inactivated with 1 M NaOH (25 µL) before adding 200 µL of the assay solution. After 30 min incubation at 37 °C, absorbance was measured at 420 nm. Reactions were also monitored by high-performance anion exchange chromatography (HPAEC), coupled to a pulsed amperometric detection (PAD) system. A Dionex ICS-3000 (Thermo Fisher Scientific) with a CarboPac PA20 pH-stable column was used. A 5-µL heat-inactivated (10 min at 95 °C) and appropriately diluted sample was analyzed at a constant flow rate of 0.5 mL/min at 30 °C. The eluent always contained 100 mM NaOH, but the concentration of sodium acetate linearly increased from 10 mM at the start to 600 mM after 18 min. This composition was maintained for 1 min, after which the acetate concentration was gradually changed back to 10 mM during 2 min. After reaching this initial composition, the run continued for 4 min.
4.6. Screening of Potential Substrates
The acceptor profile of the selected glycoside phosphorylases was evaluated by screening them on 46 potential acceptors (Table S4) using the phosphomolybdate assay. Reactions were performed with 10 mM βGlc1P, 10 mM of the acceptor and 0.1 mg/mL purified enzyme in 50 mM MOPS buffer (pH 7.0) at room temperature. For all hits, the same reaction was repeated at 30 °C and 1000 rpm and analyzed with HPAEC-PAD to dismiss any false positives.
For MmGH, nine compounds (trehalose, kojibiose, nigerose, maltose, isomaltose, isomaltulose, sucrose, turanose and melezitose) were evaluated as potential substrates. Reactions were performed with 10 mM of the substrate and 0.5 mg/mL purified enzyme in 100 mM sodium acetate buffer (pH 4.5) at room temperature. Samples were taken every 2 min for 16 min and were analyzed with the GOD-POD assay. For kojibiose, this reaction was repeated with 5 µg/mL purified enzyme to ensure measurement of the initial velocity. For all hits, the same reaction was repeated at 30 °C and 1000 rpm and analyzed with HPAEC-PAD to dismiss any false positives. To evaluate MmGH’s activity on α-1,2-oligoglucans, a mixture of kojitriose (~90%) and kojitetraose (~9%) was produced with CsKP [22]. A reaction was performed with ~50 mM kojitriose, ~5 mM kojitetraose and 0.1 mg/mL purified enzyme in 100 mM sodium acetate buffer (pH 4.5) at 30 °C and 1000 rpm. Samples were analyzed with HPAEC-PAD.
4.7. Optimal pH and Temperature and Kinetic Properties of MmGH
The influence of pH on enzyme activity was determined in 100 mM McIlvaine buffer (pH 2.5–4.0), 100 mM sodium acetate buffer (pH 4.5–5.0), 100 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer (pH 5.5–6.5), 100 mM MOPS buffer (pH 7.0–7.5) or 100 mM tris(hydroxymethyl)aminomethane-hydrochloride (Tris-HCl) buffer (pH 8.0–8.5) at 30 °C. The optimal temperature was determined in 100 mM sodium acetate buffer (pH 4.5). For each reaction, 5 µg/mL purified enzyme was incubated with 50 mM kojibiose. Samples were taken every 2 min for 16 min and analyzed with the GOD-POD assay.
The apparent kinetic parameters of MmGH for kojibiose were determined at the optimal temperature (30 °C) and pH (4.5) in 100 mM sodium acetate buffer. Michaelis–Menten curves were obtained using 5 µg/mL purified enzyme and varying kojibiose concentrations (0.25–10 mM). Parameters were calculated by non-linear regression of the Michaelis–Menten equation using SigmaPlot 13. The molecular weight (73.7 kDa) was used to calculate the turnover number kcat. All tests were performed in triplicate. One unit of enzyme activity was defined as the amount of enzyme hydrolyzing one µmol of substrate per minute under the specified conditions.
Supplementary Materials
The following are available online, Figure S1: Phylogenetic tree of family GH65 with a visualization of the amino acids present at 24 correlated positions, Figure S2: Simplified phylogenetic tree with sequence logo of 24 correlated positions, Figure S3: Reaction profile of selected GH65 glycoside phosphorylases in the synthetic direction of the reversible reaction, Figure S4: Glycoside phosphorylases and hydrolases in family GH65, Figure S5: Hydrolysis of α-1,2-oligoglucans by MmGH, Table S1: Acceptor profile of six GH65 glycoside phosphorylases in the synthetic direction of the reversible reaction, Table S2: Comparison of the substrate profile of all α-glucosidases with hydrolytic activity on kojibiose reported in the BRENDA database and MmGH, Table S3: Nucleotide sequences encoding the enzymes expressed in this work, Table S4: Overview of all compounds tested as possible acceptor for the selected GH65 glycoside phosphorylases [64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88].
Author Contributions
Conceptualization, E.D.B., J.F. and T.D.; investigation, E.D.B. and A.J.; writing—original draft preparation, E.D.B.; writing—review and editing, J.F. and T.D.; visualization, E.D.B.; supervision, T.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Research Foundation—Flanders (FWO), grant number 1S17721N (doctoral scholarship for E.D.B.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data are available within this article and its Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Sample Availability
The plasmid for expression of MmGH has been deposited at BCCM/GeneCorner Plasmid Collec-tion (accession number: LMBP 13040) and is thus publicly available.
References
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Sun, S.; You, C. Disaccharide phosphorylases: Structure, catalytic mechanisms and directed evolution. Synth. Syst. Biotechnol. 2021, 6, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Desmet, T.; Soetaert, W. Enzymatic glycosyl transfer: Mechanisms and applications. Biocatal. Biotransform. 2011, 29, 1–18. [Google Scholar] [CrossRef]
- Okada, S.; Yamamoto, T.; Watanabe, H.; Nishimoto, T.; Chaen, H.; Fukuda, S.; Wakagi, T.; Fushinobu, S. Structural and mutational analysis of substrate recognition in kojibiose phosphorylase. FEBS J. 2014, 281, 778–786. [Google Scholar] [CrossRef] [PubMed]
- Nihira, T.; Nishimoto, M.; Nakai, H.; Ohtsubo, K.; Kitaoka, M. Characterization of Two α-1,3-Glucoside Phosphorylases from Clostridium phytofermentans. J. Appl. Glycosci. 2014, 61, 59–66. [Google Scholar] [CrossRef]
- Hamazaki, H.; Hamazaki, M.H. Catalytic site of human protein-glucosylgalactosylhydroxylysine glucosidase: Three crucial carboxyl residues were determined by cloning and site-directed mutagenesis. Biochem. Biophys. Res. Commun. 2016, 469, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Hüwel, S.; Haalck, L.; Conrath, N.; Spener, F. Maltose phosphorylase from Lactobacillus brevis: Purification, characterization, and application in a biosensor for ortho-phosphate. Enzyme Microb. Technol. 1997, 21, 413–420. [Google Scholar] [CrossRef]
- Ehrmann, M.A.; Vogel, R.F. Maltose metabolism of Lactobacillus sanfranciscensis: Cloning and heterologous expression of the key enzymes, maltose phosphorylase and phosphoglucomutase. FEMS Microbiol. Lett. 1998, 169, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Yasutake, N.; Oshima, Y.; Yamamoto, Y.; Tomita, T.; Miyoshi, S.; Yatake, T. Cloning of the Maltose Phosphorylase Gene from Bacillus sp. Strain RK-1 and Efficient Production of the Cloned Gene and the Trehalose Phosphorylase Gene from Bacillus stearothermophilus SK-1 in Bacillus subtilis. Biosci. Biotechnol. Biochem. 2002, 66, 2594–2599. [Google Scholar] [CrossRef]
- Hidaka, Y.; Hatada, Y.; Akita, M.; Yoshida, M.; Nakamura, N.; Takada, M.; Nakakuki, T.; Ito, S.; Horikoshi, K. Maltose phosphorylase from a deep-sea Paenibacillus sp.: Enzymatic properties and nucleotide and amino-acid sequences. Enzyme Microb. Technol. 2005, 37, 185–194. [Google Scholar] [CrossRef]
- Nakai, H.; Baumann, M.J.; Petersen, B.O.; Westphal, Y.; Schols, H.; Dilokpimol, A.; Hachem, M.A.; Lahtinen, S.J.; Duus, J.Ø.; Svensson, B. The maltodextrin transport system and metabolism in Lactobacillus acidophilus NCFM and production of novel α-glucosides through reverse phosphorolysis by maltose phosphorylase. FEBS J. 2009, 276, 7353–7365. [Google Scholar] [CrossRef] [PubMed]
- Nihira, T.; Saito, Y.; Kitaoka, M.; Otsubo, K.; Nakai, H. Identification of Bacillus selenitireducens MLS10 maltose phosphorylase possessing synthetic ability for branched α-D-glucosyl trisaccharides. Carbohydr. Res. 2012, 360, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Mokhtari, A.; Blancato, V.S.; Repizo, G.D.; Henry, C.; Pikis, A.; Bourand, A.; de Fátima Álvarez, M.; Immel, S.; Mechakra-Maza, A.; Hartke, A.; et al. Enterococcus faecalis utilizes maltose by connecting two incompatible metabolic routes via a novel maltose 6′-phosphate phosphatase (MapP). Mol. Microbiol. 2013, 88, 234–253. [Google Scholar] [CrossRef]
- Chaen, H.; Nakada, T.; Nishimoto, T.; Kuroda, N.; Fukuda, S.; Sugimoto, T.; Kurimoto, M.; Yoshio, T. Purification and characterization of thermostable trehalose phosphorylase from Thermoanaerobium brockii. J. Appl. Glycosci. 1999, 46, 399–405. [Google Scholar] [CrossRef]
- Inoue, Y.; Ishii, K.; Tomita, T.; Yatake, T.; Fukui, F. Characterization of trehalose phosphorylase from Bacillus stearothermophilus SK-1 and nucleotide sequence of the corresponding gene. Biosci. Biotechnol. Biochem. 2002, 66, 1835–1843. [Google Scholar] [CrossRef] [PubMed]
- Van der Borght, J.; Chen, C.; Hoflack, L.; Van Renterghem, L.; Desmet, T.; Soetaert, W. Enzymatic properties and substrate specificity of the trehalose phosphorylase from Caldanaerobacter subterraneus. Appl. Environ. Microbiol. 2011, 77, 6939–6944. [Google Scholar] [CrossRef]
- Nihira, T.; Saito, Y.; Chiku, K.; Kitaoka, M.; Ohtsubo, K.; Nakai, H. Potassium ion-dependent trehalose phosphorylase from halophilic Bacillus selenitireducens MLS10. FEBS Lett. 2013, 587, 3382–3386. [Google Scholar] [CrossRef]
- Andersson, U.; Levander, F.; Rådström, P. Trehalose-6-phosphate Phosphorylase Is Part of a Novel Metabolic Pathway for Trehalose Utilization in Lactococcus lactis. J. Biol. Chem. 2001, 276, 42707–42713. [Google Scholar] [CrossRef] [PubMed]
- Nihira, T.; Nakai, H.; Chiku, K.; Kitaoka, M. Discovery of nigerose phosphorylase from Clostridium phytofermentans. Appl. Microbiol. Biotechnol. 2012, 93, 1513–1522. [Google Scholar] [CrossRef]
- Chaen, H.; Yamamoto, T.; Nishimoto, T.; Nakada, T.; Fukuda, S.; Sugimoto, T.; Kurimoto, M.; Tsujisaka, Y. Purification and Characterization of a Novel Phosphorylase, Kojibiose Phosphorylase, from Thermoanaerobium brockii. J. Appl. Glycosci. 1999, 46, 423–429. [Google Scholar] [CrossRef][Green Version]
- Yamamoto, T.; Maruta, K.; Mukai, K.; Yamashita, H.; Nishimoto, T.; Kubota, M.; Fukuda, S.; Kurimoto, M.; Tsujisaka, Y. Cloning and sequencing of kojibiose phosphorylase gene from Thermoanaerobacter brockii ATCC35047. J. Biosci. Bioeng. 2004, 98, 99–106. [Google Scholar] [CrossRef]
- Yamamoto, T.; Nishio-Kosaka, M.; Izawa, S.; Aga, H.; Nishimoto, T.; Chaen, H.; Fukuda, S. Enzymatic properties of recombinant kojibiose phosphorylase from Caldicellulosiruptor saccharolyticus ATCC43494. Biosci. Biotechnol. Biochem. 2011, 75, 1208–1210. [Google Scholar] [CrossRef]
- Jung, J.H.; Seo, D.H.; Holden, J.F.; Park, C.S. Identification and characterization of an archaeal kojibiose catabolic pathway in the hyperthermophilic Pyrococcus sp. strain ST04. J. Bacteriol. 2014, 196, 1122–1131. [Google Scholar] [CrossRef]
- Mukherjee, K.; Narindoshvili, T.; Raushel, F.M. Discovery of a Kojibiose Phosphorylase in Escherichia coli K-12. Biochemistry 2018, 57, 2857–2867. [Google Scholar] [CrossRef] [PubMed]
- Nihira, T.; Nakai, H.; Kitaoka, M. 3-O-α-D-glucopyranosyl-L-rhamnose phosphorylase from Clostridium phytofermentans. Carbohydr. Res. 2012, 350, 94–97. [Google Scholar] [CrossRef] [PubMed]
- Nihira, T.; Saito, Y.; Ohtsubo, K.; Nakai, H.; Kitaoka, M. 2-O-α-D-Glucosylglycerol Phosphorylase from Bacillus selenitireducens MLS10 Possessing Hydrolytic Activity on β-D-Glucose 1-Phosphate. PLoS ONE 2014, 9, e86548. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, P.; Klionsky, D.J. Purification and biochemical characterization of the ATH1 gene product, vacuolar acid trehalase, from Saccharomyces cerevisiae. FEBS Lett. 1996, 391, 273–278. [Google Scholar] [CrossRef]
- D’Enfert, C.; Fontaine, T. Molecular characterization of the Aspergillus nidulans treA gene encoding an acid trehalase required for growth on trehalose. Mol. Microbiol. 1997, 24, 203–216. [Google Scholar] [CrossRef]
- Weinstock, K.G.; Bush, D. Nucleic Acid Sequences Relating to Candida albicans for Diagnostics and Therapeutics. U.S. Patent US6747137, 12 February 1999. [Google Scholar]
- Pedreño, Y.; Maicas, S.; Argüelles, J.C.; Sentandreu, R.; Valentin, E. The ATC1 gene encodes a cell wall-linked acid trehalase required for growth on trehalose in Candida albicans. J. Biol. Chem. 2004, 279, 40852–40860. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, Z.; Yin, Y.; Cao, Y.; Zhao, H.; Xia, Y. Expression, purification, and characterization of recombinant Metarhizium anisopliae acid trehalase in Pichia pastoris. Protein Expr. Purif. 2007, 54, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Fresneda, R.; Martínez-Esparza, M.; Maicas, S.; Argüelles, J.-C.; Valentín, E. In Candida parapsilosis the ATC1 Gene Encodes for an Acid Trehalase Involved in Trehalose Hydrolysis, Stress Resistance and Virulence. PLoS ONE 2014, 9, e99113. [Google Scholar] [CrossRef]
- Zilli, D.M.W.; Lopes, R.G.; Alves, S.L.; Barros, L.M.; Miletti, L.C.; Stambuk, B.U. Secretion of the acid trehalase encoded by the CgATH1 gene allows trehalose fermentation by Candida glabrata. Microbiol. Res. 2015, 179, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Yamashita, H.; Mukai, K.; Watanabe, H.; Kubota, M.; Chaen, H.; Fukuda, S. Construction and characterization of chimeric enzymes of kojibiose phosphorylase and trehalose phosphorylase from Thermoanaerobacter brockii. Carbohydr. Res. 2006, 341, 2350–2359. [Google Scholar] [CrossRef] [PubMed]
- Nakai, H.; Petersen, B.O.; Westphal, Y.; Dilokpimol, A.; Abou Hachem, M.; Duus, J.Ø.; Schols, H.A.; Svensson, B. Rational engineering of Lactobacillus acidophilus NCFM maltose phosphorylase into either trehalose or kojibiose dual specificity phosphorylase. Protein Eng. Des. Sel. 2010, 23, 781–787. [Google Scholar] [CrossRef]
- Franceus, J.; Verhaeghe, T.; Desmet, T. Correlated positions in protein evolution and engineering. J. Ind. Microbiol. Biotechnol. 2017, 44, 687–695. [Google Scholar] [CrossRef]
- Kuipers, R.K.P.; Joosten, H.-J.; Verwiel, E.; Paans, S.; Akerboom, J.; van der Oost, J.; Leferink, N.G.H.; van Berkel, W.J.H.; Vriend, G.; Schaap, P.J. Correlated mutation analyses on super-family alignments reveal functionally important residues. Proteins Struct. Funct. Bioinform. 2009, 76, 608–616. [Google Scholar] [CrossRef]
- Forsberg, Z.; Bissaro, B.; Gullesen, J.; Dalhus, B.; Vaaje-Kolstad, G.; Eijsink, V.G.H. Structural determinants of bacterial lytic polysaccharide monooxygenase functionality. J. Biol. Chem. 2018, 293, 1397–1412. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, Z.; Stepnov, A.A.; Nærdal, G.K.; Klinkenberg, G.; Eijsink, V.G.H. Engineering lytic polysaccharide monooxygenases (LPMOs). Methods Enzymol. 2020, 644, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, M.; Gevaert, O.; Van Overtveldt, S.; Lange, J.; Joosten, H.J.; Desmet, T.; Beerens, K. Structure-function relationships in NDP-sugar active SDR enzymes: Fingerprints for functional annotation and enzyme engineering. Biotechnol. Adv. 2021, 48, 107705. [Google Scholar] [CrossRef]
- Egloff, M.P.; Uppenberg, J.; Haalck, L.; van Tilbeurgh, H. Crystal structure of maltose phosphorylase from Lactobacillus brevis: Unexpected evolutionary relationship with glucoamylases. Structure 2001, 9, 689–697. [Google Scholar] [CrossRef]
- Touhara, K.K.; Nihira, T.; Kitaoka, M.; Nakai, H.; Fushinobu, S. Structural basis for reversible phosphorolysis and hydrolysis reactions of 2-O-α-Glucosylglycerol phosphorylase. J. Biol. Chem. 2014, 289, 18067–18075. [Google Scholar] [CrossRef]
- Mittenbuhler, K.; Holzer, H. Purification and characterization of acid trehalase from the yeast suc2 mutant. J. Biol. Chem. 1988, 263, 8537–8543. [Google Scholar] [CrossRef]
- Destruelle, M.; Holzer, H.; Klionsky, D.J. Isolation and characterization of a novel yeast gene, ATH1, that is required for vacuolar acid trehalase activity. Yeast 1995, 11, 1015–1025. [Google Scholar] [CrossRef] [PubMed]
- Sternberg, M.; Spiro, R.G. Studies on the Catabolism of the Hydroxylysine-Linked Disaccharide Units of Basement Membranes and Collagens. Ren. Physiol. 1980, 3, 1–3. [Google Scholar] [CrossRef]
- Männistö, M.K.; Tiirola, M.; McConnell, J.; Häggblom, M.M. Mucilaginibacter frigoritolerans sp. nov., Mucilaginibacter lappiensis sp. nov. and Mucilaginibacter mallensis sp. nov., isolated from soil and lichen samples. Int. J. Syst. Evol. Microbiol. 2010, 60, 2849–2856. [Google Scholar] [CrossRef]
- Kitaoka, M. Diversity of phosphorylases in glycoside hydrolase families. Appl. Microbiol. Biotechnol. 2015, 99, 8377–8390. [Google Scholar] [CrossRef] [PubMed]
- Franceus, J.; Decuyper, L.; D’hooghe, M.; Desmet, T. Exploring the sequence diversity in glycoside hydrolase family 13_18 reveals a novel glucosylglycerol phosphorylase. Appl. Microbiol. Biotechnol. 2018, 102, 3183–3191. [Google Scholar] [CrossRef] [PubMed]
- Franceus, J.; Pinel, D.; Desmet, T. Glucosylglycerate Phosphorylase, an Enzyme with Novel Specificity Involved in Compatible Solute Metabolism. Appl. Environ. Microbiol. 2017, 83, e01434-17. [Google Scholar] [CrossRef] [PubMed]
- Franceus, J.; Lormans, J.; Cools, L.; D’hooghe, M.; Desmet, T. Evolution of Phosphorylases from N-Acetylglucosaminide Hydrolases in Family GH3. ACS Catal. 2021, 11, 6225–6233. [Google Scholar] [CrossRef]
- Beerens, K.; De Winter, K.; Van de Walle, D.; Grootaert, C.; Kamiloglu, S.; Miclotte, L.; Van de Wiele, T.; Van Camp, J.; Dewettinck, K.; Desmet, T. Biocatalytic synthesis of the rare sugar kojibiose: Process scale-up and application testing. J. Agric. Food Chem. 2017, 65, 6030–6041. [Google Scholar] [CrossRef] [PubMed]
- Franceus, J.; Dhaene, S.; Decadt, H.; Vandepitte, J.; Caroen, J.; Van der Eycken, J.; Beerens, K.; Desmet, T. Rational design of an improved transglucosylase for production of the rare sugar nigerose. Chem. Commun. 2019, 55, 4531–4533. [Google Scholar] [CrossRef] [PubMed]
- Van der Borght, J.; Desmet, T.; Soetaert, W. Enzymatic production of β-D-glucose-1-phosphate from trehalose. Biotechnol. J. 2010, 5, 986–993. [Google Scholar] [CrossRef]
- Huang, Y.; Niu, B.; Gao, Y.; Fu, L.; Li, W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics 2010, 26, 680–682. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinf. 2008, 9, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Lemoine, F.; Domelevo Entfellner, J.B.; Wilkinson, E.; Correia, D.; Dávila Felipe, M.; De Oliveira, T.; Gascuel, O. Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. Nature 2018, 556, 452–456. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger LLC. The PyMOL Molecular Graphics System; Version 2.4; Schrödinger, LLC: New York, NY, USA.
- Gawronski, J.D.; Benson, D.R. Microtiter assay for glutamine synthetase biosynthetic activity using inorganic phosphate detection. Anal. Biochem. 2004, 327, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Blecher, M.; Glassman, A.B. Determination of glucose in the presence of sucrose using glucose oxidase; effect of pH on absorption spectrum of oxidized o-dianisidine. Anal. Biochem. 1962, 3, 343–352. [Google Scholar] [CrossRef]
- Chang, A.; Jeske, L.; Ulbrich, S.; Hofmann, J.; Koblitz, J.; Schomburg, I.; Neumann-Schaal, M.; Jahn, D.; Schomburg, D. BRENDA, the ELIXIR core data resource in 2021: New developments and updates. Nucleic Acids Res. 2021, 49, D498–D508. [Google Scholar] [CrossRef] [PubMed]
- Fierobe, H.-P.; Clarke, A.J.; Tull, D.; Svensson, B. Enzymatic Properties of the Cysteinesulfinic Acid Derivative of the Catalytic-Base Mutant Glu400→Cys of Glucoamylase from Aspergillus awamori. Biochemistry 1998, 37, 3753–3759. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Ueda, Y.; Nakamura, N.; Abe, S. Hydrolysis of low molecular weight isomaltosaccharides by a p-nitrophenyl-α-D-glucopyranoside-hydrolyzing α-glucosidase from a thermophile, Bacillus thermoglucosidius KP 1006. Biochim. Biophys. Acta Enzymol. 1979, 566, 62–66. [Google Scholar] [CrossRef]
- Suzuki, Y.; Uchida, K. Three forms of α-glucosidase from welsh onion. Agric. Biol. Chem. 1984, 48, 1343–1345. [Google Scholar] [CrossRef]
- Wongchawalit, J.; Yamamoto, T.; Nakai, H.; Kim, Y.-M.; Sato, N.; Nishimoto, M.; Okuyama, M.; Mori, H.; Saji, O.; Chanchao, C.; et al. Purification and Characterization of α-Glucosidase I from Japanese Honeybee (Apis cerana japonica) and Molecular Cloning of Its cDNA. Biosci. Biotechnol. Biochem. 2006, 70, 2889–2898. [Google Scholar] [CrossRef]
- Kimura, A.; Takewaki, S.; Matsui, H.; Kubota, M.; Chiba, S. Allosteric properties, substrate specificity, and subsite affinities of honeybee α-glucosidase, I. J. Biochem. 1990, 107, 762–768. [Google Scholar] [CrossRef]
- Kato, N.; Suyama, S.; Shirokane, M.; Kato, M.; Kobayashi, T.; Tsukagoshi, N. Novel α-glucosidase from Aspergillus nidulans with strong transglycosylation activity. Appl. Environ. Microbiol. 2002, 68, 1250–1256. [Google Scholar] [CrossRef]
- Kita, A.; Matsui, H.; Somoto, A.; Kimura, A.; Takata, M.; Chiba, S. Substrate specificity and subsite affinities of crystalline α-glucosidase from Aspergillus niger. Agric. Biol. Chem. 2014, 55, 2327–2335. [Google Scholar] [CrossRef]
- Kim, N.R.; Jeong, D.W.; Ko, D.S.; Shim, J.H. Characterization of novel thermophilic alpha-glucosidase from Bifidobacterium longum. Int. J. Biol. Macromol. 2017, 99, 594–599. [Google Scholar] [CrossRef]
- Bravo-Torres, J.C.; Villagómez-Castro, J.C.; Calvo-Méndez, C.; Flores-Carreón, A.; López-Romero, E. Purification and biochemical characterisation of a membrane-bound α-glucosidase from the parasite Entamoeba histolytica. Int. J. Parasitol. 2004, 34, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Giudicelli, J.; Emiliozzi, R.; Vannier, C.; De Burlet, G.; Sudaka, P. Purification by affinity chromatography and characterization of a neutral α-glucosidase from horse kidney. Biochim. Biophys. Acta Enzymol. 1980, 612, 85–96. [Google Scholar] [CrossRef]
- Tanaka, Y.; Aki, T.; Hidaka, Y.; Furuya, Y.; Kawamoto, S.; Shigeta, S.; Ono, K.; Suzuki, O. Purification and characterization of a novel fungal α-glucosidase from mortierella alliacea with high starch-hydrolytic activity. Biosci. Biotechnol. Biochem. 2002, 66, 2415–2423. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, Y.; Miyake, T.; Suzuki, Y. Properties of crystalline α-glucosidase from mucor javanicus. Agric. Biol. Chem. 1973, 37, 251–259. [Google Scholar] [CrossRef]
- Matsui, H.; Ito, H.; Chiba, S. Low Molecular—Activity α-glucosidase from ungerminated rice seed. Agric. Biol. Chem. 1988, 52, 1859–1860. [Google Scholar] [CrossRef]
- Yamasaki, Y.; Fujimoto, M.; Kariya, J.; Konno, H. Purification and characterization of an α-glucosidase from germinating millet seeds. Phytochemistry 2005, 66, 851–857. [Google Scholar] [CrossRef]
- Kobayashi, I.; Tokuda, M.; Hashimoto, H.; Konda, T.; Nakano, H.; Kitahata, S. Purification and characterization of a new type of α-glucosidase from paecilomyces lilacinus that has transglucosylation activity to produce α-1,3- and α-1,2-linked…. Biosci. Biotechnol. Biochem. 2003, 67, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Okuyama, M.; Tanimoto, Y.; Ito, T.; Anzai, A.; Mori, H.; Kimura, A.; Matsui, H.; Chiba, S. Purification and characterization of the hyper-glycosylated extracellular α-glucosidase from Schizosaccharomyces pombe. Enzyme Microb. Technol. 2005, 37, 472–480. [Google Scholar] [CrossRef]
- Choi, K.-H.; Hwang, S.; Cha, J. Identification and characterization of malA in the maltose/maltodextrin operon of Sulfolobus acidocaldarius DSM639. J. Bacteriol. 2013, 195, 1789–1799. [Google Scholar] [CrossRef][Green Version]
- Sørensen, S.H.; Norén, O.; Sjöström, H.; Danielsen, E.M. Amphiphilic Pig Intestinal Microvillus Maltase/Glucoamylase. Eur. J. Biochem. 1982, 126, 559–568. [Google Scholar] [CrossRef]
- Seo, S.H.; Choi, K.H.; Hwang, S.; Kim, J.; Park, C.S.; Rho, J.R.; Cha, J. Characterization of the catalytic and kinetic properties of a thermostable Thermoplasma acidophilum α-glucosidase and its transglucosylation reaction with arbutin. J. Mol. Catal. B Enzym. 2011, 72, 305–312. [Google Scholar] [CrossRef]
- Alarico, S.; Da Costa, M.S.; Empadinhas, N. Molecular and physiological role of the trehalose-hydrolyzing α-glucosidase from Thermus thermophilus HB27. J. Bacteriol. 2008, 190, 2298–2305. [Google Scholar] [CrossRef]
- Matsui, H.; Yazawa, I.; Chiba, S. Purification and substrate specificity of sweet corn α-glucosidase. Agric. Biol. Chem. 1981, 45, 887–894. [Google Scholar] [CrossRef]
- Saburi, W.; Mori, H.; Saito, S.; Okuyama, M.; Kimura, A. Structural elements in dextran glucosidase responsible for high specificity to long chain substrate. Biochim. Biophys. Acta Proteins Proteom. 2006, 1764, 688–698. [Google Scholar] [CrossRef]
- Kurakata, Y.; Uechi, A.; Yoshida, H.; Kamitori, S.; Sakano, Y.; Nishikawa, A.; Tonozuka, T. Structural Insights into the Substrate Specificity and Function of Escherichia coli K12 YgjK, a Glucosidase Belonging to the Glycoside Hydrolase Family 63. J. Mol. Biol. 2008, 381, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Torres-Rodríguez, B.I.; Flores-Berrout, K.; Villagómez-Castro, J.C.; López-Romero, E. Purification and partial biochemical characterization of a membrane-bound type II-like α-glucosidase from the yeast morphotype of Sporothrix schenckii. Antonie Van Leeuwenhoek 2012, 101, 313–322. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).