Abstract
It is known that 4F2hc and rBAT are the heavy subunits of the heteromeric amino acid transporters (HATs). These heavy subunits are N-glycosylated proteins, with an N-terminal domain, one transmembrane domain and a bulky extracellular domain (ectodomain) that belongs to the α-amylase family. The heavy subunits are covalently linked to a light subunit from the SLC7 family, which is responsible for the amino acid transport activity, forming a heterodimer. The functions of 4F2hc and rBAT are related mainly to the stability and trafficking of the HATs in the plasma membrane of vertebrates, where they exert the transport activity. Moreover, 4F2hc is a modulator of integrin signaling, has a role in cell fusion and it is overexpressed in some types of cancers. On the other hand, some mutations in rBAT are found to cause the malfunctioning of the b0,+ transport system, leading to cystinuria. The ectodomains of 4F2hc and rBAT share both sequence and structure homology with α-amylase family members. Very recently, cryo-EM has revealed the structure of several HATs, including the ectodomains of rBAT and 4F2hc. Here, we analyze available data on the ectodomains of rBAT and 4Fhc and their relationship with the α-amylase family. The physiological relevance of this relationship remains largely unknown.
1. Introduction
rBAT (SLC3A1, also named D2 and NBAT) and 4F2hc (SLC3A2, also named CD98hc and FRP, for fusion regulatory protein) are the heavy subunits of the heteromeric amino acid transporters (HATs) [1] and they are linked to an L-amino acid transporter (LAT) subunit from the SLC7 gene family by a disulfide bridge (Figure 1a). These heavy subunits are type II membrane proteins with a cytoplasmatic N-terminus, one transmembrane domain (TM) and a bulky extracellular N-glycosylated C-terminal domain (50–60 kDa) or ectodomain (ED) (Figure 1a). In humans, six LAT transporters (LAT1, LAT2, y+LAT1, y+LAT2, asc-1, and xCT) heterodimerize with 4F2hc and two (b0,+AT and AGT1) with rBAT, thereby rendering a range of different amino acid transport activity [2]. The heavy subunits of HATs appeared in metazoans with a primitive heavy subunit and this evolved into the vertebrate proteins 4F2hc and rBAT [3,4]. These two human proteins share approximately 25% amino acid sequence identity. Surprisingly, their bulky extracellular domains share similar sequence identity or even greater, around 25% and 40% for 4F2hc-ED and rBAT, respectively, with members of α-amylase family, specifically from the GH13 family [5,6].
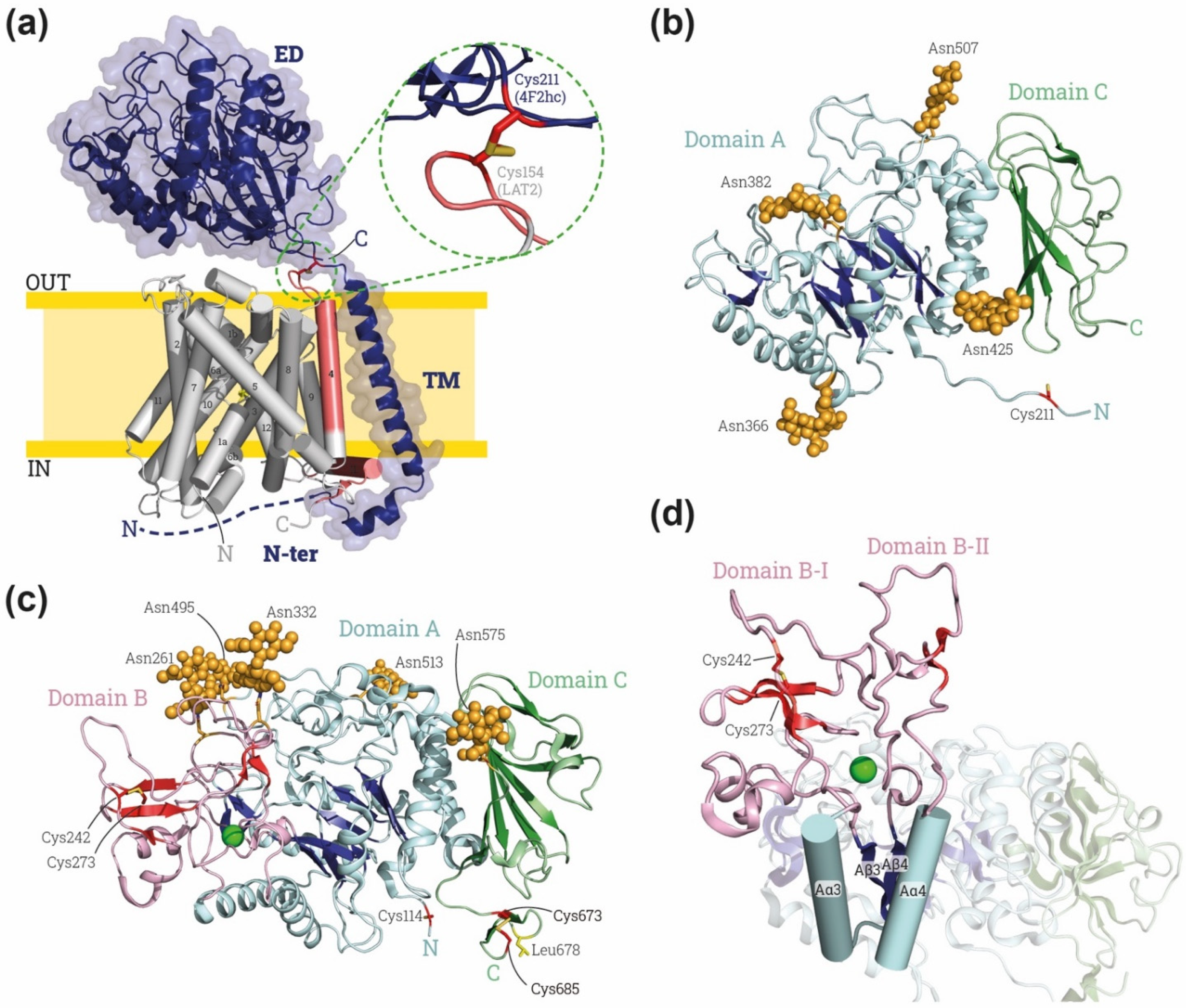
Figure 1.
Overview of HAT structures. (a) Representative HAT structure. Cryo-EM structure of human 4F2hc-LAT2 (PDB: 7CMI [7]) is shown: heavy subunit 4F2hc is indicated in dark blue and by blue cartoon with a transparent surface, and light subunit LAT2 is indicated in gray and red cylinders. The transmembrane (TM) domains of LAT2 are numbered from the N to the C-terminus. On the right, close-up for the disulfide bond between heavy and light subunit. Heavy subunit contains an ectodomain (ED), a transmembrane domain (TM) and an N-terminus domain (N-ter). Domains of LAT-2 interacting with heavy subunit are colored in red. (b) Human 4F2hc-ED structure, PDB: 7DSK [8]. The four glycosylation moieties are shown in light orange spheres, and subdomains A and C in blues and greens, respectively, with brighter colors indicating the β-sheets. The cysteine involved in the disulfide bridge with LATs (Cys 211 in human 4F2hc) is shown in red at the N-ter. (c) Human rBAT-ED structure, PDB: 6LI9 [9]. The five glycosylation moieties are labelled by light orange spheres, and subdomains A, B and C in blue, red and green, respectively, with brighter colors indicating the β-sheets. The cysteine involved in the disulfide bridge with LATs (Cys 114 in human rBAT) is shown in red at the N-terminus of rBAT-ED. L678 is colored in yellow and Ca2+ atom is represented by green ball. (d) Detailed view of the domain B with subdomains BI and BII (loops Aβ3-Aα3 and Aβ4 and Aα4, respectively) stabilized by a Ca2+ (green ball). Images were created with the PyMol Molecular Graphics System, Version 2.1.4., Schrödinger, LLC.
The heavy subunit in HATs is required for the correct localization of the functional holotransporter to the plasma membrane, whereas the light subunit is responsible for the transport function (reviewed in [10,11]). Nevertheless, the heavy subunit can modulate the transport function. In this regard, 4F2hc-LAT1, 4F2hc-LAT2, LAT1 and LAT2 expressed in Pichia pastoris demonstrate that the presence of 4F2hc modulates substrate affinity and specificity in the light subunit-associated transport [12]. Additionally, one report showed differences in the requirement of the ectodomain of 4F2hc (4F2hc-ED) for the plasma membrane localization of some light subunits. Working with domain truncations of 4F2hc revealed that the presence of the ED is essential for LAT2 and y+LAT2 but not for LAT1 [13]. Finally, studies with detergent-solubilized LAT2 revealed increased stability when the transporter was incubated with the recombinant 4F2hc-ED [14].
Furthermore, 4F2hc, with its associated light chains, is an almost ubiquitously expressed protein playing a central role in cellular nutrition, redox homeostasis and nucleotide availability, being a modulator of cell proliferation [15,16,17]. Indeed, 4F2hc together with light subunits LAT1 and xCT is overexpressed in different types of cancers [18,19,20] and in activated lymphocytic cells [21], thereby supporting that this heavy subunit is involved in cell growth. Moreover, there is a functional interaction between 4F2hc and β1A and β3 integrins, which modulates integrin-related signaling and is essential for integrin-dependent cell spreading, migration and tumor progression [22,23,24,25,26]. Mapping of the interactions of 4F2hc with integrins using protein chimeras with the unrelated glycoprotein CD69 demonstrated that the key protein parts are the TM and the cytosolic N-terminal domain [22,24]. Nevertheless, the 4F2hc-ED might also have a role modulating the β1 integrin function and tumorigenicity [27]. Additionally, the interaction of lectin galectin-3 with the extracellular domain of 4F2hc has been described to play a role in trophoblast formation by cell fusion in the placenta [28].
The heavy subunit rBAT has not been related to functions other than the targeting of b0,+AT to the plasma membrane, where the holotransporter transports neutral (0) and basic (+) amino acids. rBAT/b0,+AT expression is restricted to the apical domain of the plasma membrane of the epithelial cells in the small intestine and in the renal proximal tubule, whereas 4F2hc, together with corresponding LAT transporters, is located in the basolateral plasma membrane in epithelial cells [29], thereby pointing to the participation of the heavy subunit in the location of the holotransporter in these polarized epithelial cells. Importantly, some mutations in rBAT cause type A cystinuria, an inherited aminoaciduria caused by the loss of function of the rBAT/b0,+AT transporter in the epithelial cells [30] of the kidney and intestine. The studied mutations in rBAT-ED cause a defect in the biogenesis, stability and arrival to the plasma membrane of the holotransporter [31].
Another feature associated with heavy subunits is that, in native tissues, 4F2hc-associated transporters are single heterodimers, whereas rBAT/b0,+AT is a dimer of heterodimers ([rBAT/b0,+AT]2), despite the heterodimer being the functional transport unit in the membrane [32]. Experiments with chimeras joining rBAT to 4F2hc-associated light subunits show the formation of a functional dimer of heterodimers, indicating that the heavy subunit governs the oligomerization state [32]. In this regard, cryo-EM structures have demonstrated that the interactions of dimers of heterodimers in rBAT-b0,+AT occur through rBAT-ED (Figure 2a,b) [9,33,34]. The cytosolic N-terminus of rBAT has not been solved, leaving open the possibility of further interactions in this protein part. Although homodimers of 4F2hc can be found upon overexpression in cultured cells and in one of the crystal structures of recombinant 4F2hc-ED [5], a dimer of heterodimers never has been described in vivo or in any further 4F2hc structures (Table 1).
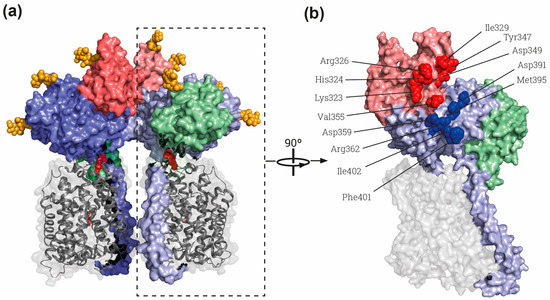
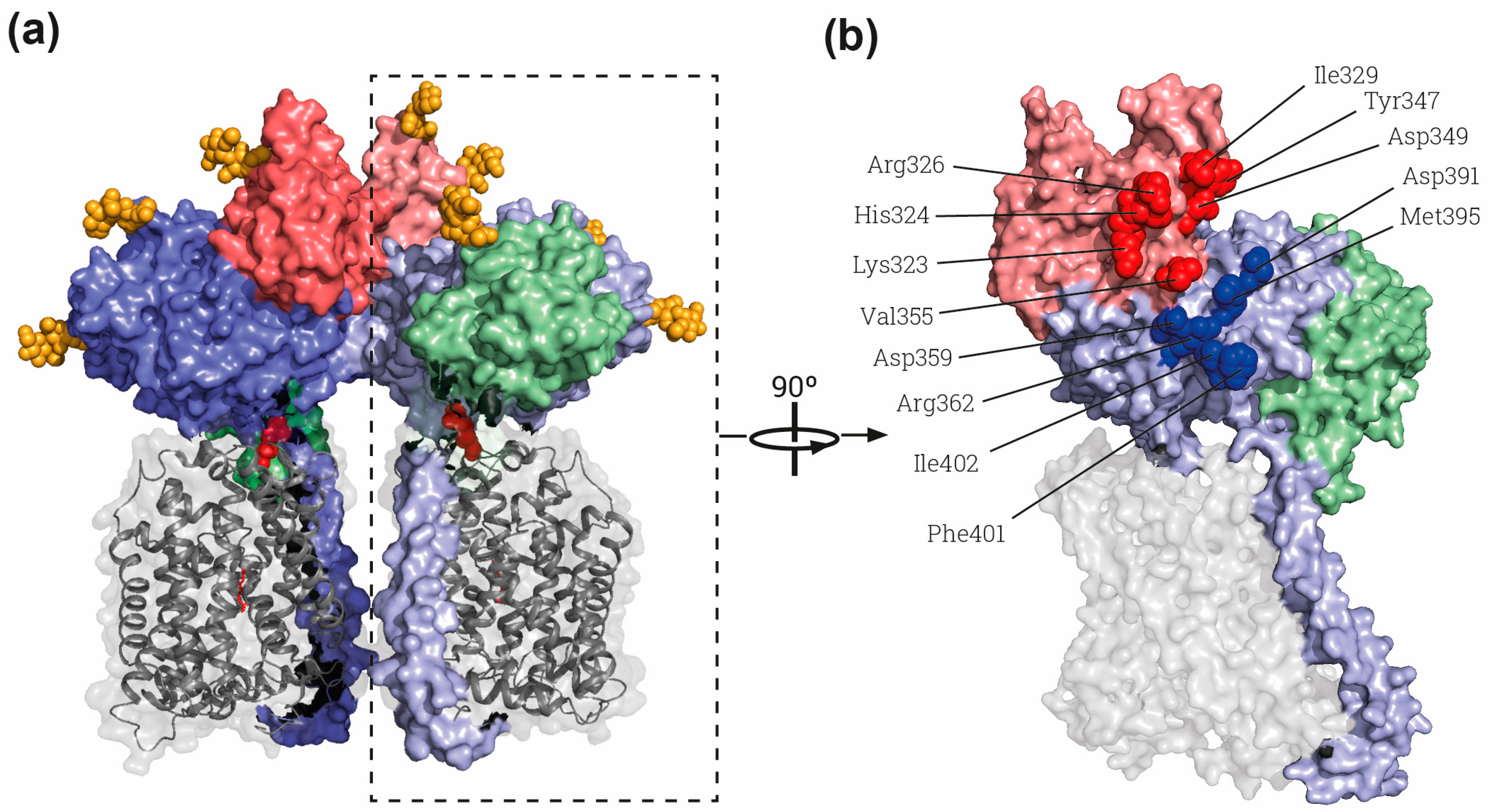
Figure 2.
The rBAT-ED is responsible for dimerization. (a) General view of the whole superdimer of rBAT-b0,+AT (PDB: 6LI9) [9]. Dashed rectangle indicates the heterodimer represented in B). (b) Lateral view of the interaction side in the heterodimer. Residues involved in the rBAT-rBAT homodimerization within the domains B and A of the ED are labelled by red and blue spheres, respectively. Each heterodimer dimer is shown in different intensity of same color, gray for b0,+AT and blue, red and green for rBAT domains TM and A, B and C, respectively. The cysteines involved in the disulfide bridge between subunits and glycosylation moieties are represented by red and orange spheres respectively. Images were created with the PyMol Molecular Graphics System, Version 2.1.4., Schrödinger, LLC, New York, USA.

Table 1.
HAT heavy subunit structures deposited in the Protein Data Bank (RCSB). Structures are grouped by the protein solved. The Uniprot code, solved region (aa), technique of resolution (Tech.), overall resolution (Å), Protein Data Bank entry codes (PDB), details of the structure (such as ligands, antibodies (Fab), nanodisc (ND) reconstitution) and references (Ref.) are also indicated.
The ectodomains of 4F2hc and rBAT show sequence homology with α-amylase enzymes, specifically with the oligo-1,6-glucosidase subfamily in glycosyl hydrolases family 13 (GH13) [4,40,41]. The large α-amylase family comprises more than 30 distinct substrate specificities, but its members share defining common features: (i) they catalyze the action on α-glucosidic bonds through a retaining reaction mechanism, (ii) the catalytic domain is a (β/α)8-barrel fold (i.e., TIM-barrel), (iii) they have between four and seven conserved regions mainly in the β strands of the barrel and (iv) the conserved catalytic site is formed by an aspartate (nucleophile), a glutamate (proton donor) and another aspartate (transition-state stabilizer) placed in the vicinities of β4, β5 and β7 strands [40,42,43,44,45,46,47,48,49]. The GH13 family includes members from several enzyme activities as α-amylases, α-glucosidases, α-1,4-glucan branching enzymes, pullulanases, cyclodextrin glucanotransferases, 4-α-glucanotransferases, oligo-α-1,6-glucosidases. Due to sequence and structural homology the ectodomains 4F2hc and rBAT have been classified in the Carbohydrate-Active Enzymes database into the GH13 family [41] in the oligo-1,6-glucosidase subfamily [50] and, more specifically, 4F2hc in GH13_34 and rBAT in GH13_35 [51].
In this review, we update the structural and functional information about the 4F2hc-ED and rBAT-ED, their relationship with α-glucosidases and we also discuss their possible physiological roles.
2. Structural Information about 4F2hc and rBAT
The cryo-EM structures of 4F2hc and rBAT (Table 1) within the respective holotransporters revealed some clues about the relationship between the two subunits. Regarding the heterodimer interface, the heavy subunit interacts extensively with its respective light subunit via several regions. A disulfide bond is present in the neck, between the TM and the ED, in the heavy subunit and the extracellular loop two (EL2), between TM3 and TM4, in the light subunit (Figure 1a and Figure 2a). In addition, there is a tight hydrophobic interaction in the membrane region between the TM of the heavy subunit and the TM4 in the light subunit (Figure 1a). In the cytoplasm, there is an interaction between the α-helix located in the N-terminal of the heavy subunit and the C-terminal of the light subunit (Figure 1a). Due to the lack of structural information for the first residues of the N-terminus of the heavy subunits other interactions cannot be disregarded.
The ED of the heavy subunits is located above the light subunit in all the solved structures (Table 1 and Figure 1a and Figure 2a), showing many polar interactions in addition to the disulfide bond between the two subunits. However, some interfaces between the N-terminal (β/α)8-barrel loops and extracellular loops of the light subunits are variable. In the 4F2hc-LAT2 structure, with LAT2 in an inward-facing conformation (i.e., the substrate vestibule open to the cytosol), these interactions are not present. On the other hand, the interfaces between 4F2hc-ED and LAT1 in an inward-facing conformation [37] are only partially conserved when LAT1 adopts a different outward-facing conformation [8]. Overall, the position of the 4F2-ED with respect to the different light subunits is very similar in 4F2hc-LAT1 and 4F2-LAT2, where the light subunits show an inward-open conformation (reviewed in [52]), but rotates approximately 5.3° in structures of 4F2-LAT1 in outward-facing conformations [8]. Moreover, in the 4F2hc-xCT structure, with xCT showing an inward-facing conformation, there is a displacement of ~20° vertically away from the entrance of the amino acid binding site in the light subunit [38]. The relative position of the bulky ED above the light subunit has been proposed using a low-resolution structure of the heterodimer 4F2hc-LAT2, with docking and crosslinking experiments between both subunits [14]. The crosslinking results do not fit with the 4F2hc-LAT2 cryo-EM structure [7] or any structure of 4F2hc with xCT or 4f2hc-LAT1. However, 4F2hc in membranes of mammalian cells might explore different conformations than in cryo-EM studies, where the transporter is solubilized in detergent micelles. More structures of different 4F2hc holotransporters and in a lipid environment are needed to define the conformational landscape of 4F2hc-ED interactions with the light subunits in HATs.
The 4F2hc-ED is linked to the TM through a neck of 11 residues (Figure 1a,b). This neck interacts with the globular ED in a very similar way in all the solved structures. This neck is shorter in rBAT-ED, having only six residues (Figure 1c and Figure 2a). This implies differences between the heterodimerization surface of the rBAT-ED and 4F2hc-ED with their respective light subunits. Cryo-EM structures showed a displacement of ~40 Å between them, creating different interfaces [34]. Moreover, the bottom surface of the rBAT-ED is mostly negatively charged, whereas that of 4F2hc-ED is positively charged, thus pointing to distinct electrostatic interactions [5,34].
In summary, the relative position of the ED of heavy subunits with respect to the light subunit in HATs is seen to be slightly variable in the structures, depending on the light subunit and also depending on the conformational state of this light subunit. In this regard, the differences in interaction interfaces suggest a possible stabilizer role of the ED but allowing a certain degree of flexibility between the two subunits of the heterodimer, which may be needed for protein function.
3. Structure of the Ectodomains of 4F2hc and rBAT
The first available structure for the HAT family was the ectodomain of 4F2hc (Table 1) [5]. As predicted by sequence homology, the structure of 4F2hc-ED shows the topology and multidomain organization of α-amylases (Figure 1b). To date, seventeen 4F2hc structures have been deposited in the RCSB database (Table 1). Five 4F2hc-ED structures have been solved by X-ray crystallography and one by cryo-EM, and eleven structures of full 4F2hc heterodimerizing with different LAT transporters and ligands have been achieved by cryo-EM (Table 1). Superimposition of all the available atomic structures of the EDs of 4F2hc (Table 1) reveals high similarity for this extracellular domain, with a calculated Rooth Mean Square Distance (RMSD) below 0.7 A2, and without significant changes in any region of the structure.
Furthermore, 4F2hc-ED contains the so-called domain A in α-amylases, with (β/α)8-barrel fold, linked to the TM domain by the eleven-residue neck indicated above (Figure 1a,b). At the end of domain A, a six-residue helix connects to the domain C (Figure 1b) that is consistently present in α-amylases, although presenting a variable β-sandwich topology [40].
The first structures of human rBAT forming a dimer of heterodimers with b0,+AT and a single homodimer of rBAT (Table 1, Figure 2) have also recently been deposited in the Protein Data Bank. The ectodomain of rBAT has a similar domain composition to 4F2hc with domain A and C, but, additionally, it contains domain B, which is also common in the loop between Aβ3 and Aα3 in the (β/α)8-barrel of the α-amylase family (Figure 1d). This loop has three β-strands and one α-helix, and it contacts domain A through a cluster of hydrophobic residues. Finally, rBAT contain a C-terminus cyclic C-terminal β-hairpin stabilized by a disulfide bridge (Cys673-Cys685) protruding from domain C (Figure 1c,d). This C-terminal small hairpin is key for the maturation of human rBAT protein as tested in mammal cells [53].
The domain B of rBAT is similar to that of oligo-1,6-glucosidases as predicted by sequence homology [40]. The loop Aβ3-Aα3 is stabilized by a disulfide bridge between Cys242 and Cys273, which had already been predicted by modeling based on oligo-1,6-glucosidase from B. cereus [5] and biochemically demonstrated [54]. Interestingly, a Ca2+ binding site is present in this loop in the cryo-EM structures (Figure 1c,d) [9,33,34]. Indeed, binding to Ca2+ is important for the function and stability of many members of the α-amylase family [55,56]. Moreover, Anoxybacillus sp. SK3-4 amylase (ASKA) has a conserved Ca2+ site in the same position as rBAT, stabilizing domain B [57]. In detail, the Ca2+ binding site in rBAT-ED stabilizes the interaction of the loop Aβ3-Aα3 with the next one in the (β/α)8-barrel, the Aβ4-Aα4 (Figure 1d). Due to these strong interactions, domain B could be considered to be formed by both loops as subdomains B-I and B-II, since Ca2+ interacts with and stabilizes them as a single domain above the (β/α)8-barrel (Figure 1c,d) [34]. Subdomain B-II is partially responsible for the homodimerization between two molecules of rBAT, as shown in the structures of the dimers of heterodimers or superdimers and in the homodimeric rBAT structure (Table 1, Figure 2a and red spheres in b) [9,33,34]. This superdimerization is crucial for heterodimer maturation, as demonstrated in experiments using mutations in residues in the homodimerization interphase (Figure 2b) [34]. In this regard, mutations that affect the stability of domain B, such as cystinuria-causing mutation T216M or the residues responsible of Ca2+ binding, confirm that this domain B is crucial for superdimerization and therefore heterodimer maturation [31,34]. There is another homodimerization interface localized in Aα4 and the loop between Aβ5 and Aα5 in domain A (blue spheres in Figure 2b), but no other interactions are present in the structure between both rBAT molecules outside of the ectodomain.
Most of the mutations in rBAT described to cause cystinuria are located in the ED [2]. Biogenesis of human rBAT-b0,+AT with mutated versions on rBAT protein T216M, R365W, M467T or M467K showed impaired biogenesis being expressed in mammal cells [31]. In this regard, recombinant rBAT-b0,+AT with mutations V183A, T216M, M467T and L678P in rBAT-ED showed decreased stability and the recombinant protein could not be purified properly [9]. With respect to the structure, T216 is located in the middle of the hydrophobic interaction between domains A and B and M467 is at the interface of domains A and C, explaining why changes in these residues might destabilize the protein. On the other hand, R365 is stabilizing loop Aα4-β5, which interacts with the light subunit, thus explaining why the R365W mutation might affect the transport characteristics of the b0,+AT system [58]. Finally, L678 is in the C-terminal β-hairpin (Figure 1c), which interacts with the light subunit and the lipid bilayer in structures [9,33,34] and is also shown to be important for the biogenesis of the rBAT-b0,+AT heterodimer [53].
The cryo-EM structures of rBAT and 4F2hc expressed in mammal cells showed the characteristic glycosylations in some Asn residues of the ED. The 4F2hc-ED shows four N-glycosylated positions in domain A (Asn264 in Aα4, Asn280 just before Aα5, Asn323 in Aα6, and Asn405 in loop Aβ8-Aα8) (Figure 1b). In contrast, human rBAT has five N-glycans (Asn261 in domain B, Asn332 in loop Aβ4-Aα4, Asn495 and Asn513 in the loop Aβ8-Aα8 in the domain A and Asn575 in the domain C) (Figure 1c). These positions are not conserved between both proteins (Figure 1b,c). Indeed, orthologous rBAT proteins have different N-glycosylation patterns, as reflected by the structure of ovine rBAT, which has six N-glycosylation sites, of which only three are shared with the human protein [34]. Unexpectedly, rBAT N-glycosylation in Asn575 is the only one that is needed for the biogenesis of the heterodimer [53]. However, the same glycosylation in the Asn575 residue is not conserved in the ovine (non-glycosylated) as in human (glycosylated) rBAT-ED [34]. N-glycosylation is also observed in some α-amylases of mammals [59], plants [60], and even fungi [61]. N-glycosylation in these enzymes is related to stability and also serves as a modulators of the enzymatic activity, but no structural pattern has been found between them [62].
The structures of 4F2hc-ED produced in E. coli (without N-glycosylations) or in mammalian cells (N-glycosilated), do not show large differences, even near the glycosylation sites, thus indicating that N-glycosilations in 4F2hc are not needed for proper protein folding. Moreover, 4F2hc-ED produced in E. coli shows high stability [63] and even confers stability to LAT2 when it is solubilized in detergent [14].
4. Sequence and Structure Relationship of rBAT-ED and 4F2-ED with α-Glucosidases
The rBAT-ED (residues 117–685) shares around 40% sequence identity (SI) with maltase-1 isoform-X3 from Cryptotermes secundus and 4F2hc-ED (residues 219–631) shares 26% SI with α-amylase of Pseudomonas indica (α-amylases with higher scores in BLAST-P against nr-database). The structure conservation between these ectodomains and α-amylases is even higher, as can be seen superimposing the structures of rBAT-ED and of the α-glucosidase Cqm1 (PDB ID 6K5P) [64],which that show only a difference of 1.06 Å2 RMSD, sharing 40% SI. Indeed, the crystal structure of 4F2-ED was solved by molecular replacement using as a model an oligo-1,6-glucosidase structure (PDB-ID 1UOK), sharing above only 25% sequence identity [5]. Detailed sequence evolutionary studies supports a greater bigger distance of 4F2hc-ED from an ancient glucosidase-like protein closer to rBAT and members of the GH13 family [4].
Importantly, most of the defining features of the α-amylase family are present in 4F2hc-ED and especially in rBAT-ED. The EDs of both proteins have a (β/α)8-barrel fold in domain A followed by a domain C with a β-sandwich fold (Figure 1b,c). rBAT-ED also has a domain B similar to that of oligo-1,6-glucosidases [40] (Figure 1c,d). In addition, rBAT-ED conserves the common catalytic site formed by a β4-strand aspartate (nucleophile), a β5-strand glutamate (proton donor) and a β7-strand aspartate (transition-state stabilizer) in positions Asp314, Glu384 and Asp449 (Figure 3d), as can be seen in comparison with the structures of the catalytic site of Halomonas sp. H11 α-glucosidase (HaG) complexed with maltose and oligo-1,6-glucosidase from B. cereus (Figure 3d). Moreover, other conserved important residues for fixing the glucose residue at subsite −1, such as His105,Asp62 and Asp333 in HaG, are conserved, whereas Arg200 or His332 (in HaG) are not conserved in human rBAT-ED (Figure 3d). Phe297 in HaG, which is also not conserved in rBAT-ED, has been related to the recognition of the nonreducing-end glucosyl residue [65]. Finally, the location of Ala315 in rBAT-ED (not shown in Figure 3d) is hypothesized to be compatible with the specific hydrolysis of α-1,4-glycosidic bonds [66].
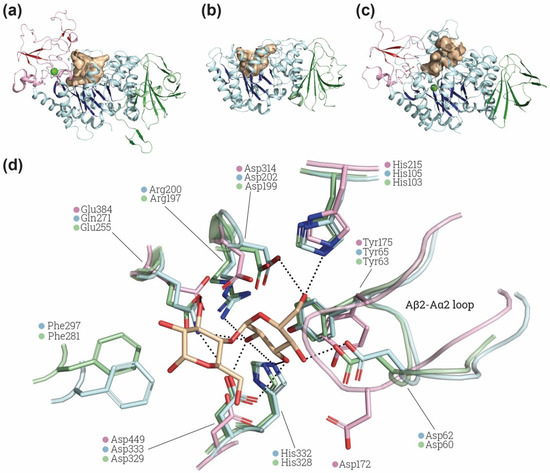
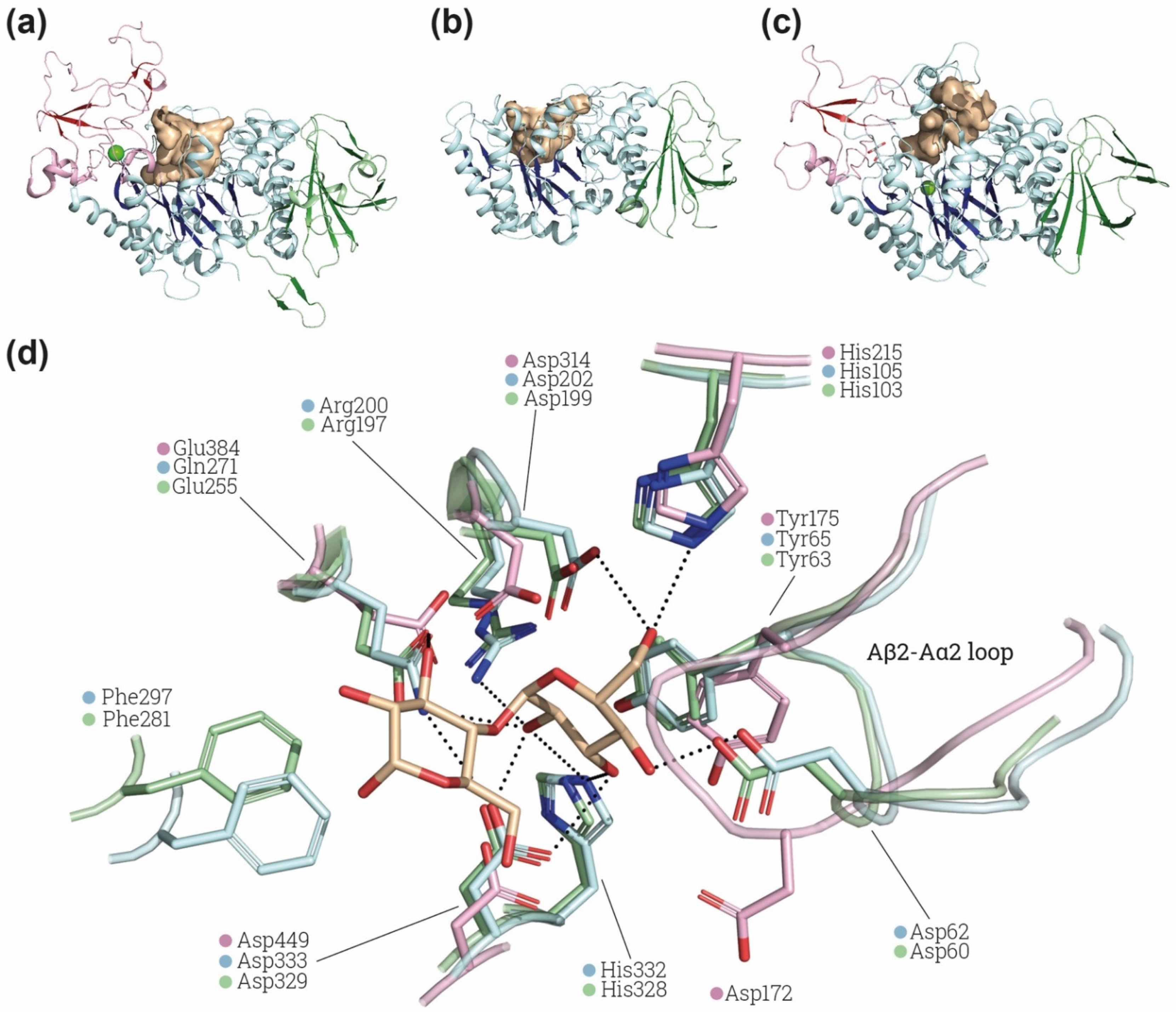
Figure 3.
α-glucosidase active site comparison. Structural comparison of the substrate cavity between rBAT-ED (PDB: 6LI9 [9]) (a), 4F2hc-ED (PDB: 6IRT [37]) (b) and Halomonas sp. H11 α-glucosidase (HaG) (PDB: 3WY4 [65]) (c). The cavities of all the structures are shown in light brown and domains A, B and C in blue, red and green, respectively, β-sheets are more brightly colored. Ions (Ca2+ and Mg2+ for rBAT and HaG respectively) are indicated in light green. Cavity area and volume were also measured using the Computed Atlas of Surface Topography of proteins (CASTp) plugin [67] on PyMol. Calculated areas for cavities are 633.32 Å2, 802.78 Å2 and 802.78 Å2 for 4F2hc, rBAT and HaG, respectively. (d) Detailed view of HaG (light blue) (PDB: 3WY4) and oligo-1,6-glucosidase of B. cereus (light green) (PDB: 1UOK [68]) binding pocket and the conserved residues found in rBAT (light pink) (PDB: 6LI9). Maltose is labelled in light brown. Non-conserved residues of rBAT are not shown. Images were created with the PyMol Molecular Graphics System, Version 2.1.4., Schrödinger, LLC.
Interestingly, the loop Aβ2-Aα2 in rBAT, where is located Asp62 in HaG, has a conformation not compatible with the position of maltose in the binding site of HaG (Figure 3d). The conformation of this loop is similar in structures of HaG with or without substrates (PDB: 3WY4 and 3WY1, respectively) and even in oligo-1,6-glucosidase of B. cereus (Figure 3d), which has different substrate specificity. In contrast, all the available structures of rBAT (Table 1) present the displaced conformation in the Aβ2-Aα2 shown in Figure 3d, suggesting a structural difference in the site of rBAT with respect to α-glucosidases and oligo-1,6-glicosidases. Furthermore, the catalytic triad is not fully conserved in all the putative rBAT orthologues [4]. Indeed, other conserved sequence regions (CSR) are more conserved than these catalytic residues among the vertebrate rBAT homologues [4]. The fact that these key residues are not preserved by negative selection suggests that the α-amylase catalytic activity is not important for rBAT function, at least in some vertebrates species [6]. All these differences might reflect changes in substrate recognition and specificity in rBAT-ED that can even lead to a total lack of α-glucosidase or oligo-1,6-glucosidase activity. Computational studies of molecular dynamics, docking and binding with different glycosidic substrates would be pertinent to predict whether this special conformation in the loop Aβ2-Aα2 can allow some flexibility or other substrates to bind.
Finally, the catalytic cleft in 4Fhc-ED and rBAT-ED is similar to that present in α-glucosidase HaG (Figure 3a–c). As expected, in 4F2hc, the accessibility of the cleft is higher because of the lack of domain B above domain A. The presence of a similar cavity suggests that some α-glucosidase-related function could be retained by these ectodomains, with or without catalytic activity.
5. Efforts to Prove Any rBAT-ED and 4F2-ED Glucosidase-Related Activity
Despite the strong evidence of the sequence and structural homology of the EDs of rBAT and 4F2hc with α-amylases, no α-glucosidase activity has been demonstrated for either protein [5,33]. In 4F2hc-ED, the three catalytic amino acids (Asp206, Glu230 and Asp297 in Taka amylase A), common in all the α-amylase family, are missing. Therefore, 4F2hc-ED is not expected to have α-glucosidase activity. In this regard, null detection of activity was found in a detailed screening using d-glucose, d-galactose or d-mannose derivatives of 4-methylumbelliferone as substrates [5]. In all, these results support the notion that 4F2hc-ED does not have catalytic α-glucosidase activity but might still retain some glycosidase-like binding activity, as is shown for some other α-amylase family members, such as Chitinase-like protein 3 from GH18 [69] and Edem1 (ER degradation-enhancing α-mannosidase-like protein 1) from the GH47 family [70]. Despite this, studies of carbohydrate binding with commercial glycochips from Glycominds (48 different glycans) and from the Glycomics Consortium (260 different glycans) failed to detect carbohydrate binding by human 4F2hc-ED [5]. It has also been proposed that some proteins or glycoproteins bind to the wide cleft of 4F2hc-ED, especially Ig-like containing proteins because of the homology with tendamistat, an α-amylase inhibitor that interacts with the catalytic cleft [71]. Moreover, several proteins with Ig-like domains, such as galectin-3 [72], ICAM-1 [73], CD-147 [74] and CEA-CAM-1 [75], have been proposed to interact with 4F2hc-ED. However, no structural information for these complexes has been published to date.
In contrast to 4F2hc-ED, rBAT-ED conserves the three catalytic residues of the α-amylase family (Figure 3d). Nevertheless, heterodimer rBAT-b0,+AT solubilized in detergent (glyco diosgenin 0.02%) failed to demonstrate α-glucosidase activity [9]. Studies with wider range of substrate species should be conducted in order to disregard any other GH13-related activity. To the best of our knowledge, no other functions for rBAT-ED, such as binding to glycans, or any other in relation to the α-amylase structure, have been proposed or tested in rBAT so far.
6. Conclusions
In conclusion, 4F2hc and rBAT, the heavy subunits of heteromeric amino acid transporters, have an ED with sequence and structure homology with α-glucosidases that can be classified as members of the GH13 family.
rBAT-ED and 4F2hc-ED have a (β/α)8 barrel as a domain A followed by domain C with a β-sandwich fold. In addition, rBAT has also domain B protruding from domain A and formed by two different loops, Aβ3-Aα3 and Aβ4-Aα4. This domain B is stabilized by a disulfide bridge and a Ca2+ ion, similarly to in α-amylases. This domain B is key for the functional oligomeric state of rBAT-b0,+AT as superdimers.
The characteristic active site of α-amylases, including key catalytic residues, is fully conserved in rBAT-ED and not in 4F2hc-ED. However, α-glucosidase-related activity (catalytic or lectin-like) has not yet been proven for any of these two heavy subunits. Further experiments are needed to unravel the physiological role of the homology of rBAT-ED and 4F2hc-ED with α-amylase family members.
Author Contributions
J.F., writing—original draft preparation; J.F. and M.P., writing—review and editing; A.N.-A., table and figure preparation; M.P., funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
Recent work in amino acid transporters in our lab has been supported by the Spanish Ministry of Science, Innovation and Universities (MSIU) (SAF2015-64869-R-FEDER, RTI2018-094211-B-I00), and the Fundació La Marató-TV3 and La Caixa Health Research (LCF/PR/HR20/52400017). We gratefully acknowledge institutional funding from MSIU to IRB Barcelona with the Centres of Excellence Severo Ochoa Award.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Palacín, M.; Nunes, V.; Font-Llitjós, M.; Jiménez-Vidal, M.; Fort, J.; Gasol, E.; Pineda, M.; Feliubadaló, L.; Chillarón, J.; Zorzano, A. The genetics of heteromeric amino acid transporters. Physiology 2005, 20, 112–124. [Google Scholar] [CrossRef]
- Chillarón, J.; Roca, R.; Valencia, A.; Zorzano, A.; Palacín, M. Heteromeric Amino Acid Transporters: Biochemistry, Genetics, and Physiology. Am. J. Physiol. Ren. Physiol. 2001, 281, F995–F1018. [Google Scholar] [CrossRef]
- Fotiadis, D.; Kanai, Y.; Palacín, M. The SLC3 and SLC7 Families of Amino Acid Transporters. Mol. Asp. Med. 2013, 34, 139–158. [Google Scholar] [CrossRef]
- Gabrisko, M.; Janecek, S. Looking for the Ancestry of the Heavy-Chain Subunits of Heteromeric Amino Acid Transporters RBAT and 4F2hc within the GH13 Alpha-Amylase Family. FEBS J. 2009, 276, 7265–7278. [Google Scholar] [CrossRef] [PubMed]
- Fort, J.; de la Ballina, L.R.; Burghardt, H.E.; Ferrer-Costa, C.; Turnay, J.; Ferrer-Orta, C.; Usón, I.; Zorzano, A.; Fernández-Recio, J.; Orozco, M.; et al. The Structure of Human 4F2hc Ectodomain Provides a Model for Homodimerization and Electrostatic Interaction with Plasma Membrane. J. Biol. Chem. 2007, 282, 31444–31452. [Google Scholar] [CrossRef] [PubMed]
- Janeček, Š.; Gabriško, M. Remarkable Evolutionary Relatedness among the Enzymes and Proteins from the α-Amylase Family. Cell. Mol. Life Sci. 2016, 73, 2707–2725. [Google Scholar] [CrossRef]
- Yan, R.; Zhou, J.; Li, Y.; Lei, J.; Zhou, Q. Structural Insight into the Substrate Recognition and Transport Mechanism of the Human LAT2-4F2hc Complex. Cell Discov. 2020, 6, 82. [Google Scholar] [CrossRef]
- Yan, R.; Li, Y.; Müller, J.; Zhang, Y.; Singer, S.; Xia, L.; Zhong, X.; Gertsch, J.; Altmann, K.H.; Zhou, Q. Mechanism of Substrate Transport and Inhibition of the Human LAT1-4F2hc Amino Acid Transporter. Cell Discov. 2021, 7, 16. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Li, Y.; Shi, Y.; Zhou, J.; Lei, J.; Huang, J.; Zhou, Q. Cryo-EM Structure of the Human Heteromeric Amino Acid Transporter b0,+AT-RBAT. Sci. Adv. 2020, 6, eaay6379. [Google Scholar] [CrossRef] [PubMed]
- Palacín, M.; Kanai, Y. The Ancillary Proteins of HATs: SLC3 Family of Amino Acid Transporters. Pflug. Arch. Eur. J. Physiol. 2004, 447, 490–494. [Google Scholar] [CrossRef]
- Verrey, F.; Closs, E.I.; Wagner, C.A.; Palacin, M.; Endou, H.; Kanai, Y. CATs and HATs: The SLC7 Family of Amino Acid Transporters. Pflug. Arch. Eur. J. Physiol. 2004, 447, 532–542. [Google Scholar] [CrossRef]
- Kantipudi, S.; Jeckelmann, J.-M.; Ucurum, Z.; Bosshart, P.D.; Fotiadis, D. The Heavy Chain 4F2hc Modulates the Substrate Affinity and Specificity of the Light Chains LAT1 and LAT2. Int. J. Mol. Sci. 2020, 21, 7573. [Google Scholar] [CrossRef] [PubMed]
- Bröer, A.; Friedrich, B.; Wagner, C.A.; Fillon, S.; Ganapathy, V.; Lang, F.; Bröer, S. Association of 4F2hc with Light Chains LAT1, LAT2 or Y+LAT2 Requires Different Domains. Biochem. J. 2001, 355, 725–731. [Google Scholar] [CrossRef]
- Rosell, A.; Meury, M.; Álvarez-Marimon, E.; Costa, M.; Pérez-Cano, L.; Zorzano, A.; Fernández-Recio, J.; Palacín, M.; Fotiadis, D. Structural Bases for the Interaction and Stabilization of the Human Amino Acid Transporter LAT2 with Its Ancillary Protein 4F2hc. Proc. Natl. Acad. Sci. USA 2014, 111, 2966–2971. [Google Scholar] [CrossRef] [PubMed]
- Cantor, J.; Browne, C.D.; Ruppert, R.; Féral, C.C.; Fässler, R.; Rickert, R.C.; Ginsberg, M.H. CD98hc Facilitates B Cell Proliferation and Adaptive Humoral Immunity. Nat. Immunol. 2009, 10, 412–419. [Google Scholar] [CrossRef]
- Fogelstrand, P.; Féral, C.C.; Zargham, R.; Ginsberg, M.H. Dependence of Proliferative Vascular Smooth Muscle Cells on CD98hc (4F2hc, SLC3A2). J. Exp. Med. 2009, 206, 2397–2406. [Google Scholar] [CrossRef]
- Cano-Crespo, S.; Chillarón, J.; Junza, A.; Fernández-Miranda, G.; García, J.; Polte, C.; de la Ballina, R.L.; Ignatova, Z.; Yanes, Ó.; Zorzano, A.; et al. CD98hc (SLC3A2) Sustains Amino Acid and Nucleotide Availability for Cell Cycle Progression. Sci. Rep. 2019, 9, 1–19. [Google Scholar] [CrossRef]
- del Amo, E.M.; Urtti, A.; Yliperttula, M. Pharmacokinetic Role of L-Type Amino Acid Transporters LAT1 and LAT2. Eur. J. Pharm. Sci. 2008, 35, 161–174. [Google Scholar] [CrossRef]
- Savaskan, N.E.; Eyüpoglu, I.Y. XCT Modulation in Gliomas: Relevance to Energy Metabolism and Tumor Microenvironment Normalization. Ann. Anat. 2010, 192, 309–313. [Google Scholar] [CrossRef]
- Maimaiti, M.; Sakamoto, S.; Sugiura, M.; Kanesaka, M.; Fujimoto, A.; Matsusaka, K.; Xu, M.; Ando, K.; Saito, S.; Wakai, K.; et al. The Heavy Chain of 4F2 Antigen Promote Prostate Cancer Progression via SKP-2. Sci. Rep. 2021, 11, 1–14. [Google Scholar] [CrossRef]
- Haynes, B.F. Human T Lymphocyte Antigens as Defined by Monoclonal Antibodies. Immunol. Rev. 1981, 57, 127–161. [Google Scholar] [CrossRef] [PubMed]
- Feral, C.C.; Nishiya, N.; Fenczik, C.A.; Stuhlmann, H.; Slepak, M.; Ginsberg, M.H. CD98hc (SLC3A2) Mediates Integrin Signaling. Proc. Natl. Acad. Sci. USA 2005, 102, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Takesono, A.; Moger, J.; Faroq, S.; Cartwright, E.; Dawid, I.B.; Wilson, S.W.; Kudoh, T. Solute Carrier Family 3 Member 2 (Slc3a2) Controls Yolk Syncytial Layer (YSL) Formation by Regulating Microtubule Networks in the Zebrafish Embryo. Proc. Natl. Acad. Sci. USA 2012, 109, 3371–3376. [Google Scholar] [CrossRef] [PubMed]
- Fenczik, C.; Zent, R.; Dellos, M.; Calderwood, D.; Satriano, J.; Kelly, C.; Ginsberg, M. Distinct Domains of CD98hc Regulate Integrins and Amino Acid Transport. J. Biol. Chem. 2001, 276, 8746–8752. [Google Scholar] [CrossRef] [PubMed]
- Zent, R.; Fenczik, C.A.; Calderwood, D.A.; Liu, S.; Dellos, M.; Ginsberg, M.H. Class- and Splice Variant-Specific Association of CD98 with Integrin Beta Cytoplasmic Domains. J. Biol. Chem. 2000, 275, 5059–5064. [Google Scholar] [CrossRef]
- Poettler, M.; Unseld, M.; Braemswig, K.; Haitel, A.; Zielinski, C.C.; Prager, G.W. CD98hc (SLC3A2) Drives Integrin-Dependent Renal Cancer Cell Behavior. Mol. Cancer 2013, 12, 1–12. [Google Scholar] [CrossRef]
- Hara, K.; Kudoh, H.; Enomoto, T.; Hashimoto, Y.; Masuko, T. Enhanced Tumorigenicity Caused by Truncation of the Extracellular Domain of GP125/CD98 Heavy Chain. Oncogene 2000, 19, 6209–6215. [Google Scholar] [CrossRef]
- Dalton, P.; Christian, H.C.; Redman, C.W.; Sargent, I.L.; Boyd, C.A. Membrane Trafficking of CD98 and Its Ligand Galectin 3 in BeWo Cells—Implication for Placental Cell Fusion. FEBS J. 2007, 274, 2715–2727. [Google Scholar] [CrossRef]
- Bröer, S.; Palacín, M. The Role of Amino Acid Transporters in Inherited and Acquired Diseases. Biochem. J. 2011, 436, 193–211. [Google Scholar] [CrossRef] [PubMed]
- Calonge, M.J.; Gasparini, P.; Chillarón, J.; Chillón, M.; Gallucci, M.; Rousaud, F.; Zelante, L.; Testar, X.; Dallapiccola, B.; di Silverio, F. Cystinuria Caused by Mutations in RBAT, a Gene Involved in the Transport of Cystine. Nat. Genet. 1994, 6, 420–425. [Google Scholar] [CrossRef]
- Bartoccioni, P.; Rius, M.; Zorzano, A.; Palacín, M.; Chillarón, J. Distinct Classes of Trafficking RBAT Mutants Cause the Type I Cystinuria Phenotype. Hum. Mol. Genet. 2008, 17, 1845–1854. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fernández, E.; Jiménez-Vidal, M.; Calvo, M.; Zorzano, A.; Tebar, F.; Palacín, M.; Chillarón, J. The Structural and Functional Units of Heteromeric Amino Acid Transporters. The Heavy Subunit RBAT Dictates Oligomerization of the Heteromeric Amino Acid Transporters. J. Biol. Chem. 2006, 281, 26552–26561. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Grund, T.N.; Welsch, S.; Mills, D.J.; Michel, M.; Safarian, S.; Michel, H. Structural Basis for Amino Acid Exchange by a Human Heteromeric Amino Acid Transporter. Proc. Natl. Acad. Sci. USA 2020, 117, 21281–21287. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Wiriyasermkul, P.; Moriyama, S.; Mills, D.J.; Kühlbrandt, W.; Nagamori, S. Ca2+-Mediated Higher-Order Assembly of b0,+AT–RBAT Is a Key Step for System b0,+ Biogenesis and Cystinuria. bioRxiv 2021. [Google Scholar] [CrossRef]
- Deuschle, F.C.; Morath, V.; Schiefner, A.; Brandt, C.; Ballke, S.; Reder, S.; Steiger, K.; Schwaiger, M.; Weber, W.; Skerra, A. Development of a High Affinity Anticalin ® Directed against Human CD98hc for Theranostic Applications. Theranostics 2020, 10, 2172–2187. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Wiriyasermkul, P.; Jin, C.; Quan, L.; Ohgaki, R.; Okuda, S.; Kusakizako, T.; Nishizawa, T.; Oda, K.; Ishitani, R.; et al. Cryo-EM Structure of the Human L-Type Amino Acid Transporter 1 in Complex with Glycoprotein CD98hc. Nat. Struct. Mol. Biol. 2019, 26, 510–517. [Google Scholar] [CrossRef]
- Yan, R.; Zhao, X.; Lei, J.; Zhou, Q. Structure of the Human LAT1–4F2hc Heteromeric Amino Acid Transporter Complex. Nature 2019, 568, 127–130. [Google Scholar] [CrossRef]
- Parker, J.L.; Deme, J.C.; Kolokouris, D.; Kuteyi, G.; Biggin, P.C.; Lea, S.M.; Newstead, S. Molecular Basis for Redox Control by the Human Cystine/Glutamate Antiporter System Xc−. bioRxiv 2021. [Google Scholar] [CrossRef]
- Oda, K.; Lee, Y.; Wiriyasermkul, P.; Tanaka, Y.; Takemoto, M.; Yamashita, K.; Nagamori, S.; Nishizawa, T.; Nureki, O. Consensus Mutagenesis Approach Improves the Thermal Stability of System Xc− Transporter, XCT, and Enables Cryo-EM Analyses. Protein Sci. 2020, 29, 2398–2407. [Google Scholar] [CrossRef]
- Janecek, S.; Svensson, B.; Henrissat, B. Domain Evolution in the Alpha-Amylase Family. J. Mol. Evol. 1997, 45, 322–331. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The Carbohydrate-Active Enzymes Database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Svensson, B. Protein Engineering in the α-Amylase Family: Catalytic Mechanism, Substrate Specificity, and Stability. Plant Mol. Biol. 1994, 25, 141–157. [Google Scholar] [CrossRef]
- Janeček, Š. How many conserved sequence regions are there in the alpha-amylase family? Biologia 2002, 57 (Suppl. S11), 29–41. [Google Scholar]
- Kuriki, T.; Imanaka, T. The Concept of the Alpha-Amylase Family: Structural Similarity and Common Catalytic Mechanism. J. Biosci. Bioeng. 1999, 87, 557–565. [Google Scholar] [CrossRef]
- MacGregor, E.A.; Janecek, S.; Svensson, B. Relationship of Sequence and Structure to Specificity in the Alpha-Amylase Family of Enzymes. Biochim. Biophys. Acta 2001, 1546, 1–20. [Google Scholar] [CrossRef]
- Seo, E.S.; Christiansen, C.; Abou Hachem, M.; Nielsen, M.M.; Fukuda, K.; Bozonnet, S.; Blennow, A.; Aghajari, N.; Haser, R.; Svensson, B. An Enzyme Family Reunion—Similarities, Differences and Eccentricities in Actions on α-Glucans. Biologia 2008, 63, 967–979. [Google Scholar] [CrossRef]
- MacGregor, E.A. An Overview of Clan GH-H and Distantly-Related Families. Biologia 2005, 60, 5–12. [Google Scholar]
- van der Maarel, M.J.; van der Veen, B.; Uitdehaag, J.C.; Leemhuis, H.; Dijkhuizen, L. Properties and Applications of Starch-Converting Enzymes of the Alpha-Amylase Family. J. Biotechnol. 2002, 94, 137–155. [Google Scholar] [CrossRef]
- Kuriki, T.; Takata, H.; Yanase, M.; Ohdan, K.; Fujii, K.; Terada, Y.; Takaha, T.; Hondoh, H.; Matsuura, Y.; Imanaka, T. The Concept of the ALPHA-Amylase Family: A Rational Tool for Interconverting Glucanohydrolases/Glucanotransferases, and Their Specificities. J. Appl. Glycosci. 2006, 53, 155–161. [Google Scholar] [CrossRef][Green Version]
- Majzlová, K.; Pukajová, Z.; Janeček, S. Tracing the Evolution of the α-Amylase Subfamily GH13_36 Covering the Amylolytic Enzymes Intermediate between Oligo-1,6-Glucosidases and Neopullulanases. Carbohydr. Res. 2013, 367, 48–57. [Google Scholar] [CrossRef]
- Stam, M.R.; Danchin, E.G.J.; Rancurel, C.; Coutinho, P.M.; Henrissat, B. Dividing the Large Glycoside Hydrolase Family 13 into Subfamilies: Towards Improved Functional Annotations of α-Amylase-Related Proteins. Protein Eng. Des. Sel. 2006, 19, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Nicolàs-Aragó, A.; Fort, J.; Palacín, M.; Errasti-Murugarren, E. Rush Hour of LATs towards Their Transport Cycle. Membranes 2021, 11, 602. [Google Scholar] [CrossRef] [PubMed]
- Rius, M.; Sala, L.; Chillarón, J. The Role of N-Glycans and the C-Terminal Loop of the Subunit RBAT in the Biogenesis of the Cystinuria-Associated Transporter. Biochem. J. 2016, 473, 233–244. [Google Scholar] [CrossRef]
- Rius, M.; Chillarón, J. Carrier Subunit of Plasma Membrane Transporter Is Required for Oxidative Folding of Its Helper Subunit. J. Biol. Chem. 2012, 287, 18190–18200. [Google Scholar] [CrossRef] [PubMed]
- Declerck, N.; Machius, M.; Joyet, P.; Wiegand, G.; Huber, R.; Gaillardin, C. Hyperthermostabilization of Bacillus Licheniformis Alpha-Amylase and Modulation of Its Stability over a 50 Degrees C Temperature Range. Protein Eng. 2003, 16, 287–293. [Google Scholar] [CrossRef]
- Torrance, J.W.; Macarthur, M.W.; Thornton, J.M. Evolution of Binding Sites for Zinc and Calcium Ions Playing Structural Roles. Proteins 2008, 71, 813–830. [Google Scholar] [CrossRef]
- Chai, K.P.; Othman, N.F.; Teh, A.H.; Ho, K.L.; Chan, K.G.; Shamsir, M.S.; Goh, K.M.; Ng, C.L. Crystal Structure of Anoxybacillus α-Amylase Provides Insights into Maltose Binding of a New Glycosyl Hydrolase Subclass. Sci. Rep. 2016, 6, 23126. [Google Scholar] [CrossRef]
- Pineda, M.; Wagner, C.A.; Bröer, A.; Stehberger, P.A.; Kaltenbach, S.; Gelpí, J.L.; Martín Del Río, R.; Zorzano, A.; Palacín, M.; Lang, F.; et al. Cystinuria-Specific RBAT(R365W) Mutation Reveals Two Translocation Pathways in the Amino Acid Transporter RBAT-b0,+AT. Biochem. J. 2004, 377, 665–674. [Google Scholar] [CrossRef]
- Takashima, S.; Amano, J.; Takashima, S.; Amano, J. Glycosylation and Secretion of Human α-Amylases. Adv. Biol. Chem. 2012, 2, 10–19. [Google Scholar] [CrossRef][Green Version]
- Ochiai, A.; Sugai, H.; Harada, K.; Tanaka, S.; Ishiyama, Y.; Ito, K.; Tanaka, T.; Uchiumi, T.; Taniguchi, M.; Mitsui, T. Crystal Structure of α-Amylase from Oryza sativa: Molecular Insights into Enzyme Activity and Thermostability. OUP 2014, 78, 989–997. [Google Scholar] [CrossRef]
- Eriksen, S.H.; Jensen, B.; Olsen, J. Effect of N-Linked Glycosylation on Secretion, Activity, and Stability of Alpha-Amylase from Aspergillus oryzae. Curr. Microbiol. 1998, 37, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.; Xu, B.; Bai, Y.; Luo, H.; Ma, R.; Shi, P.; Yao, B. Role of N-Linked Glycosylation in the Enzymatic Properties of a Thermophilic GH 10 Xylanase from Aspergillus fumigatus Expressed in Pichia pastoris. PLoS ONE 2017, 12, e0171111. [Google Scholar] [CrossRef] [PubMed]
- Turnay, J.; Fort, J.; Olmo, N.; Santiago-Gómez, A.; Palacín, M.; Lizarbe, M.A.M.A. Structural Characterization and Unfolding Mechanism of Human 4F2hc Ectodomain. Biochim. Biophys. Acta Proteins Proteom. 2011, 1814, 536–544. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Kumar, V. Crystal Structure of BinAB Toxin Receptor (Cqm1) Protein and Molecular Dynamics Simulations Reveal the Role of Unique Ca(II) Ion. Int. J. Biol. Macromol. 2019, 140, 1315–1325. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Saburi, W.; Gai, Z.; Kato, K.; Ojima-Kato, T.; Yu, J.; Komoda, K.; Kido, Y.; Matsui, H.; Mori, H.; et al. Structural Analysis of the α-Glucosidase HaG Provides New Insights into Substrate Specificity and Catalytic Mechanism. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 1382–1391. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Miyake, H.; Kusunoki, M.; Osaki, S. Steric Hindrance by 2 Amino Acid Residues Determines the Substrate Specificity of Isomaltase from Saccharomyces cerevisiae. J. Biosci. Bioeng. 2011, 112, 545–550. [Google Scholar] [CrossRef]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed Atlas of Surface Topography of Proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef]
- Watanabe, K.; Hata, Y.; Kizaki, H.; Katsube, Y.; Suzuki, Y. The Refined Crystal Structure of Bacillus cereus Oligo-1,6-Glucosidase at 2.0 Å Resolution: Structural Characterization of Proline-Substitution Sites for Protein Thermostabilization. J. Mol. Biol. 1997, 269, 142–153. [Google Scholar] [CrossRef]
- Chang, N.C.; Hung, S.I.; Hwa, K.Y.; Kato, I.; Chen, J.E.; Liu, C.H.; Chang, A.C. Macrophage Protein, Ym1, Transiently Expressed during Inflammation Is a Novel Mammalian Lectin. J. Biol. Chem. 2001, 276, 17497–17506. [Google Scholar] [CrossRef]
- Hosokawa, N.; Wada, I.; Hasegawa, K.; Yorihuzi, T.; Tremblay, L.O.; Herscovics, A.; Nagata, K. A Novel ER α-Mannosidase-like Protein Accelerates ER-Associated Degradation. EMBO Rep. 2001, 2, 415. [Google Scholar] [CrossRef]
- Wiegand, G.; Epp, O.; Huber, R. The Crystal Structure of Porcine Pancreatic Alpha-Amylase in Complex with the Microbial Inhibitor Tendamistat. J. Mol. Biol. 1995, 247, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Hughes, R.C. Macrophage Surface Glycoproteins Binding to Galectin-3 (Mac-2-Antigen). Glycoconj. J. 1997, 14, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Charrier, L.; Gewirtz, A.; Sitaraman, S.; Merlin, D. CD98 and Intracellular Adhesion Molecule I Regulate the Activity of Amino Acid Transporter LAT-2 in Polarized Intestinal Epithelia. J. Biol. Chem. 2003, 278, 23672–23677. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Hemler, M.E. Metabolic Activation-Related CD147-CD98 Complex. Mol. Cell. Proteom. 2005, 4, 1061–1071. [Google Scholar] [CrossRef] [PubMed]
- Kakugawa, K.; Hattori, M.; Beauchemin, N.; Minato, N. Activation of CEA-CAM-1-Mediated Cell Adhesion via CD98: Involvement of PKCdelta. FEBS Lett. 2003, 552, 184–188. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).