Rapid Identification between Two Fish Species Using UV-Vis Spectroscopy for Substitution Detection
Abstract
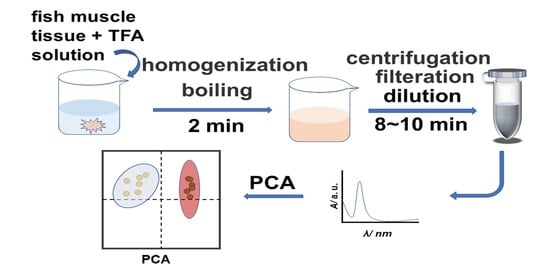
:1. Introduction
2. Results and Discussion
2.1. Impact of Dilution on Classification of UV-Vis Spectra of Different Fish Samples
2.2. Reproducibility of the Utilized Pretreatment Method
2.3. Testing the UV-Vis Technique in Fish Species Classification
2.4. Demonstration of Applicability of UV-Vis Spectroscopy in Fish Species Authenticity
2.5. Distinguishing of Fish Samples at Different Level
2.6. Classification of Fish Sample on the Market
2.7. Identification of Incorrectly Labeled Fish Sample for Substitution Detection
3. Materials and Methods
3.1. Chemicals and Fish Samples
3.2. Preparation and UV-Vis Detection of Extract of Fish Muscle Samples
3.3. Genetic Test of Fish Samples
3.4. Principal Component Analysis (PCA) of the UV-Vis Spectral Data
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- de Santana, F.B.; Borges Neto, W.; Poppi, R.J. Random forest as one-class classifier and infrared spectroscopy for food adulteration detection. Food Chem. 2019, 293, 323–332. [Google Scholar] [CrossRef]
- Amane, D.; Ananthanarayan, L. Detection of adulteration in black gram-based food products using DNA barcoding. Food Control 2019, 104, 193–200. [Google Scholar] [CrossRef]
- Wielogorska, E.; Chevallier, O.; Black, C.; Galvin-King, P.; Delêtre, M.; Kelleher, C.T.; Haughey, S.A.; Elliott, C.T. Development of a comprehensive analytical platform for the detection and quantitation of food fraud using a biomarker approach. The oregano adulteration case study. Food Chem. 2018, 239, 32–39. [Google Scholar] [CrossRef] [Green Version]
- Acutis, P.L.; Cambiotti, V.; Riina, M.V.; Meistro, S.; Maurella, C.; Massaro, M.; Stacchini, P.; Gili, S.; Malandra, R.; Pezzolato, M.; et al. Detection of fish species substitution frauds in Italy: A targeted National Monitoring Plan. Food Control 2019, 101, 151–155. [Google Scholar] [CrossRef]
- Al-Zahrani, F.A.M.; Abu Mellah, K.; El-Shishtawy, R.M.; Al-Soliemy, A.M.; Asiri, A.M. Synthesis and photophysical studies on a new fluorescent phenothiazine-based derivative. Luminescence 2020, 35, 608–617. [Google Scholar] [CrossRef] [PubMed]
- Puertas, G.; Vázquez, M. Fraud detection in hen housing system declared on the eggs’ label: An accuracy method based on UV-VIS-NIR spectroscopy and chemometrics. Food Chem. 2019, 288, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Hu, H.; Wu, T.; Tong, P.; Liu, B.; Zhu, B.; Du, Y. Rapid and sensitive determination of plasticizer diethylhexyl phthalate in drink by diffuse reflectance UV spectroscopy coupled with membrane filtration. Food Control 2014, 35, 218–222. [Google Scholar] [CrossRef]
- Bi, H.Y.; Fernandes, A.C.; Cardoso, S.; Freitas, P. Interference-blind microfluidic sensor for ascorbic acid determination by UV/vis spectroscopy. Sens. Actuators B 2016, 224, 668–675. [Google Scholar] [CrossRef] [Green Version]
- Bi, H.Y.; Duarte, C.M.; Brito, M.; Vilas-Boas, V.; Cardoso, S.; Freitas, P. Performance enhanced UV/vis spectroscopic microfluidic sensor for ascorbic acid quantification in human blood. Biosens. Bioelectron. 2016, 85, 568–572. [Google Scholar] [CrossRef]
- Alamprese, C.; Casale, M.; Sinelli, N.; Lanteri, S.; Casiraghi, E. Detection of minced beef adulteration with turkey meat by UV–vis, NIR and MIR spectroscopy. LWT-Food Sci. Technol. 2013, 53, 225–232. [Google Scholar] [CrossRef]
- El Darra, N.; Rajha, H.N.; Saleh, F.; Al-Oweini, R.; Maroun, R.G.; Louka, N. Food fraud detection in commercial pomegranate molasses syrups by UV–VIS spectroscopy, ATR-FTIR spectroscopy and HPLC methods. Food Control 2017, 78, 132–137. [Google Scholar] [CrossRef]
- Yang, L.; Wu, T.; Liu, Y.; Zou, J.; Huang, Y.M.; Babu, V.S.; Lin, L. Rapid Identification of Pork Adulterated in the Beef and Mutton by Infrared Spectroscopy. J. Spectrosc. 2018, 2018, 2413874. [Google Scholar] [CrossRef]
- Dong, D.; Zheng, W.; Jiao, L.; Lang, Y.; Zhao, X. Chinese vinegar classification via volatiles using long-optical-path infrared spectroscopy and chemometrics. Food Chem. 2016, 194, 95–100. [Google Scholar] [CrossRef]
- Ríos-Reina, R.; García-González, D.L.; Callejón, R.M.; Amigo, J.M. NIR spectroscopy and chemometrics for the typification of Spanish wine vinegars with a protected designation of origin. Food Control 2018, 89, 108–116. [Google Scholar] [CrossRef]
- Alves, F.; Coqueiro, A.; Março, P.H.; Valderrama, P. Evaluation of olive oils from the Mediterranean region by UV-Vis spectroscopy and Independent Component Analysis. Food Chem. 2019, 273, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Karunathilaka, S.R.; Farris, S.; Mossoba, M.M.; Moore, J.C.; Yakes, B.J. Characterising variances of milk powder and instrumentation for the development of a non-targeted, Raman spectroscopy and chemometrics detection method for the evaluation of authenticity. Food Addit. Contam. Part A 2016, 33, 921–932. [Google Scholar] [CrossRef] [Green Version]
- Ghidini, S.; Varrà, M.O.; Zanardi, E. Approaching Authenticity Issues in Fish and Seafood Products by Qualitative Spectroscopy and Chemometrics. Molecules 2019, 24, 1812. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.Y.; Hu, W.; Teng, J.; Peng, H.H.; Gan, J.H.; Wang, X.C.; Sun, S.Q.; Xu, C.H.; Liu, Y. Rapid recognition of marine fish surimi by one-step discriminant analysis based on near-infrared diffuse reflectance spectroscopy. Int. J. Food Prop. 2017, 20, 2932–2943. [Google Scholar] [CrossRef] [Green Version]
- Velioğlu, H.M.; Temiz, H.T.; Boyaci, I.H. Differentiation of fresh and frozen-thawed fish samples using Raman spectroscopy coupled with chemometric analysis. Food Chem. 2015, 172, 283–290. [Google Scholar] [CrossRef]
- Wang, C.; Li, X.; Zhao, X.; Bi, H.; Xie, J. The investigation of storage situation of fish muscle via the analysis of its exudate by MALDI-TOF MS. Food Chem 2021, 373, 131450. [Google Scholar] [CrossRef]
- Bi, H.; Zhong, C.; Shao, M.; Wang, C.; Yi, J.; Qiao, L.; Zhang, J. Differentiation and Authentication of Fishes at Species Level Through Analysis of Fish Skin by MALDI TOF MS. Rapid Commun. Mass Spectrom. 2019, 33, 1336–1343. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Bi, H. Direct identification of fish species by surface molecular transferring. Analyst 2020, 145, 4148–4155. [Google Scholar] [CrossRef]
- Wang, C.; Bi, H.; Xie, J. Visualization of the Distance among Fishes by MALDI MS for Rapid Determination of the Taxonomic Status of Fish Fillets. J. Agric. Food Chem. 2020, 68, 8438–8446. [Google Scholar] [CrossRef]
- Chengyu, W.; Hongyan, B. Super-fast seafood authenticity analysis by One-step pretreatment and comparison of mass spectral patterns. Food Control 2021, 123, 107751. [Google Scholar]
- Mazzeo, M.F.; Giulio, B.D.; Guerriero, G.; Ciarcia, G.; Malorni, A.; Russo, G.L.; Siciliano, R.A. Fish Authentication by MALDI-TOF Mass Spectrometry. J. Agric. Food Chem. 2008, 56, 11071–11076. [Google Scholar] [CrossRef] [PubMed]
- Abid, A.; Zhang, M.J.; Bagaria, V.K.; Zou, J. Exploring patterns enriched in a dataset with contrastive principal component analysis. Nat. Commun. 2018, 9, 2134. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.J.; Fu, M.Y.; Huang, M.H.; Cui, X.W.; Li, Y.; Cao, M.; Wang, L.B.; Xiong, X.H.; Xiong, X. Duplex real-time PCR combined with melting curve analysis for rapid detection of Atlantic salmon (Salmo salar) and rainbow trout (Oncorhynchus mykiss). J. Food Compos. Anal. 2021, 97, 103765. [Google Scholar] [CrossRef]
- Yu, X.; Li, L.Q.; Wang, H.H.; Song, G.S.; Wang, J.; Li, S.Y.; Wang, Y.; Shen, Q. Lipidomics study of rainbow trout (Oncorhynchus mykiss) and salmons (Oncorhynchus tshawytscha and Salmo salar) using hydrophilic interaction chromatography and mass spectrometry. LWT-Food Sci. Technol. 2020, 121, 108988. [Google Scholar] [CrossRef]
- Ardura, A.; Pola, I.G.; Ginuino, I.; Gomes, V.; Garcia-Vazquez, E. Application of barcoding to Amazonian commercial fish labelling. Food Res. Int. 2010, 43, 1549–1552. [Google Scholar] [CrossRef]
- Cline, E. Marketplace substitution of Atlantic salmon for Pacific salmon in Washington State detected by DNA barcoding. Food Res. Int. 2012, 45, 388–393. [Google Scholar] [CrossRef]
- Xing, B.; Zhang, Z.; Sun, R.; Wang, Y.; Lin, M.; Wang, C. Mini-DNA barcoding for the identification of commercial fish sold in the markets along the Taiwan Strait. Food Control 2020, 112, 107143. [Google Scholar] [CrossRef]
- Chen, C.; Ding, Y.; Jiang, Z.; Lu, C.; Zhang, L.; Chen, Z.; Zhu, C. DNA Barcoding of Yellow croakers (Larimichthys spp.) and Morphologically Similar Fish Species for Authentication. Food Control 2021, 127, 108087. [Google Scholar] [CrossRef]






| Run | LC | LP | SJ | SN | SS | OM | |
|---|---|---|---|---|---|---|---|
| 1 | λmax/nm | 249 ± 0.5 | 249 ± 0.5 | 248 ± 1.0 | 248 ± 1.0 | 249 ± 0.5 | 249 ± 0.5 |
| A/a. u. | 0.111 ± 0.003 | 0.110 ± 0.005 | 0.393 ± 0.005 | 0.361 ± 0.008 | 0.181 ± 0.005 | 0.204 ± 0.006 | |
| 2 | λmax/nm | 249 ± 0.5 | 249 ± 0.5 | 248 ± 1.0 | 248 ± 1.0 | 249 ± 0.5 | 249 ± 0.5 |
| A/a. u. | 0.109 ± 0.003 | 0.113 ± 0.004 | 0.396 ± 0.004 | 0.358 ± 0.008 | 0.176 ± 0.006 | 0.208 ± 0.007 | |
| 3 | λmax/nm | 249 ± 0.5 | 249 ± 0.5 | 248 ± 1.0 | 248 ± 1.0 | 249 ± 0.5 | 249 ± 0.5 |
| A/a. u. | 0.108 ± 0.006 | 0.113 ± 0.006 | 0.392 ± 0.006 | 0.368 ± 0.004 | 0.175 ± 0.005 | 0.203 ± 0.006 | |
| avg | λmax/nm | 249 ± 0.5 | 249 ± 0.5 | 248 ± 1.0 | 248 ± 1.0 | 249 ± 0.5 | 249 ± 0.5 |
| A/a. u. | 0.109 ± 0.006 | 0.112 ± 0.004 | 0.394 ± 0.005 | 0.362 ± 0.009 | 0.177 ± 0.006 | 0.205 ± 0.005 | |
| RSD (%) of A | 5.5 | 3.6 | 1.3 | 2.5 | 3.4 | 2.4 | |
| Fish Samples | Belonging to the Same | |
|---|---|---|
| Pampus argenteus (PaA) | Pseudaspius leptocephalus (PL) | class |
| Epinephelus rivulatus (ER) | Pagrosomus major (PM) | order |
| Scomberomorus niphonius (SN) | Scomber japonicus (SJ) | family |
| Larimichthys polyactis (LP) | Larimichthys crocea (LC) | genus |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chai, Z.; Wang, C.; Bi, H. Rapid Identification between Two Fish Species Using UV-Vis Spectroscopy for Substitution Detection. Molecules 2021, 26, 6529. https://doi.org/10.3390/molecules26216529
Chai Z, Wang C, Bi H. Rapid Identification between Two Fish Species Using UV-Vis Spectroscopy for Substitution Detection. Molecules. 2021; 26(21):6529. https://doi.org/10.3390/molecules26216529
Chicago/Turabian StyleChai, Zhaoliang, Chengyu Wang, and Hongyan Bi. 2021. "Rapid Identification between Two Fish Species Using UV-Vis Spectroscopy for Substitution Detection" Molecules 26, no. 21: 6529. https://doi.org/10.3390/molecules26216529
APA StyleChai, Z., Wang, C., & Bi, H. (2021). Rapid Identification between Two Fish Species Using UV-Vis Spectroscopy for Substitution Detection. Molecules, 26(21), 6529. https://doi.org/10.3390/molecules26216529