Biological Effects of Quinolones: A Family of Broad-Spectrum Antimicrobial Agents
Abstract
:1. Introduction
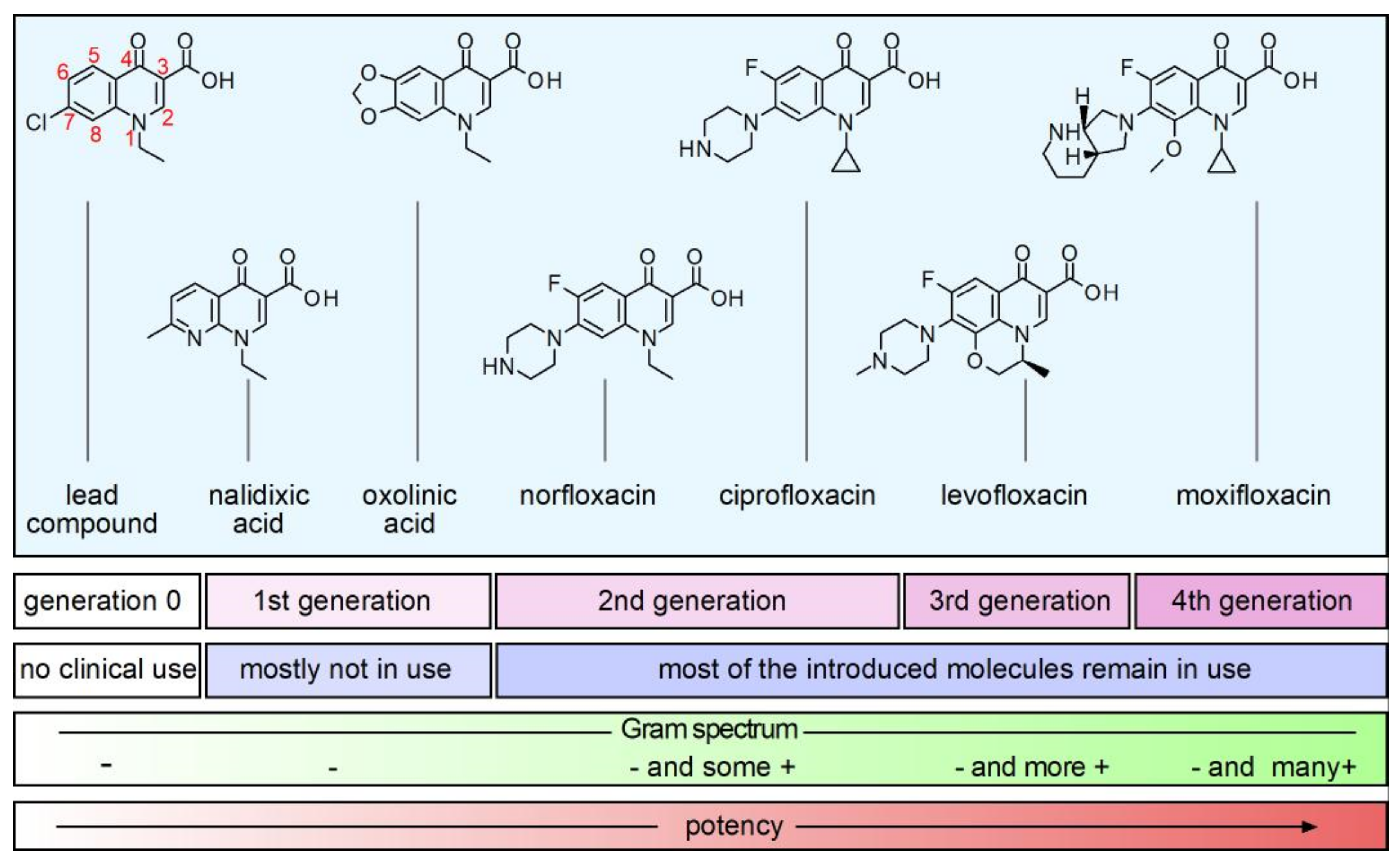
2. Structure and Anti-Bacterial Activity
3. Physicochemical Properties of Quinolones
4. Synthetic and Natural-Occurring Quinolones
5. Structure-Activity Relationship (SAR) of Quinolones
6. Other Biological Activities
7. Clinical Importance of Quinolones
8. Mechanism of Antimicrobial Activity of Quinolones
9. Mechanisms of Resistance to Quinolones
10. Veterinary Use of Quinolones and Animal Production
11. Impact of Quinolones in the Environment
12. Considerations for the Use of Quinolones: Human and Veterinary Medicine
13. Recent Finding about Quinolones
14. Future of Quinolones, Final remarks, and Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Price, J.R. Alkaloids of the Australian Rutaceae: Melicope fareana. I. Isolation of the Constituent Alkaloids. Aust. J. Chem. 1949, 2, 249–254. [Google Scholar] [CrossRef]
- Drummond, L.J.; Lahey, F.N. Alkaloids of the Australian Rutaceae: Acronychia baueri. III. The Structure of Acronycine. Aust. J. Chem. 1949, 2, 630–638. [Google Scholar] [CrossRef]
- Crow, W.D.; Price, J.R. Alkaloids of the Australian Rutaceae: Melicope fareana. V. The Structure of the Alkaloids. Aust. J. Chem. 1949, 2, 282–306. [Google Scholar] [CrossRef]
- Bisacchi, G.S. Origins of the Quinolone Class of Antibacterials: An Expanded “Discovery Story”. J. Med. Chem. 2015, 58, 4874–4882. [Google Scholar] [CrossRef] [Green Version]
- Mitscher, L.A. Bacterial Topoisomerase Inhibitors: Quinolone and Pyridone Antibacterial Agents. Chem. Rev. 2005, 105, 559–592. [Google Scholar] [CrossRef] [PubMed]
- Redgrave, L.S.; Sutton, S.B.; Webber, M.A.; Piddock, L.J.V. Fluoroquinolone Resistance: Mechanisms, Impact on Bacteria, and Role in Evolutionary Success. Trends Microbiol. 2014, 22, 438–445. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.I. Development of the Quinolones. J. Antimicrob. Chemother. 2003, 51, 1–11. [Google Scholar] [CrossRef]
- Mandell, L.; Tillotson, G. Safety of Fluoroquinolones: An Update. Can. J. Infect. Dis. 2002, 13, 54–61. [Google Scholar] [CrossRef] [Green Version]
- Nagaoka, S. (Ed.) Drug Discovery in Japan: Investigating the Sources of Innovation; Springer: Singapore, 2019; ISBN 9789811389054. [Google Scholar]
- Pavithra, B.H.; Prakash, N.; Jayakumar, K. PK-PD Modelling of Norfloxacin after Oral Administration in Rabbits. Vet. World 2010, 3, 546–548. [Google Scholar]
- Appelbaum, P.C. Quinolone Activity Against Anaerobes. Drugs 1999, 58, 60–64. [Google Scholar] [CrossRef]
- Prabhala, R.H.; Rao, B.; Marshall, R.; Bansal, M.B.; Thadepalli, H. In Vitro Susceptibility of Anaerobic Bacteria to Ciprofloxacin (Bay o 9867). Antimicrob. Agents Chemother. 1984, 26, 785–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldstein, E.J.; Citron, D.M. Comparative Activity of Ciprofloxacin, Ofloxacin, Sparfloxacin, Temafloxacin, CI-960, CI-990, and WIN 57273 against Anaerobic Bacteria. Antimicrob. Agents Chemother. 1992, 36, 1158–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naeem, A.; Badshah, S.L.; Muska, M.; Ahmad, N.; Khan, K. The Current Case of Quinolones: Synthetic Approaches and Antibacterial Activity. Molecules 2016, 21, 268. [Google Scholar] [CrossRef] [Green Version]
- King, D.E.; Malone, R.; Lilley, S.H. New Classification and Update on the Quinolone Antibiotics. Am. Fam. Physician 2000, 61, 2741–2748. [Google Scholar]
- Wexler, H.M.; Molitoris, E.; Reeves, D.; Finegold, S.M. In Vitro Activity of DU-6859a against Anaerobic Bacteria. Antimicrob. Agents Chemother. 1994, 38, 2504–2509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balwan, A.; Nicolau, D.P.; Wungwattana, M.; Zuckerman, J.B.; Waters, V. Clinafloxacin for Treatment of Burkholderia Cenocepacia Infection in a Cystic Fibrosis Patient. Antimicrob. Agents Chemother. 2015, 60, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Lauderdale, T.-L.; Shiau, Y.-R.; Lai, J.-F.; Chen, H.-C.; King, C.-H.R. Comparative In Vitro Activities of Nemonoxacin (TG-873870), a Novel Nonfluorinated Quinolone, and Other Quinolones against Clinical Isolates. Antimicrob. Agents Chemother. 2010, 54, 1338–1342. [Google Scholar] [CrossRef] [Green Version]
- Stubbings, W.; Labischinski, H. New Antibiotics for Antibiotic-Resistant Bacteria. F1000 Biol. Rep. 2009, 1, 40. [Google Scholar] [CrossRef] [Green Version]
- Roychoudhury, S.; Twinem, T.L.; Makin, K.M.; McIntosh, E.J.; Ledoussal, B.; Catrenich, C.E. Activity of Non-Fluorinated Quinolones (NFQs) against Quinolone-Resistant Escherichia coli and Streptococcus pneumoniae. J. Antimicrob. Chemother. 2001, 48, 29–36. [Google Scholar] [CrossRef] [Green Version]
- Adam, H.J.; Laing, N.M.; King, C.R.; Lulashnyk, B.; Hoban, D.J.; Zhanel, G.G. In Vitro Activity of Nemonoxacin, a Novel Nonfluorinated Quinolone, against 2440 Clinical Isolates. Antimicrob. Agents Chemother. 2009, 53, 4915–4920. [Google Scholar] [CrossRef] [Green Version]
- Qin, X.; Huang, H. Review of Nemonoxacin with Special Focus on Clinical Development. Drug Des. Dev. Ther. 2014, 8, 765–774. [Google Scholar] [CrossRef] [Green Version]
- Borges, A.; Ferreira, C.; Saavedra, M.J.; Simões, M. Antibacterial Activity and Mode of Action of Ferulic and Gallic Acids Against Pathogenic Bacteria. Microb. Drug Resist. 2013, 19, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Ibitoye, O.B.; Ajiboye, T.O. Ferulic Acid Potentiates the Antibacterial Activity of Quinolone-Based Antibiotics against Acinetobacter baumannii. Microb. Pathog. 2019, 126, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Courtney, C.M.; Goodman, S.M.; Nagy, T.A.; Levy, M.; Bhusal, P.; Madinger, N.E.; Detweiler, C.S.; Nagpal, P.; Chatterjee, A. Potentiating Antibiotics in Drug-Resistant Clinical Isolates via Stimuli-Activated Superoxide Generation. Sci. Adv. 2017, 3, e1701776. [Google Scholar] [CrossRef] [Green Version]
- Neu, H.C. Synergy and Antagonism of Combinations with Quinolones. Eur. J. Clin. Microbiol. Infect. Dis. 1991, 10, 255–261. [Google Scholar] [CrossRef]
- Tintino, S.R.; de Souza, V.C.A.; da Silva, J.M.A.; Oliveira-Tintino, C.D.d.M.; Pereira, P.S.; Leal-Balbino, T.C.; Pereira-Neves, A.; Siqueira-Junior, J.P.; da Costa, J.G.M.; Rodrigues, F.F.G.; et al. Effect of Vitamin K3 Inhibiting the Function of NorA Efflux Pump and Its Gene Expression on Staphylococcus aureus. Membranes 2020, 10, 130. [Google Scholar] [CrossRef]
- Lim, C.S.Q.; Ha, K.P.; Clarke, R.S.; Gavin, L.-A.; Cook, D.T.; Hutton, J.A.; Sutherell, C.L.; Edwards, A.M.; Evans, L.E.; Tate, E.W.; et al. Identification of a Potent Small-Molecule Inhibitor of Bacterial DNA Repair That Potentiates Quinolone Antibiotic Activity in Methicillin-Resistant Staphylococcus aureus. Bioorgan. Med. Chem. 2019, 27, 114962. [Google Scholar] [CrossRef]
- Wilkinson, M.; Troman, L.; Wan Nur Ismah, W.A.; Chaban, Y.; Avison, M.B.; Dillingham, M.S.; Wigley, D.B. Structural Basis for the Inhibition of RecBCD by Gam and Its Synergistic Antibacterial Effect with Quinolones. eLife 2016, 5, e22963. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.-H.; Otsuka, N.; Noyori, K.; Shiota, S.; Ogawa, W.; Kuroda, T.; Hatano, T.; Tsuchiya, T. Synergistic Effect of Kaempferol Glycosides Purified from Laurus Nobilis and Fluoroquinolones on Methicillin-Resistant Staphylococcus aureus. Biol. Pharm. Bull. 2009, 32, 489–492. [Google Scholar] [CrossRef] [Green Version]
- Cannalire, R.; Machado, D.; Felicetti, T.; Santos Costa, S.; Massari, S.; Manfroni, G.; Barreca, M.L.; Tabarrini, O.; Couto, I.; Viveiros, M.; et al. Natural Isoflavone Biochanin A as a Template for the Design of New and Potent 3-Phenylquinolone Efflux Inhibitors against Mycobacterium avium. Eur. J. Med. Chem. 2017, 140, 321–330. [Google Scholar] [CrossRef]
- Duan, X.; Huang, X.; Wang, X.; Yan, S.; Guo, S.; Abdalla, A.E.; Huang, C.; Xie, J. L-Serine Potentiates Fluoroquinolone Activity against Escherichia Coli by Enhancing Endogenous Reactive Oxygen Species Production. J. Antimicrob. Chemother. 2016, 71, 2192–2199. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Hu, L.; Hao, Z. Palmatine Is a Plasmid-Mediated Quinolone Resistance (PMQR) Inhibitor That Restores the Activity of Ciprofloxacin Against QnrS and AAC(6ʹ)-Ib-Cr-Producing Escherichia coli. Infect. Drug Resist. 2020, 13, 749–759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradfute, S.B.; Ye, C.; Clarke, E.C.; Kumar, S.; Timmins, G.S.; Deretic, V. Ambroxol and Ciprofloxacin Show Activity Against SARS-CoV2 in Vero E6 Cells at Clinically-Relevant Concentrations. bioRxiv 2020. [Google Scholar] [CrossRef]
- Wei, W.; Yang, H.; Hu, L.; Ye, Y.; Li, J. Activity of Levofloxacin in Combination with Colistin against Acinetobacter baumannii: In Vitro and in a Galleria mellonella Model. J. Microbiol. Immunol. Infect. 2017, 50, 821–830. [Google Scholar] [CrossRef] [Green Version]
- Gorityala, B.K.; Guchhait, G.; Fernando, D.M.; Deo, S.; McKenna, S.A.; Zhanel, G.G.; Kumar, A.; Schweizer, F. Adjuvants Based on Hybrid Antibiotics Overcome Resistance in Pseudomonas aeruginosa and Enhance Fluoroquinolone Efficacy. Angew. Chem. Int. Ed. Engl. 2015, 55, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Gorityala, B.K.; Guchhait, G.; Goswami, S.; Fernando, D.M.; Kumar, A.; Zhanel, G.G.; Schweizer, F. Hybrid Antibiotic Overcomes Resistance in P. aeruginosa by Enhancing Outer Membrane Penetration and Reducing Efflux. J. Med. Chem. 2016, 59, 8441–8455. [Google Scholar] [CrossRef]
- Domalaon, R.; Idowu, T.; Zhanel, G.G.; Schweizer, F. Antibiotic Hybrids: The Next Generation of Agents and Adjuvants against Gram-Negative Pathogens? Clin. Microbiol. Rev. 2018, 31, e00077-17. [Google Scholar] [CrossRef] [Green Version]
- Park, H.-R.; Kim, T.H.; Bark, K.-M. Physicochemical Properties of Quinolone Antibiotics in Various Environments. Eur. J. Med. Chem. 2002, 37, 443–460. [Google Scholar] [CrossRef]
- Ross, D.L.; Riley, C.M. Physicochemical Properties of the Fluoroquinolone Antimicrobials. II. Acid Ionization Constants and Their Relationship to Structure. Int. J. Pharm. 1992, 83, 267–272. [Google Scholar] [CrossRef]
- Cramariuc, O.; Rog, T.; Javanainen, M.; Monticelli, L.; Polishchuk, A.V.; Vattulainen, I. Mechanism for Translocation of Fluoroquinolones across Lipid Membranes. Biochim. Biophys. Acta BBA Biomembr. 2012, 1818, 2563–2571. [Google Scholar] [CrossRef] [Green Version]
- Van Bambeke, F. Delafloxacin, a Non-Zwitterionic Fluoroquinolone in Phase III of Clinical Development: Evaluation of Its Pharmacology, Pharmacokinetics, Pharmacodynamics and Clinical Efficacy. Future Microbiol. 2015, 10, 1111–1123. [Google Scholar] [CrossRef] [Green Version]
- Lodise, T.; Corey, R.; Hooper, D.; Cammarata, S. Safety of Delafloxacin: Focus on Adverse Events of Special Interest. Open Forum Infect. Dis. 2018, 5, ofy220. [Google Scholar] [CrossRef]
- Rusu, A.; Lungu, I.-A.; Moldovan, O.-L.; Tanase, C.; Hancu, G. Structural Characterization of the Millennial Antibacterial (Fluoro)Quinolones—Shaping the Fifth Generation. Pharmaceutics 2021, 13, 1289. [Google Scholar] [CrossRef] [PubMed]
- Kłosińska-Szmurło, E.; Pluciński, F.A.; Grudzień, M.; Betlejewska-Kielak, K.; Biernacka, J.; Mazurek, A.P. Experimental and Theoretical Studies on the Molecular Properties of Ciprofloxacin, Norfloxacin, Pefloxacin, Sparfloxacin, and Gatifloxacin in Determining Bioavailability. J. Biol. Phys. 2014, 40, 335–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kłosińska-Szmurło, E.; Grudzień, M.; Betlejewska-Kielak, K.; Pluciński, F.; Biernacka, J.; Mazurek, A.P. Physicochemical Properties of Lomefloxacin, Levofloxacin, and Moxifloxacin Relevant to the Biopharmaceutics Classification System. Acta Chim. Slov. 2014, 61, 827–834. [Google Scholar] [PubMed]
- Uivarosi, V. Metal Complexes of Quinolone Antibiotics and Their Applications: An Update. Molecules 2013, 18, 11153–11197. [Google Scholar] [CrossRef] [PubMed]
- Ross, D.L.; Riley, C.M. Physicochemical Properties of the Fluoroquinolone Antimicrobials V. Effect of Fluoroquinolone Structure and PH on the Complexation of Various Fluoroquinolones with Magnesium and Calcium Ions. Int. J. Pharm. 1993, 93, 121–129. [Google Scholar] [CrossRef]
- Pitman, S.K.; Hoang, U.T.P.; Wi, C.H.; Alsheikh, M.; Hiner, D.A.; Percival, K.M. Revisiting Oral Fluoroquinolone and Multivalent Cation Drug-Drug Interactions: Are They Still Relevant? Antibiotics 2019, 8, 108. [Google Scholar] [CrossRef] [Green Version]
- Stass, H.; Kubitza, D. Effects of Iron Supplements on the Oral Bioavailability of Moxifloxacin, a Novel 8-Methoxyfluoroquinolone, in Humans. Clin. Pharm. 2001, 40, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Djurdjevic, P.; Jakovljevic, I.; Joksovic, L.; Ivanovic, N.; Jelikic-Stankov, M. The Effect of Some Fluoroquinolone Family Members on Biospeciation of Copper(II), Nickel(II) and Zinc(II) Ions in Human Plasma. Molecules 2014, 19, 12194–12223. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Dahyabhai Prajapati, J.; Kleinekathöfer, U.; Winterhalter, M. Dynamic Interaction of Fluoroquinolones with Magnesium Ions Monitored Using Bacterial Outer Membrane Nanopores. Chem. Sci. 2020, 11, 10344–10353. [Google Scholar] [CrossRef] [PubMed]
- Valisena, S.; Palumbo, M.; Parolin, C.; Palú, G.; Meloni, G.A. Relevance of Ionic Effects on Norfloxacin Uptake by Escherichia coli. Biochem. Pharmacol. 1990, 40, 431–436. [Google Scholar] [CrossRef]
- Mortimer, P.; Piddock, L. A comparison of methods used for measuring the accumulation of quinolones by Enterobacteriaceae, Pseudomonas aeruginosa and Staphylococcus aureus. J. Antimicrob. Chemother. 1991, 28, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Furet, Y.X.; Deshusses, J.; Pechère, J.C. Transport of Pefloxacin across the Bacterial Cytoplasmic Membrane in Quinolone-Susceptible Staphylococcus aureus. Antimicrob. Agents Chemother. 1992, 36, 2506–2511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, D.L.; Elkinton, S.K.; Riley, C.M. Physicochemical Properties of the Fluoroquinolone Antimicrobials. III. 1-Octanol/Water Partition Coefficients and Their Relationships to Structure. Int. J. Pharm. 1992, 88, 379–389. [Google Scholar] [CrossRef]
- Ferreira, M.; Bessa, L.J.; Sousa, C.F.; Eaton, P.; Bongiorno, D.; Stefani, S.; Campanile, F.; Gameiro, P. Fluoroquinolone Metalloantibiotics: A Promising Approach against Methicillin-Resistant Staphylococcus aureus. Int. J. Environ. Res. Public Health 2020, 17, 3127. [Google Scholar] [CrossRef]
- Feio, M.J.; Sousa, I.; Ferreira, M.; Cunha-Silva, L.; Saraiva, R.G.; Queirós, C.; Alexandre, J.G.; Claro, V.; Mendes, A.; Ortiz, R.; et al. Fluoroquinolone–Metal Complexes: A Route to Counteract Bacterial Resistance? J. Inorg. Biochem. 2014, 138, 129–143. [Google Scholar] [CrossRef]
- Yang, S.; Tian, M.; Yuan, L.; Deng, H.; Wang, L.; Li, A.; Hou, Z.; Li, Y.; Zhang, Y. Analysis of E. rutaecarpa Alkaloids Constituents In Vitro and In Vivo by UPLC-Q-TOF-MS Combined with Diagnostic Fragment. Available online: https://www.hindawi.com/journals/jamc/2016/4218967/ (accessed on 25 September 2020).
- Heeb, S.; Fletcher, M.P.; Chhabra, S.R.; Diggle, S.P.; Williams, P.; Cámara, M. Quinolones: From Antibiotics to Autoinducers. FEMS Microbiol. Rev. 2011, 35, 247–274. [Google Scholar] [CrossRef] [Green Version]
- Dekker, K.A.; Inagaki, T.; Gootz, T.D.; Huang, L.H.; Kojima, Y.; Kohlbrenner, W.E.; Matsunaga, Y.; McGuirk, P.R.; Nomura, E.; Sakakibara, T.; et al. New Quinolone Compounds from Pseudonocardia Sp. with Selective and Potent Anti-Helicobacter pylori Activity: Taxonomy of Producing Strain, Fermentation, Isolation, Structural Elucidation and Biological Activities. J. Antibiot. 1998, 51, 145–152. [Google Scholar] [CrossRef]
- Geddis, S.M.; Coroama, T.; Forrest, S.; Hodgkinson, J.T.; Welch, M.; Spring, D.R. Synthesis and Biological Evaluation of 1,2-Disubstituted 4-Quinolone Analogues of Pseudonocardia Sp. Natural Products. Beilstein J. Org. Chem. 2018, 14, 2680–2688. [Google Scholar] [CrossRef] [Green Version]
- Albrecht, R. Development of antibacterial agents of the nalidixic acid type. In Progress in Drug Research/Fortschritte der Arzneimittelforschung/Progrès des Rechersches Pharmaceutiques; Jucker, E., Ed.; Birkhäuser: Basel, Switzerland, 1977; pp. 9–104. ISBN 978-3-0348-7098-6. [Google Scholar]
- Schaumann, R.; Rodloff, A. Activities of Quinolones Against Obligately Anaerobic Bacteria. Anti Infect. Agents Med. Chem. 2006, 6, 49–56. [Google Scholar] [CrossRef]
- Bryskier, A. Antimicrobial Agents: Antibacterials and Antifungals; ASM Press: Washington, DC, USA, 2005; ISBN 978-1-55581-237-9. [Google Scholar]
- Domagala, J.M. Structure-Activity and Structure-Side-Effect Relationships for the Quinolone Antibacterials. J. Antimicrob. Chemother. 1994, 33, 685–706. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Hayakawa, I.; Akimoto, T. The history of the development and changes of quinolone antibacterial agents. Yakushigaku Zasshi 2003, 38, 161–179. [Google Scholar] [PubMed]
- Emmerson, A.M. The Quinolones: Decades of Development and Use. J. Antimicrob. Chemother. 2003, 51, 13–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rohlfing, S.R.; Gerster, J.F.; Kvam, D.C. Bioevaluation of the Antibacterial Flumequine for Urinary Tract Use. Antimicrob. Agents Chemother. 1976, 10, 20–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wise, R.; Andrews, J.M.; Edwards, L.J. In Vitro Activity of Bay 09867, a New Quinoline Derivative, Compared with Those of Other Antimicrobial Agents. Antimicrob. Agents Chemother. 1983, 23, 559–564. [Google Scholar] [CrossRef] [Green Version]
- Chin, N.X.; Neu, H.C. Ciprofloxacin, a Quinolone Carboxylic Acid Compound Active against Aerobic and Anaerobic Bacteria. Antimicrob. Agents Chemother. 1984, 25, 319–326. [Google Scholar] [CrossRef] [Green Version]
- Hoshino, K.; Sato, K.; Une, T.; Osada, Y. Inhibitory Effects of Quinolones on DNA Gyrase of Escherichia coli and Topoisomerase II of Fetal Calf Thymus. Antimicrob. Agents Chemother. 1989, 33, 1816–1818. [Google Scholar] [CrossRef] [Green Version]
- Shah, P.; Westwell, A.D. The Role of Fluorine in Medicinal Chemistry. J Enzym. Inhib. Med. Chem. 2007, 22, 527–540. [Google Scholar] [CrossRef] [Green Version]
- Sato, K.; Matsuura, Y.; Inoue, M.; Une, T.; Osada, Y.; Ogawa, H.; Mitsuhashi, S. In Vitro and in Vivo Activity of DL-8280, a New Oxazine Derivative. Antimicrob. Agents Chemother. 1982, 22, 548–553. [Google Scholar] [CrossRef] [Green Version]
- Woodcock, J.M.; Andrews, J.M.; Boswell, F.J.; Brenwald, N.P.; Wise, R. In Vitro Activity of BAY 12-8039, a New Fluoroquinolone. Antimicrob. Agents Chemother. 1997, 41, 101–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, T.; Zhao, X.; Li, X.; Drlica-Wagner, A.; Wang, J.-Y.; Domagala, J.; Drlica, K. Enhancement of Fluoroquinolone Activity by C-8 Halogen and Methoxy Moieties: Action against a Gyrase Resistance Mutant of Mycobacterium smegmatis and a Gyrase-Topoisomerase IV Double Mutant of Staphylococcus aureus. Antimicrob. Agents Chemother. 2001, 45, 2703–2709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz, J. Transferable Mechanisms of Quinolone Resistance from 1998 Onward. Clin. Microbiol. Rev. 2019, 32, e00007-19. [Google Scholar] [CrossRef] [PubMed]
- Klomp, F.; Wenzel, C.; Drozdzik, M.; Oswald, S. Drug–Drug Interactions Involving Intestinal and Hepatic CYP1A Enzymes. Pharmaceutics 2020, 12, 1201. [Google Scholar] [CrossRef] [PubMed]
- Bridle, M.J.; Janesko, B.G. Computational Study of Fluoroquinolone Binding to Mg(H2O)N2+ and Its Applicability to Future Drug Design. Int. J. Quantum Chem. 2017, 117, e25428. [Google Scholar] [CrossRef]
- Panda, S.S.; Liaqat, S.; Girgis, A.S.; Samir, A.; Hall, C.D.; Katritzky, A.R. Novel Antibacterial Active Quinolone–Fluoroquinolone Conjugates and 2D-QSAR Studies. Bioorgan. Med. Chem. Lett. 2015, 25, 3816–3821. [Google Scholar] [CrossRef]
- Hou, Y.; Zhao, Y.; Li, Y. Environmentally Friendly Fluoroquinolone Derivatives with Lower Plasma Protein Binding Rate Designed Using 3D-QSAR, Molecular Docking and Molecular Dynamics Simulation. Int. J. Environ. Res. Public Health 2020, 17, 6626. [Google Scholar] [CrossRef]
- Zhao, X.; Zhao, Y.; Ren, Z.; Li, Y. Combined QSAR/QSPR and Molecular Docking Study on Fluoroquinolones to Reduce Biological Enrichment. Comput. Biol. Chem. 2019, 79, 177–184. [Google Scholar] [CrossRef]
- Molecular Design of Lower Photodegradation Fluoroquinolone Antibiotics and Their Photolysis Paths Inference. Available online: http://www.cjcu.jlu.edu.cn/EN/10.7503/cjcu20180475 (accessed on 28 October 2021).
- Yu, W.; MacKerell, A.D. Computer-Aided Drug Design Methods. In Antibiotics; Methods in Molecular Biology; Sass, P., Ed.; Springer: New York, NY, USA, 2017; Volume 1520, pp. 85–106. ISBN 978-1-4939-6632-5. [Google Scholar]
- Sarma, P.S. Norfloxacin: A New Drug in the Treatment of Falciparum Malaria. Ann. Intern. Med. 1989, 111, 336–337. [Google Scholar] [CrossRef]
- Tripathi, K.D.; Sharma, A.K.; Valecha, N.; Kulpati, D.D. Curative Efficacy of Norfloxacin in Falciparum Malaria. Indian J. Med. Res. 1993, 97, 176–178. [Google Scholar]
- Dalhoff, A. Antiviral, Antifungal, and Antiparasitic Activities of Fluoroquinolones Optimized for Treatment of Bacterial Infections: A Puzzling Paradox or a Logical Consequence of Their Mode of Action? Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Ridley, R.G.; Hofheinz, W.; Matile, H.; Jaquet, C.; Dorn, A.; Masciadri, R.; Jolidon, S.; Richter, W.F.; Guenzi, A.; Girometta, M.A.; et al. 4-Aminoquinoline Analogs of Chloroquine with Shortened Side Chains Retain Activity against Chloroquine-Resistant Plasmodium falciparum. Antimicrob. Agents Chemother. 1996, 40, 1846–1854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, L.S. Malaria. Clin. Lab. Med. 2010, 30, 93–129. [Google Scholar] [CrossRef] [PubMed]
- Cross, R.M.; Monastyrskyi, A.; Mutka, T.S.; Burrows, J.N.; Kyle, D.E.; Manetsch, R. Endochin Optimization: Structure-Activity and Structure-Property Relationship Studies of 3-Substituted 2-Methyl-4(1H)-Quinolones with Antimalarial Activity. J. Med. Chem. 2010, 53, 7076–7094. [Google Scholar] [CrossRef] [PubMed]
- Kurasawa, Y.; Yoshida, K.; Yamazaki, N.; Kaji, E.; Sasaki, K.; Zamami, Y.; Sakai, Y.; Fujii, T.; Ito, H. Quinolone Analogs 11: Synthesis of Novel 4-Quinolone-3-Carbohydrazide Derivatives with Antimalarial Activity. J. Heterocycl. Chem. 2012, 49, 288–292. [Google Scholar] [CrossRef]
- Martins, D.A.; Gouvea, L.R.; Muniz, G.S.V.; Louro, S.R.W.; Batista, D.d.G.J.; Soeiro, M.d.N.C.; Teixeira, L.R. Norfloxacin and N-Donor Mixed-Ligand Copper(II) Complexes: Synthesis, Albumin Interaction, and Anti-Trypanosoma cruzi Activity. Bioinorg. Chem. Appl. 2016, 2016, 5027404. [Google Scholar] [CrossRef] [Green Version]
- Hiltensperger, G.; Hecht, N.; Kaiser, M.; Rybak, J.-C.; Hoerst, A.; Dannenbauer, N.; Müller-Buschbaum, K.; Bruhn, H.; Esch, H.; Lehmann, L.; et al. Quinolone Amides as Antitrypanosomal Lead Compounds with In Vivo Activity. Antimicrob. Agents Chemother. 2016, 60, 4442–4452. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Damu, G.L.V.; Lv, J.-S.; Geng, R.-X.; Yang, D.-C.; Zhou, C.-H. Design, Synthesis and Evaluation of Clinafloxacin Triazole Hybrids as a New Type of Antibacterial and Antifungal Agents. Bioorgan. Med. Chem. Lett. 2012, 22, 5363–5366. [Google Scholar] [CrossRef]
- El-Halim, H.F.A.; Mohamed, G.G.; El-Dessouky, M.M.I.; Mahmoud, W.H. Ligational Behaviour of Lomefloxacin Drug towards Cr(III), Mn(II), Fe(III), Co(II), Ni(II), Cu(II), Zn(II), Th(IV) and UO2(VI) Ions: Synthesis, Structural Characterization and Biological Activity Studies. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2011, 82, 8–19. [Google Scholar] [CrossRef]
- Sultana, N.; Naz, A.; Arayne, M.S.; Mesaik, M.A. Synthesis, Characterization, Antibacterial, Antifungal and Immunomodulating Activities of Gatifloxacin–Metal Complexes. J. Mol. Struct. 2010, 969, 17–24. [Google Scholar] [CrossRef]
- Debnath, A.; Mogha, N.K.; Masram, D.T. Metal Complex of the First-Generation Quinolone Antimicrobial Drug Nalidixic Acid: Structure and Its Biological Evaluation. Appl Biochem Biotechnol 2015, 175, 2659–2667. [Google Scholar] [CrossRef]
- Khamkhenshorngphanuch, T.; Kulkraisri, K.; Janjamratsaeng, A.; Plabutong, N.; Thammahong, A.; Manadee, K.; Na Pombejra, S.; Khotavivattana, T. Synthesis and Antimicrobial Activity of Novel 4-Hydroxy-2-Quinolone Analogs. Molecules 2020, 25, 3059. [Google Scholar] [CrossRef]
- Xi, X.G. Helicases as Antiviral and Anticancer Drug Targets. Curr. Med. Chem. 2007, 14, 883–915. [Google Scholar] [CrossRef]
- Portolani, M.; Pietrosemoli, P.; Cermelli, C.; Mannini-Palenzona, A.; Grossi, M.P.; Paolini, L.; Barbanti-Brodano, G. Suppression of BK Virus Replication and Cytopathic Effect by Inhibitors of Prokaryotic DNA Gyrase. Antivir. Res. 1988, 9, 205–218. [Google Scholar] [CrossRef]
- Koukoulaki, M.; Apostolou, T.; Hadjiconstantinou, V.; Drakopoulos, S. Impact of Prophylactic Administration of Ciprofloxacin on BK Polyoma Virus Replication. Transpl. Infect. Dis. 2008, 10, 449–451. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.H.; Chandraker, A.; DeCaprio, J.A. Inhibition of Simian Virus 40 Large T Antigen Helicase Activity by Fluoroquinolones. Antivir. Ther. 2007, 12, 1–6. [Google Scholar] [PubMed]
- Khan, I.A.; Siddiqui, S.; Rehmani, S.; Kazmi, S.U.; Ali, S.H. Fluoroquinolones Inhibit HCV by Targeting Its Helicase. Antivir. Ther. 2012, 17, 467–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habtemariam, S.; Nabavi, S.F.; Banach, M.; Berindan-Neagoe, I.; Sarkar, K.; Sil, P.C.; Nabavi, S.M. Should We Try SARS-CoV-2 Helicase Inhibitors for COVID-19 Therapy? Arch. Med Res. 2020, 51, 733–735. [Google Scholar] [CrossRef]
- Marciniec, K.; Beberok, A.; Pęcak, P.; Boryczka, S.; Wrześniok, D. Ciprofloxacin and Moxifloxacin Could Interact with SARS-CoV-2 Protease: Preliminary in Silico Analysis. Pharm. Rep. 2020, 72, 1553–1561. [Google Scholar] [CrossRef] [PubMed]
- Tauber, S.C.; Nau, R. Immunomodulatory Properties of Antibiotics. Curr. Mol. Pharm. 2008, 1, 68–79. [Google Scholar]
- Dalhoff, A.; Shalit, I. Immunomodulatory Effects of Quinolones. Lancet Infect. Dis. 2003, 3, 359–371. [Google Scholar] [CrossRef]
- Ogino, H.; Fujii, M.; Ono, M.; Maezawa, K.; Hori, S.; Kizu, J. In Vivo and in Vitro Effects of Fluoroquinolones on Lipopolysaccharide-Induced pro-Inflammatory Cytokine Production. J. Infect. Chemother. 2009, 15, 168–173. [Google Scholar] [CrossRef]
- Zhang, J.-Z.; Ward, K.W. Besifloxacin, a Novel Fluoroquinolone Antimicrobial Agent, Exhibits Potent Inhibition of pro-Inflammatory Cytokines in Human THP-1 Monocytes. J. Antimicrob. Chemother. 2008, 61, 111–116. [Google Scholar] [CrossRef] [Green Version]
- Zusso, M.; Lunardi, V.; Franceschini, D.; Pagetta, A.; Lo, R.; Stifani, S.; Frigo, A.C.; Giusti, P.; Moro, S. Ciprofloxacin and Levofloxacin Attenuate Microglia Inflammatory Response via TLR4/NF-KB Pathway. J. Neuroinflamm. 2019, 16, 148. [Google Scholar] [CrossRef] [Green Version]
- Araujo, F.G.; Slifer, T.L.; Remington, J.S. Effect of Moxifloxacin on Secretion of Cytokines by Human Monocytes Stimulated with Lipopolysaccharide. Clin. Microbiol. Infect. 2002, 8, 26–30. [Google Scholar] [CrossRef] [Green Version]
- Shalit, I.; Halperin, D.; Haite, D.; Levitov, A.; Romano, J.; Osherov, N.; Fabian, I. Anti-Inflammatory Effects of Moxifloxacin on IL-8, IL-1beta and TNF-Alpha Secretion and NFkappaB and MAP-Kinase Activation in Human Monocytes Stimulated with Aspergillus fumigatus. J. Antimicrob. Chemother. 2006, 57, 230–235. [Google Scholar] [CrossRef] [Green Version]
- Fabian, I.; Reuveni, D.; Levitov, A.; Halperin, D.; Priel, E.; Shalit, I. Moxifloxacin Enhances Antiproliferative and Apoptotic Effects of Etoposide but Inhibits Its Proinflammatory Effects in THP-1 and Jurkat Cells. Br. J. Cancer 2006, 95, 1038–1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batalha, P.N.; Vieira de Souza, M.C.B.; Peña-Cabrera, E.; Cruz, D.C.; da Costa Santos Boechat, F. Quinolones in the Search for New Anticancer Agents. Curr. Pharm. Des. 2016, 22, 6009–6020. [Google Scholar] [CrossRef] [PubMed]
- Beberok, A.; Wrześniok, D.; Rok, J.; Rzepka, Z.; Respondek, M.; Buszman, E. Ciprofloxacin Triggers the Apoptosis of Human Triple-Negative Breast Cancer MDA-MB-231 Cells via the P53/Bax/Bcl-2 Signaling Pathway. Int. J. Oncol. 2018, 52, 1727–1737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beberok, A.; Wrześniok, D.; Minecka, A.; Rok, J.; Delijewski, M.; Rzepka, Z.; Respondek, M.; Buszman, E. Ciprofloxacin-Mediated Induction of S-Phase Cell Cycle Arrest and Apoptosis in COLO829 Melanoma Cells. Pharmacol. Rep. 2018, 70, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Nishi, K.; Kato, M.; Sakurai, S.; Matsumoto, A.; Iwase, Y.; Yumita, N. Enoxacin with UVA Irradiation Induces Apoptosis in the AsPC1 Human Pancreatic Cancer Cell Line Through ROS Generation. Anticancer Res. 2017, 37, 6211–6214. [Google Scholar]
- Beberok, A.; Rzepka, Z.; Respondek, M.; Rok, J.; Sierotowicz, D.; Wrześniok, D. GSH Depletion, Mitochondrial Membrane Breakdown, Caspase-3/7 Activation and DNA Fragmentation in U87MG Glioblastoma Cells: New Insight into the Mechanism of Cytotoxicity Induced by Fluoroquinolones. Eur. J. Pharmacol. 2018, 835, 94–107. [Google Scholar] [CrossRef] [PubMed]
- Patitungkho, S.; Adsule, S.; Dandawate, P.; Padhye, S.; Ahmad, A.; Sarkar, F.H. Synthesis, Characterization and Anti-Tumor Activity of Moxifloxacin-Copper Complexes against Breast Cancer Cell Lines. Bioorgan. Med. Chem. Lett. 2011, 21, 1802–1806. [Google Scholar] [CrossRef] [PubMed]
- Nakai, S.; Imaizumi, T.; Watanabe, T.; Iwase, Y.; Nishi, K.; Okudaira, K.; Yumita, N. Photodynamically-Induced Apoptosis Due to Ultraviolet A in the Presence of Lomefloxacin in Human Promyelocytic Leukemia Cells. Anticancer Res. 2017, 37, 6407–6413. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.; Talwar, P. Repositioning of Fluoroquinolones from Antibiotic to Anti-Cancer Agents: An Underestimated Truth. Biomed. Pharmacother. 2019, 111, 934–946. [Google Scholar] [CrossRef]
- Berninger, M.; Erk, C.; Fuß, A.; Skaf, J.; Al-Momani, E.; Israel, I.; Raschig, M.; Güntzel, P.; Samnick, S.; Holzgrabe, U. Fluorine Walk: The Impact of Fluorine in Quinolone Amides on Their Activity against African Sleeping Sickness. Eur. J. Med. Chem. 2018, 152, 377–391. [Google Scholar] [CrossRef]
- Kraus, J.M.; Tatipaka, H.B.; McGuffin, S.A.; Chennamaneni, N.K.; Karimi, M.; Arif, J.; Verlinde, C.L.M.J.; Buckner, F.S.; Gelb, M.H. Second Generation Analogues of the Cancer Drug Clinical Candidate Tipifarnib for Anti-Chagas Disease Drug Discovery. J. Med. Chem. 2010, 53, 3887–3898. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Zhao, S.-J.; Lv, Z.-S.; Gao, F.; Wang, Y.; Zhang, F.; Bai, L.; Deng, J.-L. Fluoroquinolone-Isatin Hybrids and Their Biological Activities. Eur. J. Med. Chem. 2019, 162, 396–406. [Google Scholar] [CrossRef]
- Sato, M.; Motomura, T.; Aramaki, H.; Matsuda, T.; Yamashita, M.; Ito, Y.; Kawakami, H.; Matsuzaki, Y.; Watanabe, W.; Yamataka, K.; et al. Novel HIV-1 Integrase Inhibitors Derived from Quinolone Antibiotics. J. Med. Chem. 2006, 49, 1506–1508. [Google Scholar] [CrossRef]
- Kljun, J.; Bytzek, A.K.; Kandioller, W.; Bartel, C.; Jakupec, M.A.; Hartinger, C.G.; Keppler, B.K.; Turel, I. Physicochemical Studies and Anticancer Potency of Ruthenium H6-p-Cymene Complexes Containing Antibacterial Quinolones. Organometallics 2011, 30, 2506–2512. [Google Scholar] [CrossRef]
- Regmi, N.L.; Abd El-Aty, A.M.; Kubota, R.; Shah, S.S.; Shimoda, M. Lack of Inhibitory Effects of Several Fluoroquinolones on Cytochrome P-450 3A Activities at Clinical Dosage in Dogs. J. Vet. Pharm. 2007, 30, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-J.; Jin, P.; Wang, S.-B.; Li, F.-N.; Guan, L.-P.; Quan, Z.-S. Synthesis and Anticonvulsant Activity Evaluation of 4-Phenyl-[1,2,4]Triazolo[4,3-a]Quinazolin-5(4H)-One and Its Derivatives. Arch. Pharm. 2015, 348, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Hu, J.; Yang, X.; Feng, X.; Li, X.; Huang, L.; Chan, A.S.C. Design, Synthesis, and Evaluation of Orally Bioavailable Quinoline–Indole Derivatives as Innovative Multitarget-Directed Ligands: Promotion of Cell Proliferation in the Adult Murine Hippocampus for the Treatment of Alzheimer’s Disease. J. Med. Chem. 2018, 61, 1871–1894. [Google Scholar] [CrossRef]
- Ukrainets, I.V.; Bereznyakova, N.L.; Mospanova, E.V. 4-Hydroxy-2-Quinolones 121. Synthesis and Biological Properties of 1-Hydroxy-3-Oxo-5,6-Dihydro-3h-Pyrrolo[3,2,1-Ij]Quino-Line-2-Carboxylic Acid Alkylamides. Chem. Heterocycl. Compd. 2007, 43, 856–862. [Google Scholar] [CrossRef]
- Van Bambeke, F.; Michot, J.-M.; Van Eldere, J.; Tulkens, P.M. Quinolones in 2005: An Update. Clin. Microbiol. Infect. 2005, 11, 256–280. [Google Scholar] [CrossRef] [Green Version]
- Goa, K.L.; Bryson, H.M.; Markham, A. Sparfloxacin. A Review of Its Antibacterial Activity, Pharmacokinetic Properties, Clinical Efficacy and Tolerability in Lower Respiratory Tract Infections. Drugs 1997, 53, 700–725. [Google Scholar] [CrossRef] [PubMed]
- Oliphant, C.M.; Green, G. Quinolones: A Comprehensive Review. Am. Fam. Physician 2002, 65, 455. [Google Scholar]
- Nicolau, D.; Quintiliani, R.; Nightingale, C.H. Ofloxacin vs. Ciprofloxacin: A Comparison. Connect. Med. 1992, 56, 261–263. [Google Scholar]
- Bidell, M.R.; Lodise, T.P. Fluoroquinolone-Associated Tendinopathy: Does Levofloxacin Pose the Greatest Risk? Pharmacotherapy 2016, 36, 679–693. [Google Scholar] [CrossRef]
- Rubinstein, E. History of Quinolones and Their Side Effects. Chemotherapy 2001, 47 (Suppl. 3), 3–8; discussion 44–48. [Google Scholar] [CrossRef]
- Haria, M.; Lamb, H.M. Trovafloxacin. Drugs 1997, 54, 435–445; discussion 446. [Google Scholar] [CrossRef] [PubMed]
- Cunha, B.A. Antibiotic Side Effects. Med. Clin. N. Am. 2001, 85, 149–185. [Google Scholar] [CrossRef]
- Stewardson, A.J.; Gaïa, N.; François, P.; Malhotra-Kumar, S.; Delémont, C.; Martinez de Tejada, B.; Schrenzel, J.; Harbarth, S.; Lazarevic, V. SATURN WP1 and WP3 Study Groups. Collateral Damage from Oral Ciprofloxacin vs. Nitrofurantoin in Outpatients with Urinary Tract Infections: A Culture-Free Analysis of Gut Microbiota. Clin. Microbiol. Infect. 2015, 21, 344.e1–344.e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fox, M.T.; Melia, M.T.; Same, R.G.; Conley, A.T.; Tamma, P.D. A Seven-Day Course of Trimethoprim-Sulfamethoxazole May Be as Effective as a Seven-Day Course of Ciprofloxacin for the Treatment of Pyelonephritis. Am. J. Med. 2017, 130, 842–845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.-T.G.; Lee, S.-H.; Chang, S.-S.; Lee, S.-H.; Lee, M.; Fang, C.-C.; Chen, S.-C.; Lee, C.-C. Comparative Effectiveness of Different Oral Antibiotics Regimens for Treatment of Urinary Tract Infection in Outpatients: An Analysis of National Representative Claims Database. Medicine 2014, 93, e304. [Google Scholar] [CrossRef]
- Hooper, D.C. New Uses for New and Old Quinolones and the Challenge of Resistance. Clin. Infect. Dis. 2000, 30, 243–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kreling, V.; Falcone, F.H.; Kehrenberg, C.; Hensel, A. Campylobacter Sp.: Pathogenicity Factors and Prevention Methods—New Molecular Targets for Innovative Antivirulence Drugs? Appl. Microbiol. Biotechnol. 2020, 104, 10409–10436. [Google Scholar] [CrossRef]
- Berning, M.; Krasz, S.; Miehlke, S. Should Quinolones Come First in Helicobacter Pylori Therapy? Ther. Adv. Gastroenterol. 2011, 4, 103–114. [Google Scholar] [CrossRef] [Green Version]
- Wentzell, L.M.; Maxwell, A. The Complex of DNA Gyrase and Quinolone Drugs on DNA Forms a Barrier to the T7 DNA Polymerase Replication Complex. J. Mol. Biol. 2000, 304, 779–791. [Google Scholar] [CrossRef]
- Willmott, C.J.R.; Critchlow, S.E.; Eperon, I.C.; Maxwell, A. The Complex of DNA Gyrase and Quinolone Drugs with DNA Forms a Barrier to Transcription by RNA Polymerase. J. Mol. Biol. 1994, 242, 351–363. [Google Scholar] [CrossRef]
- Hiasa, H. DNA Topoisomerases as Targets for Antibacterial Agents. In DNA Topoisomerases: Methods and Protocols; Methods in Molecular Biology; Drolet, M., Ed.; Springer: New York, NY, USA, 2018; pp. 47–62. ISBN 978-1-4939-7459-7. [Google Scholar]
- Ashley, R.E.; Dittmore, A.; McPherson, S.A.; Turnbough, C.L.; Neuman, K.C.; Osheroff, N. Activities of Gyrase and Topoisomerase IV on Positively Supercoiled DNA. Nucleic Acids Res. 2017, 45, 9611–9624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugino, A.; Peebles, C.L.; Kreuzer, K.N.; Cozzarelli, N.R. Mechanism of Action of Nalidixic Acid: Purification of Escherichia coli NalA Gene Product and Its Relationship to DNA Gyrase and a Novel Nicking-Closing Enzyme. Proc. Natl. Acad. Sci. USA 1977, 74, 4767–4771. [Google Scholar] [CrossRef] [Green Version]
- Forterre, P.; Gadelle, D. Phylogenomics of DNA Topoisomerases: Their Origin and Putative Roles in the Emergence of Modern Organisms. Nucleic Acids Res. 2009, 37, 679–692. [Google Scholar] [CrossRef] [Green Version]
- Colgan, A.M.; Quinn, H.J.; Kary, S.C.; Mitchenall, L.A.; Maxwell, A.; Cameron, A.D.S.; Dorman, C.J. Negative Supercoiling of DNA by Gyrase Is Inhibited in Salmonella enterica Serovar Typhimurium during Adaptation to Acid Stress. Mol. Microbiol. 2018, 107, 734–746. [Google Scholar] [CrossRef] [Green Version]
- Aldred, K.J.; Kerns, R.J.; Osheroff, N. Mechanism of Quinolone Action and Resistance. Biochemistry 2014, 53, 1565–1574. [Google Scholar] [CrossRef]
- Kreuzer, K.N.; Cozzarelli, N.R. Escherichia coli Mutants Thermosensitive for Deoxyribonucleic Acid Gyrase Subunit A: Effects on Deoxyribonucleic Acid Replication, Transcription, and Bacteriophage Growth. J. Bacteriol. 1979, 140, 424–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khodursky, A.B.; Zechiedrich, E.L.; Cozzarelli, N.R. Topoisomerase IV Is a Target of Quinolones in Escherichia coli. Proc. Natl. Acad. Sci. USA 1995, 92, 11801–11805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gellert, M.; Mizuuchi, K.; O’Dea, M.H.; Itoh, T.; Tomizawa, J.-I. Nalidixic Acid Resistance: A Second Genetic Character Involved in DNA Gyrase Activity. Proc. Natl. Acad. Sci. USA 1977, 74, 4772–4776. [Google Scholar] [CrossRef] [Green Version]
- Hooper, D.C. Bacterial Topoisomerases, Anti-Topoisomerases, and Anti-Topoisomerase Resistance. Clin. Infect. Dis. 1998, 27, S54–S63. [Google Scholar] [CrossRef]
- Muñoz, R.; De La Campa, A.G. ParC Subunit of DNA Topoisomerase IV of Streptococcus pneumoniae Is a Primary Target of Fluoroquinolones and Cooperates with DNA Gyrase A Subunit in Forming Resistance Phenotype. Antimicrob. Agents Chemother. 1996, 40, 2252–2257. [Google Scholar] [CrossRef] [Green Version]
- Pan, X.S.; Ambler, J.; Mehtar, S.; Fisher, L.M. Involvement of Topoisomerase IV and DNA Gyrase as Ciprofloxacin Targets in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 1996, 40, 2321–2326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fournier, B.; Zhao, X.; Lu, T.; Drlica, K.; Hooper, D.C. Selective Targeting of Topoisomerase IV and DNA Gyrase in Staphylococcus aureus: Different Patterns of Quinolone- Induced Inhibition of DNA Synthesis. Antimicrob. Agents Chemother. 2000, 44, 2160–2165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ince, D.; Zhang, X.; Silver, L.C.; Hooper, D.C. Dual Targeting of DNA Gyrase and Topoisomerase IV: Target Interactions of Garenoxacin (BMS-284756, T-3811ME), a New Desfluoroquinolone. Antimicrob. Agents Chemother. 2002, 46, 3370–3380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nilius, A.M.; Shen, L.L.; Hensey-Rudloff, D.; Almer, L.S.; Beyer, J.M.; Balli, D.J.; Cai, Y.; Flamm, R.K. In Vitro Antibacterial Potency and Spectrum of ABT-492, a New Fluoroquinolone. Antimicrob. Agents Chemother. 2003, 47, 3260–3269. [Google Scholar] [CrossRef] [Green Version]
- Markham, A. Delafloxacin: First Global Approval. Drugs 2017, 77, 1481–1486. [Google Scholar] [CrossRef] [Green Version]
- Cambau, E.; Matrat, S.; Pan, X.-S.; Roth Dit Bettoni, R.; Corbel, C.; Aubry, A.; Lascols, C.; Driot, J.-Y.; Fisher, L.M. Target Specificity of the New Fluoroquinolone Besifloxacin in Streptococcus pneumoniae, Staphylococcus aureus and Escherichia coli. J. Antimicrob. Chemother. 2009, 63, 443–450. [Google Scholar] [CrossRef] [Green Version]
- Pham, T.D.M.; Ziora, Z.M.; Blaskovich, M.A.T. Quinolone Antibiotics. Med. Chem. Commun. 2019, 10, 1719–1739. [Google Scholar] [CrossRef]
- Elshaboury, R.H.; Dilworth, T.J.; Rotschafer, J.C. Pharmacodynamics of Fluoroquinolones. In Antibiotic Pharmacodynamics; Methods in Pharmacology and Toxicology; Rotschafer, J.C., Andes, D.R., Rodvold, K.A., Eds.; Springer: New York, NY, USA, 2016; pp. 177–198. ISBN 978-1-4939-3323-5. [Google Scholar]
- Cuprys, A.; Pulicharla, R.; Brar, S.K.; Drogui, P.; Verma, M.; Surampalli, R.Y. Fluoroquinolones Metal Complexation and Its Environmental Impacts. Coord. Chem. Rev. 2018, 376, 46–61. [Google Scholar] [CrossRef]
- Drlica, K.; Zhao, X.; Malik, M.; Salz, T.; Kerns, R. Fluoroquinolone Resistance: Mechanisms, Restrictive Dosing, and Anti-Mutant Screening Strategies for New Compounds. In Antibiotic Discovery and Development; Dougherty, T.J., Pucci, M.J., Eds.; Springer: Boston, MA, USA, 2012; pp. 485–514. ISBN 978-1-4614-1400-1. [Google Scholar]
- Mustaev, A.; Malik, M.; Zhao, X.; Kurepina, N.; Luan, G.; Oppegard, L.M.; Hiasa, H.; Marks, K.R.; Kerns, R.J.; Berger, J.M.; et al. Fluoroquinolone-Gyrase-DNA Complexes. J. Biol. Chem. 2014, 289, 12300–12312. [Google Scholar] [CrossRef] [Green Version]
- Wohlkonig, A.; Chan, P.F.; Fosberry, A.P.; Homes, P.; Huang, J.; Kranz, M.; Leydon, V.R.; Miles, T.J.; Pearson, N.D.; Perera, R.L.; et al. Structural Basis of Quinolone Inhibition of Type IIA Topoisomerases and Target-Mediated Resistance. Nat. Struct. Mol. Biol. 2010, 17, 1152–1153. [Google Scholar] [CrossRef]
- Malik, M.; Mustaev, A.; Schwanz, H.A.; Luan, G.; Shah, N.; Oppegard, L.M.; de Souza, E.C.; Hiasa, H.; Zhao, X.; Kerns, R.J.; et al. Suppression of Gyrase-Mediated Resistance by C7 Aryl Fluoroquinolones. Nucleic Acids Res. 2016, 44, 3304–3316. [Google Scholar] [CrossRef] [Green Version]
- Marians, K.J.; Hiasa, H. Mechanism of Quinolone Action: A drug-induced structural perturbation of the DNA precedes strand cleavage by Topoisomerase IV. J. Biol. Chem. 1997, 272, 9401–9409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kampranis, S.C.; Maxwell, A. The DNA Gyrase-Quinolone Complex. ATP Hydrolysis and the Mechanism of DNA Cleavage. J. Biol. Chem. 1998, 273, 22615–22626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Critchlow, S.; Maxwell, A. Biochemistry; American Chemical Society: Washington, DC, USA, 1996; pp. 7387–7393. [Google Scholar]
- Drlica, K.; Hiasa, H.; Kerns, R.; Malik, M.; Mustaev, A.; Zhao, X. Quinolones: Action and Resistance Updated. Curr. Top. Med. Chem. 2009, 9, 981–998. [Google Scholar] [CrossRef]
- Hiasa, H.; Shea, M.E. DNA Gyrase-Mediated Wrapping of the DNA Strand Is Required for the Replication Fork Arrest by the DNA Gyrase-Quinolone-DNA Ternary Complex. J. Biol. Chem. 2000, 275, 34780–34786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, V.E.; Zaniewski, R.P.; Kaczmarek, F.S.; Gootz, T.D.; Osheroff, N. Quinolones Inhibit DNA Religation Mediated by Staphylococcus aureus Topoisomerase IV: Changes in drug mechanism across evolutionary boundaries. J. Biol. Chem. 1999, 274, 35927–35932. [Google Scholar] [CrossRef] [Green Version]
- Laponogov, I.; Sohi, M.K.; Veselkov, D.A.; Pan, X.-S.; Sawhney, R.; Thompson, A.W.; McAuley, K.E.; Fisher, L.M.; Sanderson, M.R. Structural Insight into the Quinolone-DNA Cleavage Complex of Type IIA Topoisomerases. Nat. Struct. Mol. Biol. 2009, 16, 667–669. [Google Scholar] [CrossRef]
- Chen, C.R.; Malik, M.; Snyder, M.; Drlica, K. DNA Gyrase and Topoisomerase IV on the Bacterial Chromosome: Quinolone-Induced DNA Cleavage. J. Mol. Biol. 1996, 258, 627–637. [Google Scholar] [CrossRef]
- Drlica, K.; Malik, M.; Kerns, R.J.; Zhao, X. Quinolone-Mediated Bacterial Death. Antimicrob. Agents Chemother. 2008, 52, 385–392. [Google Scholar] [CrossRef] [Green Version]
- Malik, M.; Hussain, S.; Drlica, K. Effect of Anaerobic Growth on Quinolone Lethality with Escherichia coli. Antimicrob. Agents Chemother. 2007, 51, 28–34. [Google Scholar] [CrossRef] [Green Version]
- Hong, Y.; Zeng, J.; Wang, X.; Drlica, K.; Zhao, X. Post-Stress Bacterial Cell Death Mediated by Reactive Oxygen Species. Proc. Natl. Acad. Sci. USA 2019, 116, 10064–10071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A Common Mechanism of Cellular Death Induced by Bactericidal Antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baquero, F.; Levin, B.R. Proximate and Ultimate Causes of the Bactericidal Action of Antibiotics. Nat. Rev. Microbiol. 2021, 19, 123–132. [Google Scholar] [CrossRef]
- Luan, G.; Hong, Y.; Drlica, K.; Zhao, X. Suppression of Reactive Oxygen Species Accumulation Accounts for Paradoxical Bacterial Survival at High Quinolone Concentration. Antimicrob. Agents Chemother. 2018, 62, e01622-17. [Google Scholar] [CrossRef] [Green Version]
- Bhatnagar, K.; Wong, A. The Mutational Landscape of Quinolone Resistance in Escherichia coli. PLoS ONE 2019, 14, e0224650. [Google Scholar] [CrossRef] [Green Version]
- Baker, S.; Duy, P.T.; Nga, T.V.T.; Dung, T.T.N.; Phat, V.V.; Chau, T.T.; Turner, A.K.; Farrar, J.; Boni, M.F. Fitness Benefits in Fluoroquinolone-Resistant Salmonella Typhi in the Absence of Antimicrobial Pressure. eLife 2013, 2, e01229. [Google Scholar] [CrossRef] [PubMed]
- Castro, R.A.D.; Ross, A.; Kamwela, L.; Reinhard, M.; Loiseau, C.; Feldmann, J.; Borrell, S.; Trauner, A.; Gagneux, S. The Genetic Background Modulates the Evolution of Fluoroquinolone-Resistance in Mycobacterium tuberculosis. Mol. Biol. Evol. 2020, 37, 195–207. [Google Scholar] [CrossRef] [Green Version]
- Vergalli, J.; Bodrenko, I.V.; Masi, M.; Moynié, L.; Acosta-Gutiérrez, S.; Naismith, J.H.; Davin-Regli, A.; Ceccarelli, M.; van den Berg, B.; Winterhalter, M.; et al. Porins and Small-Molecule Translocation across the Outer Membrane of Gram-Negative Bacteria. Nat. Rev. Microbiol. 2020, 18, 164–176. [Google Scholar] [CrossRef]
- Sousa, C.; Coimbra, J.T.S.; Gomes, I.; Franco, R.; Fernandes, P.A.; Gameiro, P. The Binding of Free and Copper-Complexed Fluoroquinolones to OmpF Porins: An Experimental and Molecular Docking Study. RSC Adv. 2017, 7, 10009–10019. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, M.; Gameiro, P. Ciprofloxacin Metalloantibiotic: An Effective Antibiotic with an Influx Route Strongly Dependent on Lipid Interaction? J. Membr. Biol. 2015, 248, 125–136. [Google Scholar] [CrossRef]
- Toro, C.S.; Lobos, S.R.; Calderón, I.; Rodríguez, M.; Mora, G.C. Clinical Isolate of a Porinless Salmonella Typhi Resistant to High Levels of Chloramphenicol. Antimicrob. Agents Chemother. 1990, 34, 1715–1719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karczmarczyk, M.; Martins, M.; Quinn, T.; Leonard, N.; Fanning, S. Mechanisms of Fluoroquinolone Resistance in Escherichia coli Isolates from Food-Producing Animals. Appl. Environ. Microbiol. 2011, 77, 7113–7120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rushdy, A.A.; Mabrouk, M.I.; Abu-Sef, F.A.-H.; Kheiralla, Z.H.; Abdel-All, S.M.; Saleh, N.M. Contribution of Different Mechanisms to the Resistance to Fluoroquinolones in Clinical Isolates of Salmonella enterica. Braz. J. Infect. Dis. 2013, 17, 431–437. [Google Scholar] [CrossRef] [Green Version]
- Ballesté-Delpierre, C.; Fàbrega, A.; Ferrer-Navarro, M.; Mathur, R.; Ghosh, S.; Vila, J. Attenuation of in Vitro Host–Pathogen Interactions in Quinolone-Resistant Salmonella Typhi Mutants. J. Antimicrob. Chemother. 2016, 71, 111–122. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Zhang, H.; Zheng, Y.; Li, A.; Wang, M.; Zhou, H.; Zhu, X.; Schneider, Z.; Chen, L.; Kreiswirth, B.N.; et al. RpoE Is a Putative Antibiotic Resistance Regulator of Salmonella enteric Serovar Typhi. Curr. Microbiol. 2016, 72, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Vidovic, S.; An, R.; Rendahl, A. Molecular and Physiological Characterization of Fluoroquinolone-Highly Resistant Salmonella Enteritidis Strains. Front. Microbiol. 2019, 10, 729. [Google Scholar] [CrossRef]
- Du, D.; Wang-Kan, X.; Neuberger, A.; van Veen, H.W.; Pos, K.M.; Piddock, L.J.V.; Luisi, B.F. Multidrug Efflux Pumps: Structure, Function and Regulation. Nat. Rev. Microbiol. 2018, 16, 523–539. [Google Scholar] [CrossRef]
- Attia, N.M.; ElBaradei, A. Fluoroquinolone Resistance Conferred by GyrA, ParC Mutations, and AbaQ Efflux Pump among Acinetobacter baumannii Clinical Isolates Causing Ventilator-Associated Pneumonia. Acta Microbiol. Immunol. Hung. 2020, 67, 234–238. [Google Scholar] [CrossRef]
- Martínez, J.L. Mechanisms of Action and of Resistance to Quinolones. Antibiot. Drug Resist. 2019, 1, 39–55. [Google Scholar] [CrossRef]
- Fàbrega, A.; Martin, R.G.; Rosner, J.L.; Tavio, M.M.; Vila, J. Constitutive SoxS Expression in a Fluoroquinolone-Resistant Strain with a Truncated SoxR Protein and Identification of a New Member of the MarA-SoxS-Rob Regulon, MdtG. Antimicrob. Agents Chemother. 2010, 54, 1218–1225. [Google Scholar] [CrossRef] [Green Version]
- Oethinger, M.; Podglajen, I.; Kern, W.V.; Levy, S.B. Overexpression of the MarA or SoxS Regulatory Gene in Clinical Topoisomerase Mutants of Escherichia coli. Antimicrob. Agents Chemother. 1998, 42, 2089–2094. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Haycocks, J.R.J.; Middlemiss, A.D.; Kettles, R.A.; Sellars, L.E.; Ricci, V.; Piddock, L.J.V.; Grainger, D.C. The Multiple Antibiotic Resistance Operon of Enteric Bacteria Controls DNA Repair and Outer Membrane Integrity. Nat. Commun. 2017, 8, 1444. [Google Scholar] [CrossRef] [Green Version]
- Villagra, N.A.; Valenzuela, L.M.; Mora, A.Y.; Millanao, A.R.; Saavedra, C.P.; Mora, G.C.; Hidalgo, A.A. Cysteine Auxotrophy Drives Reduced Susceptibility to Quinolones and Paraquat by Inducing the Expression of Efflux-Pump Systems and Detoxifying Enzymes in S. Typhimurium. Biochem. Biophys. Res. Commun. 2019, 515, 339–344. [Google Scholar] [CrossRef]
- Millanao, A.R.; Mora, A.Y.; Saavedra, C.P.; Villagra, N.A.; Mora, G.C.; Hidalgo, A.A. Inactivation of Glutamine Synthetase-Coding Gene GlnA Increases Susceptibility to Quinolones Through Increasing Outer Membrane Protein F in Salmonella enterica Serovar Typhi. Front. Microbiol. 2020, 11, 428. [Google Scholar] [CrossRef] [Green Version]
- Yeung, A.T.Y.; Bains, M.; Hancock, R.E.W. The Sensor Kinase CbrA Is a Global Regulator That Modulates Metabolism, Virulence, and Antibiotic Resistance in Pseudomonas Aeruginosa. J. Bacteriol. 2011, 193, 918–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez, J.L.; Rojo, F. Metabolic Regulation of Antibiotic Resistance. FEMS Microbiol. Rev. 2011, 35, 768–789. [Google Scholar] [CrossRef] [PubMed]
- Stokes, J.M.; Lopatkin, A.J.; Lobritz, M.A.; Collins, J.J. Bacterial Metabolism and Antibiotic Efficacy. Cell Metab. 2019, 30, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Villagra, N.A.; Fuentes, J.A.; Jofré, M.R.; Hidalgo, A.A.; García, P.; Mora, G.C. The Carbon Source Influences the Efflux Pump-Mediated Antimicrobial Resistance in Clinically Important Gram-Negative Bacteria. J. Antimicrob. Chemother. 2012, 67, 921–927. [Google Scholar] [CrossRef] [Green Version]
- Hidalgo, A.A.; Arias, Á.J.; Fuentes, J.A.; García, P.; Mora, G.C.; Villagra, N.A. Xylose Improves Antibiotic Activity of Chloramphenicol and Tetracycline against K. pneumoniae and A. baumannii in a Murine Model of Skin Infection. Available online: https://www.hindawi.com/journals/cjidmm/2018/3467219/ (accessed on 28 January 2021).
- Zhang, H.; Chang, M.; Zhang, X.; Cai, P.; Dai, Y.; Song, T.; Wu, Z.; Xu, H.; Qiao, M. Functional Identification and Evolutionary Analysis of Two Novel Plasmids Mediating Quinolone Resistance in Proteus vulgaris. Microorganisms 2020, 8, 1074. [Google Scholar] [CrossRef]
- Nishikawa, R.; Murase, T.; Ozaki, H. Plasmid-Mediated Quinolone Resistance in Escherichia coli Isolates from Commercial Broiler Chickens and Selection of Fluoroquinolone-Resistant Mutants. Poult. Sci. 2019, 98, 5900–5907. [Google Scholar] [CrossRef]
- Cheng, P.; Yang, Y.; Li, F.; Li, X.; Liu, H.; Fazilani, S.A.; Guo, W.; Xu, G.; Zhang, X. The Prevalence and Mechanism of Fluoroquinolone Resistance in Escherichia coli Isolated from Swine Farms in China. BMC Vet. Res. 2020, 16, 258. [Google Scholar] [CrossRef] [PubMed]
- Geetha, P.V.; Aishwarya, K.V.L.; Mariappan, S.; Sekar, U. Fluoroquinolone Resistance in Clinical Isolates of Klebsiella pneumonia e. J. Lab. Physicians 2020, 12, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhang, D.; Song, Q. Profiles of GyrA Mutations and Plasmid-Mediated Quinolone Resistance Genes in Shigella Isolates with Different Levels of Fluoroquinolone Susceptibility. Infect. Drug Resist. 2020, 13, 2285–2290. [Google Scholar] [CrossRef]
- Zhang, C.-Z.; Zhang, Y.; Ding, X.-M.; Lin, X.-L.; Lian, X.-L.; Trampari, E.; Thomson, N.M.; Ding, H.-Z.; Webber, M.A.; Jiang, H.-X. Emergence of Ciprofloxacin Heteroresistance in Foodborne Salmonella enterica Serovar Agona. J. Antimicrob. Chemother. 2020, 75, 2773–2779. [Google Scholar] [CrossRef] [PubMed]
- Pachanon, R.; Koide, K.; Kongsoi, S.; Nakajima, C.; Kapalamula, T.F.; Suthienkul, O.; Suzuki, Y. Interaction of the Plasmid-Encoded Quinolone Resistance Protein QnrB19 with Salmonella Typhimurium DNA Gyrase. J. Infect. Chemother. 2020, 26, 1139–1145. [Google Scholar] [CrossRef]
- Vinué, L.; Sater, M.R.A.; Herriott, I.C.; Huntley, M.H.; Wang, M.; Jacoby, G.A.; Hooper, D.C. Plasmids and Genes Contributing to High-Level Quinolone Resistance in Escherichia coli. Int. J. Antimicrob. Agents 2020, 56, 105987. [Google Scholar] [CrossRef]
- Vieira, D.C.; Lima, W.G.; de Paiva, M.C. Plasmid-Mediated Quinolone Resistance (PMQR) among Enterobacteriales in Latin America: A Systematic Review. Mol. Biol. Rep. 2020, 47, 1471–1483. [Google Scholar] [CrossRef]
- Rodríguez-Martínez, J.M.; Machuca, J.; Cano, M.E.; Calvo, J.; Martínez-Martínez, L.; Pascual, A. Plasmid-Mediated Quinolone Resistance: Two Decades On. Drug Resist. Updates 2016, 29, 13–29. [Google Scholar] [CrossRef]
- Zhang, Y.; Zheng, Z.; Chan, E.W.-C.; Dong, N.; Xia, X.; Chen, S. Molecular Characterization of QnrVC Genes and Their Novel Alleles in Vibrio Spp. Isolated from Food Products in China. Antimicrob. Agents Chemother. 2018, 62, e01882-17. [Google Scholar] [CrossRef] [Green Version]
- Xiong, X.; Bromley, E.H.C.; Oelschlaeger, P.; Woolfson, D.N.; Spencer, J. Structural Insights into Quinolone Antibiotic Resistance Mediated by Pentapeptide Repeat Proteins: Conserved Surface Loops Direct the Activity of a Qnr Protein from a Gram-Negative Bacterium. Nucleic Acids Res. 2011, 39, 3917–3927. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.S.; Chen, C.; Braun, M.; Kim, H.Y.; Okumura, R.; Wang, Y.; Jacoby, G.A.; Hooper, D.C. Interactions between QnrB, QnrB Mutants, and DNA Gyrase. Antimicrob. Agents Chemother. 2015, 59, 5413–5419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno-Switt, A.I.; Pezoa, D.; Sepúlveda, V.; González, I.; Rivera, D.; Retamal, P.; Navarrete, P.; Reyes-Jara, A.; Toro, M. Transduction as a Potential Dissemination Mechanism of a Clonal QnrB19-Carrying Plasmid Isolated from Salmonella of Multiple Serotypes and Isolation Sources. Front. Microbiol. 2019, 10, 2503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Zhang, Y.; Zhou, X.; Hu, X.; Zhou, Y.; Liu, D.; Maxwell, A.; Mi, K. The Plasmid-Borne Quinolone Resistance Protein QnrB, a Novel DnaA-Binding Protein, Increases the Bacterial Mutation Rate by Triggering DNA Replication Stress. Mol. Microbiol. 2019, 111, 1529–1543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinué, L.; Corcoran, M.A.; Hooper, D.C.; Jacoby, G.A. Mutations That Enhance the Ciprofloxacin Resistance of Escherichia coli with QnrA1. Antimicrob. Agents Chemother. 2016, 60, 1537–1545. [Google Scholar] [CrossRef] [Green Version]
- Aedo, S.; Ivanova, L.; Tomova, A.; Cabello, F.C. Plasmid-Related Quinolone Resistance Determinants in Epidemic Vibrio parahaemolyticus, Uropathogenic Escherichia coli, and Marine Bacteria from an Aquaculture Area in Chile. Microb. Ecol. 2014, 68, 324–328. [Google Scholar] [CrossRef]
- Chávez-Jacobo, V.M.; Hernández-Ramírez, K.C.; Silva-Sánchez, J.; Garza-Ramos, U.; Barrios-Camacho, H.; Ortiz-Alvarado, R.; Cervantes, C.; Meza-Carmen, V.; Ramírez-Díaz, M.I. Prevalence of the CrpP Gene Conferring Decreased Ciprofloxacin Susceptibility in Enterobacterial Clinical Isolates from Mexican Hospitals. J. Antimicrob. Chemother. 2019, 74, 1253–1259. [Google Scholar] [CrossRef]
- Yamane, K.; Wachino, J.; Suzuki, S.; Kimura, K.; Shibata, N.; Kato, H.; Shibayama, K.; Konda, T.; Arakawa, Y. New Plasmid-Mediated Fluoroquinolone Efflux Pump, QepA, Found in an Escherichia coli Clinical Isolate. Antimicrob. Agents Chemother. 2007, 51, 3354–3360. [Google Scholar] [CrossRef] [Green Version]
- Cattoir, V.; Poirel, L.; Nordmann, P. Plasmid-Mediated Quinolone Resistance Pump QepA2 in an Escherichia coli Isolate from France. Antimicrob. Agents Chemother. 2008, 52, 3801–3804. [Google Scholar] [CrossRef] [Green Version]
- Sørensen, A.H.; Hansen, L.H.; Johannesen, E.; Sørensen, S.J. Conjugative Plasmid Conferring Resistance to Olaquindox. Antimicrob. Agents Chemother. 2003, 47, 798–799. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Chen, G.; Liu, Z.; Xu, D.; Wu, Z.; Li, Z.; Hong, M. Site-Directed Mutagenesis Reveals Crucial Residues in Escherichia coli Resistance-Nodulation-Division Efflux Pump OqxB. Microb. Drug Resist. 2020, 26, 550–560. [Google Scholar] [CrossRef]
- Hansen, L.H.; Jensen, L.B.; Sørensen, H.I.; Sørensen, S.J. Substrate Specificity of the OqxAB Multidrug Resistance Pump in Escherichia coli and Selected Enteric Bacteria. J. Antimicrob. Chemother. 2007, 60, 145–147. [Google Scholar] [CrossRef]
- Yan, L.; Liu, D.; Wang, X.-H.; Wang, Y.; Zhang, B.; Wang, M.; Xu, H. Bacterial Plasmid-Mediated Quinolone Resistance Genes in Aquatic Environments in China. Sci. Rep. 2017, 7, 40610. [Google Scholar] [CrossRef]
- Recacha, E.; Machuca, J.; de Alba, P.D.; Ramos-Güelfo, M.; Docobo-Pérez, F.; Rodriguez-Beltrán, J.; Blázquez, J.; Pascual, A.; Rodríguez-Martínez, J.M. Quinolone Resistance Reversion by Targeting the SOS Response. mBio 2017, 8, e00971-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatnagar, K.; Hinz, A.; Kohlman, M.; Wong, A. An SRNA Screen for Reversal of Quinolone Resistance in Escherichia coli. G3 Genes Genomes Genet. 2020, 10, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.; Ahmad, F.; Yaqub, B.; Ramzan, A.; Imran, A.; Afzaal, M.; Mirza, S.A.; Mazhar, I.; Younus, M.; Akram, Q.; et al. Chapter 4—Current trends of antimicrobials used in food animals and aquaculture. In Antibiotics and Antimicrobial Resistance Genes in the Environment; Hashmi, M.Z., Ed.; Advances in Environmental Pollution Research Series; Elsevier: Amsterdam, The Netherlands, 2020; Volume 1, pp. 39–69. ISBN 978-0-12-818882-8. [Google Scholar]
- Committee for Medicinal Products for Veterinary Use (CVMP), European Medicines Agency (EMA). Reflection Paper on the Use of Fluoroquinolones in Food-Producing Animals in the European Union: Development of Resistance and Impact on Human and Animal Health; European Medicines Agency: Amsterdam, the Netherlands, 2006; p. 26. [Google Scholar]
- Gouvêa, R.; dos Santos, F.; de Aquino, M.; Pereira, V.L.d.A. Fluoroquinolones in Industrial Poultry Production, Bacterial Resistance and Food Residues: A Review. Braz. J. Poult. Sci. 2015, 17, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.-Q.; Ying, G.-G.; Pan, C.-G.; Liu, Y.-S.; Zhao, J.-L. Comprehensive Evaluation of Antibiotics Emission and Fate in the River Basins of China: Source Analysis, Multimedia Modeling, and Linkage to Bacterial Resistance. Environ. Sci. Technol. 2015, 49, 6772–6782. [Google Scholar] [CrossRef]
- Rakesh, K.; Ganapathi Naik, M.; Pinto, N.; Dharmakar, P.; Pai, M.; Anjusha, K. A Review on Drugs Used in Shrimp Aquaculture. Int. J. Pure App. Biosci. 2018, 6, 77–86. [Google Scholar] [CrossRef]
- Nawaz, M.; Sung, K.; Kweon, O.; Khan, S.; Nawaz, S.; Steele, R. Characterization of Novel Mutations Involved in Quinolone Resistance in Escherichia coli Isolated from Imported Shrimp. Int. J. Antimicrob. Agents 2015, 45, 471–476. [Google Scholar] [CrossRef]
- Millanao, A.R.; Barrientos, M.; Gómez, C.; Tomova, A.; Buschmann, A.; Dölz, H.; Cabello, F.C. Uso Inadecuado y Excesivo de Antibióticos: Salud Pública y Salmonicultura En Chile. Rev. Méd. Chile 2011, 139, 107–118. [Google Scholar] [CrossRef] [Green Version]
- Davis, J.L.; Smith, G.W.; Baynes, R.E.; Tell, L.A.; Webb, A.I.; Riviere, J.E. Update on Drugs Prohibited from Extralabel Use in Food Animals. J. Am. Vet. Med. Assoc. 2009, 235, 528–534. [Google Scholar] [CrossRef]
- Tyson, G.H.; Tate, H.P.; Zhao, S.; Li, C.; Dessai, U.; Simmons, M.; McDermott, P.F. Identification of Plasmid-Mediated Quinolone Resistance in Salmonella Isolated from Swine Ceca and Retail Pork Chops in the United States. Antimicrob. Agents Chemother. 2017, 61, e00653-18. [Google Scholar] [CrossRef] [Green Version]
- Roth, N.; Käsbohrer, A.; Mayrhofer, S.; Zitz, U.; Hofacre, C.; Domig, K. The Application of Antibiotics in Broiler Production and the Resulting Antibiotic Resistance in Escherichia coli: A Global Overview|Elsevier Enhanced Reader. Poult. Sci. 2019, 98, 1791–1804. [Google Scholar] [CrossRef]
- Abd El-Hamid, M.I.; Awad, N.F.S.; Hashem, Y.M.; Abdel-Rahman, M.A.; Abdelaziz, A.M.; Mohammed, I.A.A.; Abo-Shama, U.H. In Vitro Evaluation of Various Antimicrobials against Field Mycoplasma gallisepticum and Mycoplasma synoviae Isolates in Egypt. Poult. Sci. 2019, 98, 6281–6288. [Google Scholar] [CrossRef] [PubMed]
- Vanni, M.; Meucci, V.; Tognetti, R.; Cagnardi, P.; Montesissa, C.; Piccirillo, A.; Rossi, A.M.; Di Bello, D.; Intorre, L. Fluoroquinolone Resistance and Molecular Characterization of GyrA and ParC Quinolone Resistance-Determining Regions in Escherichia coli Isolated from Poultry. Poult. Sci. 2014, 93, 856–863. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Huo, S.; Li, Y.; Chen, L.; Zhang, F.; Wu, X. Molecular Epidemiological Survey on Quinolone Resistance Genotype and Phenotype of Escherichia coli in Septicemic Broilers in Hebei, China. Poult. Sci. 2014, 93, 335–339. [Google Scholar] [CrossRef]
- Nelson, J.M.; Chiller, T.M.; Powers, J.H.; Angulo, F.J. Fluoroquinolone-Resistant Campylobacter Species and the Withdrawal of Fluoroquinolones from Use in Poultry: A Public Health Success Story. Clin. Infect. Dis. 2007, 44, 977–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zawack, K.; Li, M.; Booth, J.G.; Love, W.; Lanzas, C.; Gröhn, Y.T. Monitoring Antimicrobial Resistance in the Food Supply Chain and Its Implications for FDA Policy Initiatives. Antimicrob. Agents Chemother. 2016, 60, 5302–5311. [Google Scholar] [CrossRef] [Green Version]
- Love, D.C.; Fry, J.P.; Cabello, F.; Good, C.M.; Lunestad, B.T. Veterinary Drug Use in United States Net Pen Salmon Aquaculture: Implications for Drug Use Policy. Aquaculture 2020, 518, 734820. [Google Scholar] [CrossRef]
- Cabello, F.C.; Godfrey, H.P.; Tomova, A.; Ivanova, L.; Dölz, H.; Millanao, A.; Buschmann, A.H. Antimicrobial Use in Aquaculture Re-Examined: Its Relevance to Antimicrobial Resistance and to Animal and Human Health. Environ. Microbiol. 2013, 15, 1917–1942. [Google Scholar] [CrossRef]
- Werner, N.; McEwen, S.; Kreienbrock, L. Monitoring Antimicrobial Drug Usage in Animals: Methods and Applications. In Antimicrobial Resistance in Bacteria from Livestock and Companion Animals; Wiley: Hoboken, NJ, USA, 2018; pp. 569–594. [Google Scholar] [CrossRef] [Green Version]
- WHO. Global Action Plan on Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2015; p. 28. [Google Scholar]
- OIE World Organisation for Animal Health. OIE Annual Report on Antimicrobial Agents Intended for Use in Animals; Third Report; OIE: Paris, France, 2018; p. 131. [Google Scholar]
- FDA U.S. Food and Drug Administration. Antimicrobials Sold or Distributed in 2019 for Use in Food-Producing Animals; FDA: Silver Spring, MD, USA, 2020; p. 49. [Google Scholar]
- FDA U.S. Food and Drug Administration. Antimicrobials Sold or Distributed in 2018 for Use in Food-Producing Animals; FDA: Silver Spring, MD, USA, 2019; p. 49. [Google Scholar]
- FDA U.S. Food and Drug Administration. Antimicrobials Sold or Distributed in 2013 for Use in Food-Producing Animals; FDA: Silver Spring, MD, USA, 2015; p. 57. [Google Scholar]
- FDA U.S. Food and Drug Administration. Antimicrobials Sold or Distributed in 2014 for Use in Food-Producing Animals; FDA: Silver Spring, MD, USA, 2015; p. 58. [Google Scholar]
- FDA U.S. Food and Drug Administration. Antimicrobials Sold or Distributed in 2015 for Use in Food-Producing Animals; FDA: Silver Spring, MD, USA, 2016; p. 58. [Google Scholar]
- FDA U.S. Food and Drug Administration. Antimicrobials Sold or Distributed in 2016 for Use in Food-Producing Animals; FDA: Silver Spring, MD, USA, 2017; p. 67. [Google Scholar]
- FDA U.S. Food and Drug Administration. Antimicrobials Sold or Distributed in 2017 for Use in Food-Producing Animals; FDA: Silver Spring, MD, USA, 2018; p. 52. [Google Scholar]
- European Surveillance of Veterinary Antimicrobial Consumption, EMA European Medicines Agency. Sales of Veterinary Antimicrobial Agents in 31 European Countries in 2017; (EMA/294674/2019); EMA: Amsterdam, The Netherlands, 2019; p. 109. [Google Scholar]
- European Surveillance of Veterinary Antimicrobial Consumption, EMA European Medicines Agency. Sales of Veterinary Antimicrobial Agents in 30 European Countries in 2016; (EMA/275982/2018); EMA: Amsterdam, The Netherlands, 2018; p. 184. [Google Scholar]
- European Surveillance of Veterinary Antimicrobial Consumption, EMA European Medicines Agency. Sales of Veterinary Antimicrobial Agents in 30 European Countries in 2015; (EMA/184855/2017); EMA: Amsterdam, The Netherlands, 2017; p. 184. [Google Scholar]
- European Surveillance of Veterinary Antimicrobial Consumption, EMA European Medicines Agency. Sales of Veterinary Antimicrobial Agents in 29 European Countries in 2014; (EMA/61769/2016); EMA: Amsterdam, The Netherlands, 2016; p. 176. [Google Scholar]
- European Surveillance of Veterinary Antimicrobial Consumption, EMA European Medicines Agency. Sales of Veterinary Antimicrobial Agents in 26 EU/EEA Countries in 2013; (EMA/387934/2015); EMA: Amsterdam, The Netherlands, 2015; p. 162. [Google Scholar]
- European Surveillance of Veterinary Antimicrobial Consumption, EMA European Medicines Agency. Sales of Veterinary Antimicrobial Agents in 26 EU/EEA Countries in 2012; (EMA/333921/2014); EMA: Amsterdam, The Netherlands, 2014; p. 128. [Google Scholar]
- European Surveillance of Veterinary Antimicrobial Consumption, EMA European Medicines Agency. Sales of Veterinary Antimicrobial Agents in 25 EU/EEA Countries in 2011; (EMA/236501/2013); EMA: Amsterdam, The Netherlands, 2013; p. 98. [Google Scholar]
- European Surveillance of Veterinary Antimicrobial Consumption, EMA European Medicines Agency. Sales of Veterinary Antimicrobial Agents in 31 European Countries in 2018; (EMA/24309/2020); EMA: Amsterdam, The Netherlands, 2020; p. 104. [Google Scholar]
- NORM/NORM-VET. Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway 2019; NORM—Norwegian Surveillance System for Antimicrobial Drug Resistance/NORM-VET Norwegian Veterinary Antimicrobial Resistance Monitoring Tromsø: Oslo, Norway, 2020.
- NORM/NORM-VET. Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway 2018; NORM—Norwegian Surveillance System for Antimicrobial Drug Resistance/NORM-VET—Norwegian Veterinary Antimicrobial Resistance Monitoring Tromsø: Oslo, Norway, 2019.
- ANSES. Rapport de l’Anses: Suivi Des Ventes de Médicaments Vétérinaires Contenant Des Antibiotiques En France En 2018; Anses—Agence Nationale de Sécurité Sanitaire de L’alimentation, de L’environnement et Du Travail: París, France, 2019.
- Cuong, N.V.; Padungtod, P.; Thwaites, G.; Carrique-Mas, J.J. Antimicrobial Usage in Animal Production: A Review of the Literature with a Focus on Low- and Middle-Income Countries. Antibiotics 2018, 7, 75. [Google Scholar] [CrossRef] [Green Version]
- González Pereyra, V.; Pol, M.; Pastorino, F.; Herrero, A. Quantification of Antimicrobial Usage in Dairy Cows and Preweaned Calves in Argentina. Prev. Vet. Med. 2015, 122, 273–279. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global Trends in Antimicrobial Use in Food Animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649. [Google Scholar] [CrossRef] [Green Version]
- Millanao, A.R. Estudio Cualitativo y Cuantitativo de las Quinolonas y Fluoroquinolonas Importadas y Autorizadas Para Uso y Disposición en Medicina y en Veterinaria en Chile, en el Período 1998–2001. Consideraciones Sobre su Impacto Para la Salud Pública y el Medio Ambiente. Bachelor’s Thesis, Escuela de Química y Farmacia, Universidad Austral de Chile, Valdivia, Chile, 2002. [Google Scholar]
- Barrientos, M. Estudio Cualitativo y Cuantitativo de las Quinolonas y Fluoroquinolonas Importadas y Autorizadas Para Uso y Disposición en Medicina y en Veterinaria en Chile, en el Período 2002–2005. Consideraciones Sobre su Impacto Para la Salud Pública y el Medio Ambiente. Bachelor’s Thesis, Escuela de Química y Farmacia, Universidad Austral de Chile, Valdivia, Chile, 2006. [Google Scholar]
- SERNAPESCA. Informe Sobre Uso de Antimicrobianos en la Salmonicultura Nacional Año 2015; Servicio Nacional de Pesca: Valparaíso, Chile, 2016; p. 26.
- SERNAPESCA. Informe Sobre Uso de Antimicrobianos en la Salmonicultura Nacional Año 2016; Servicio Nacional de Pesca: Valparaíso, Chile, 2017; p. 13.
- SERNAPESCA. Informe Sobre Uso de Antimicrobianos en la Salmonicultura Nacional Año 2017; Servicio Nacional de Pesca: Valparaíso, Chile, 2017; p. 10.
- SERNAPESCA. Informe Sobre Uso de Antimicrobianos en la Salmonicultura Nacional Año 2018; Servicio Nacional de Pesca: Valparaíso, Chile, 2019; p. 10.
- SAG. Declaración de Venta de Antimicrobianos; Servicio Agrícola y Ganadero—SAG: Santiago, Chile, 2014–2019. Available online: https://www.sag.gob.cl/ambitos-de-accion/declaracion-de-venta-de-antimicrobianos (accessed on 13 March 2021).
- Millanao, A.R.; Barrientos, C.; Siegel-Tike, C.; Tomova, A.; Ivanova, L.; Godfrey, H.P.; Dölz, H.J.; Buschmann, A.H.; Cabello, F. Resistencia a los antimicrobianos en Chile y el paradigma de Una Salud: Manejando los riesgos para la salud pública humana y animal resultante del uso de antimicrobianos en la acuicultura del salmón y en medicina. Rev. Chil. Infectol. 2018, 35, 299–303. [Google Scholar] [CrossRef]
- Barrientos, C. Estudio Cualitativo y Cuantitativo de las Quinolonas y Fluoroquinolonas Importadas y Autorizadas Para Uso y Disposición en Medicina y en Veterinaria en Chile, en el Período 2013–2015. Consideraciones Sobre su Impacto Para la Salud Pública y el Medio Ambiente. Bachelor’s Thesis, Escuela de Química y Farmacia, Universidad Austral de Chile, Valdivia, Chile, 2016. [Google Scholar]
- Ritchie, H.; Roser, M. Meat and Dairy Production. Available online: https://ourworldindata.org/meat-production (accessed on 21 September 2020).
- Karam, G.; Chastre, J.; Wilcox, M.H.; Vincent, J.-L. Antibiotic Strategies in the Era of Multidrug Resistance. Crit. Care 2016, 20, 136. [Google Scholar] [CrossRef] [Green Version]
- Huber, L.; Agunos, A.; Gow, S.P.; Carson, C.A.; Boeckel, T.P.V. Reduction in Antimicrobial Use and Resistance to Salmonella, Campylobacter, and Escherichia coli in Broiler Chickens, Canada, 2013–2019. Emerg. Infect. Dis. 2021, 27, 2434–2444. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Jeannot, K.; Xiao, Y.; Knapp, C.W. Editorial: Horizontal Gene Transfer Mediated Bacterial Antibiotic Resistance. Front. Microbiol. 2019, 10, 1933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landers, T.F.; Cohen, B.; Wittum, T.E.; Larson, E.L. A Review of Antibiotic Use in Food Animals: Perspective, Policy, and Potential. Public Health Rep. 2012, 127, 4–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chantziaras, I.; Boyen, F.; Callens, B.; Dewulf, J. Correlation between Veterinary Antimicrobial Use and Antimicrobial Resistance in Food-Producing Animals: A Report on Seven Countries. J. Antimicrob. Chemother. 2014, 69, 827–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jans, C.; Sarno, E.; Collineau, L.; Meile, L.; Stärk, K.D.C.; Stephan, R. Consumer Exposure to Antimicrobial Resistant Bacteria from Food at Swiss Retail Level. Front. Microbiol. 2018, 9, 362. [Google Scholar] [CrossRef]
- Eguale, T.; Birungi, J.; Asrat, D.; Njahira, M.N.; Njuguna, J.; Gebreyes, W.A.; Gunn, J.S.; Djikeng, A.; Engidawork, E. Genetic Markers Associated with Resistance to Beta-Lactam and Quinolone Antimicrobials in Non-Typhoidal Salmonella Isolates from Humans and Animals in Central Ethiopia. Antimicrob. Resist. Infect. Control. 2017, 6, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, C.; He, J. Effect of Antibiotics in the Environment on Microbial Populations. Appl. Microbiol. Biotechnol. 2010, 87, 925–941. [Google Scholar] [CrossRef] [PubMed]
- Grenni, P.; Ancona, V.; Barra Caracciolo, A. Ecological Effects of Antibiotics on Natural Ecosystems: A Review. Microchem. J. 2018, 136, 25–39. [Google Scholar] [CrossRef]
- Cycoń, M.; Mrozik, A.; Piotrowska-Seget, Z. Antibiotics in the Soil Environment—Degradation and Their Impact on Microbial Activity and Diversity. Front. Microbiol. 2019, 10, 338. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, X.; Yang, Q.; Sun, L.; Yang, X.; Zhou, M.; Deng, R.; Bi, L. Plant Growth, Antibiotic Uptake, and Prevalence of Antibiotic Resistance in an Endophytic System of Pakchoi under Antibiotic Exposure. Int. J. Env. Res. Public Health 2017, 14, 1336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leal, R.M.P.; Alleoni, L.R.F.; Tornisielo, V.L.; Regitano, J.B. Sorption of Fluoroquinolones and Sulfonamides in 13 Brazilian Soils. Chemosphere 2013, 92, 979–985. [Google Scholar] [CrossRef] [Green Version]
- Riaz, L.; Mahmood, T.; Khalid, A.; Rashid, A.; Ahmed Siddique, M.B.; Kamal, A.; Coyne, M.S. Fluoroquinolones (FQs) in the Environment: A Review on Their Abundance, Sorption and Toxicity in Soil. Chemosphere 2018, 191, 704–720. [Google Scholar] [CrossRef]
- Van Doorslaer, X.; Dewulf, J.; Van Langenhove, H.; Demeestere, K. Fluoroquinolone Antibiotics: An Emerging Class of Environmental Micropollutants. Sci. Total Environ. 2014, 500–501, 250–269. [Google Scholar] [CrossRef]
- Völgyi, G.; Vizserálek, G.; Takács-Novák, K.; Avdeef, A.; Tam, K.Y. Predicting the Exposure and Antibacterial Activity of Fluoroquinolones Based on Physicochemical Properties. Eur. J. Pharm. Sci. 2012, 47, 21–27. [Google Scholar] [CrossRef]
- Kümmerer, K. (Ed.) Pharmaceuticals in the Environment: Sources, Fate, Effects and Risks, 3rd ed.; Springer: Berlin/Heidelberg, Germany, 2008; ISBN 978-3-540-74663-8. [Google Scholar]
- Aristilde, L.; Sposito, G. Molecular Modeling of Metal Complexation by a Fluoroquinolone Antibiotic. Environ Toxicol Chem 2008, 27, 2304–2310. [Google Scholar] [CrossRef]
- Zhao, F.; Yang, L.; Chen, L.; Xiang, Q.; Li, S.; Sun, L.; Yu, X.; Fang, L. Soil Contamination with Antibiotics in a Typical Peri-Urban Area in Eastern China: Seasonal Variation, Risk Assessment, and Microbial Responses. J. Environ. Sci. 2019, 79, 200–212. [Google Scholar] [CrossRef]
- Huang, L.; Mo, Y.; Wu, Z.; Rad, S.; Song, X.; Zeng, H.; Bashir, S.; Kang, B.; Chen, Z. Occurrence, Distribution, and Health Risk Assessment of Quinolone Antibiotics in Water, Sediment, and Fish Species of Qingshitan Reservoir, South China. Sci. Rep. 2020, 10, 15777. [Google Scholar] [CrossRef]
- Baietto, L.; Corcione, S.; Pacini, G.; Di Perri, G.; D’Avolio#†, A.; Giuseppe De Rosa†, F. A 30-Years Review on Pharmacokinetics of Antibiotics: Is the Right Time for Pharmacogenetics? Curr. Drug Metab. 2014, 15, 581–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sukul, P.; Lamshöft, M.; Kusari, S.; Zühlke, S.; Spiteller, M. Metabolism and Excretion Kinetics of 14C-Labeled and Non-Labeled Difloxacin in Pigs after Oral Administration, and Antimicrobial Activity of Manure Containing Difloxacin and Its Metabolites. Environ. Res. 2009, 109, 225–231. [Google Scholar] [CrossRef]
- Slana, M.; Pahor, V.; Maričič, L.C.; Sollner-Dolenc, M. Excretion Pattern of Enrofloxacin after Oral Treatment of Chicken Broilers. J. Vet. Pharmacol. Ther. 2014, 37, 611–614. [Google Scholar] [CrossRef] [PubMed]
- De Bairros, A.V.; Pereira, D.B.; Cordeiro, E.W.F.; Paim, C.S.; da Silva, F.E.B.; Malesuik, M.D.; Paula, F.R. Evaluation of the Influence of Fluoroquinolone Chemical Structure on Stability: Forced Degradation and in Silico Studies. Braz. J. Pharm. Sci. 2018, 54, e00188. [Google Scholar] [CrossRef]
- Babić, S.; Periša, M.; Škorić, I. Photolytic Degradation of Norfloxacin, Enrofloxacin and Ciprofloxacin in Various Aqueous Media. Chemosphere 2013, 91, 1635–1642. [Google Scholar] [CrossRef] [PubMed]
- Rusch, M.; Spielmeyer, A.; Zorn, H.; Hamscher, G. Degradation and Transformation of Fluoroquinolones by Microorganisms with Special Emphasis on Ciprofloxacin. Appl Microbiol. Biotechnol 2019, 103, 6933–6948. [Google Scholar] [CrossRef]
- Ahmadzadeh, S.; Asadipour, A.; Pournamdari, M.; Behnam, B.; Rahimi, H.R.; Dolatabadi, M. Removal of Ciprofloxacin from Hospital Wastewater Using Electrocoagulation Technique by Aluminum Electrode: Optimization and Modelling through Response Surface Methodology. Process. Saf. Environ. Prot. 2017, 109, 538–547. [Google Scholar] [CrossRef]
- Castrignanò, E.; Kannan, A.M.; Feil, E.; Kasprzyk-Hordern, B. Enantioselective Fractionation of Fluoroquinolones in the Aqueous Environment Using Chiral Liquid Chromatography Coupled with Tandem Mass Spectrometry. Chemosphere 2018, 206, 376–386. [Google Scholar] [CrossRef]
- Rodrigues-Silva, C.; Porto, R.; dos Santos, S.; Schneider, J.; Rath, S. Fluoroquinolones in Hospital Wastewater: Analytical Method, Occurrence, Treatment with Ozone and Residual Antimicrobial Activity Evaluation. J. Braz. Chem. Soc. 2019, 30, 1447–1457. [Google Scholar] [CrossRef]
- Sturini, M.; Speltini, A.; Maraschi, F.; Profumo, A.; Tarantino, S.; Gualtieri, A.F.; Zema, M. Removal of Fluoroquinolone Contaminants from Environmental Waters on Sepiolite and Its Photo-Induced Regeneration. Chemosphere 2016, 150, 686–693. [Google Scholar] [CrossRef]
- Lucas, D.; Badia-Fabregat, M.; Vicent, T.; Caminal, G.; Rodríguez-Mozaz, S.; Balcázar, J.L.; Barceló, D. Fungal Treatment for the Removal of Antibiotics and Antibiotic Resistance Genes in Veterinary Hospital Wastewater. Chemosphere 2016, 152, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Wang, S.-P.; Fu, J.; Zhou, Z.-Q.; Zhang, N.; Guo, L. Influence of Ciprofloxacin on Microbial Community Structure and Function in Soils. Biol. Fertil. Soils 2014, 50, 939–947. [Google Scholar] [CrossRef]
- Leonard, A.; Möhlis, K.; Schlüter, R.; Taylor, E.; Lalk, M.; Methling, K. Exploring Metabolic Adaptation of Streptococcus pneumoniae to Antibiotics. J. Antibiot. 2020, 73, 441–454. [Google Scholar] [CrossRef]
- Frade, V.M.F.; Dias, M.; Teixeira, A.C.S.C.; Palma, M.S.A.; Frade, V.M.F.; Dias, M.; Teixeira, A.C.S.C.; Palma, M.S.A. Environmental Contamination by Fluoroquinolones. Braz. J. Pharm. Sci. 2014, 50, 41–54. [Google Scholar] [CrossRef]
- Janecko, N.; Pokludova, L.; Blahova, J.; Svobodova, Z.; Literak, I. Implications of Fluoroquinolone Contamination for the Aquatic Environment—A Review. Environ. Toxicol. Chem. 2016, 35, 2647–2656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giebułtowicz, J.; Nałęcz-Jawecki, G.; Harnisz, M.; Kucharski, D.; Korzeniewska, E.; Płaza, G. Environmental Risk and Risk of Resistance Selection Due to Antimicrobials’ Occurrence in Two Polish Wastewater Treatment Plants and Receiving Surface Water. Molecules 2020, 25, 1470. [Google Scholar] [CrossRef] [Green Version]
- Rutgersson, C.; Fick, J.; Marathe, N.; Kristiansson, E.; Janzon, A.; Angelin, M.; Johansson, A.; Shouche, Y.; Flach, C.-F.; Larsson, D.G.J. Fluoroquinolones and Qnr Genes in Sediment, Water, Soil, and Human Fecal Flora in an Environment Polluted by Manufacturing Discharges. Available online: https://pubs.acs.org/doi/pdf/10.1021/es501452a (accessed on 7 October 2020).
- Conte, D.; Palmeiro, J.K.; da Silva Nogueira, K.; de Lima, T.M.R.; Cardoso, M.A.; Pontarolo, R.; Degaut Pontes, F.L.; Dalla-Costa, L.M. Characterization of CTX-M Enzymes, Quinolone Resistance Determinants, and Antimicrobial Residues from Hospital Sewage, Wastewater Treatment Plant, and River Water. Ecotoxicol. Environ. Saf. 2017, 136, 62–69. [Google Scholar] [CrossRef]
- Kaplan, E.; Marano, R.B.M.; Jurkevitch, E.; Cytryn, E. Enhanced Bacterial Fitness Under Residual Fluoroquinolone Concentrations Is Associated with Increased Gene Expression in Wastewater-Derived Qnr Plasmid-Harboring Strains. Front. Microbiol. 2018, 9, 1176. [Google Scholar] [CrossRef]
- Tomova, A.; Ivanova, L.; Buschmann, A.H.; Rioseco, M.L.; Kalsi, R.K.; Godfrey, H.P.; Cabello, F.C. Antimicrobial Resistance Genes in Marine Bacteria and Human Uropathogenic Escherichia coli from a Region of Intensive Aquaculture. Environ. Microbiol. Rep. 2015, 7, 803–809. [Google Scholar] [CrossRef]
- Tomova, A.; Ivanova, L.; Buschmann, A.H.; Godfrey, H.P.; Cabello, F.C. Plasmid-Mediated Quinolone Resistance (PMQR) Genes and Class 1 Integrons in Quinolone-Resistant Marine Bacteria and Clinical Isolates of Escherichia coli from an Aquacultural Area. Microb. Ecol. 2018, 75, 104–112. [Google Scholar] [CrossRef]
- Cabello, F.C.; Godfrey, H.P.; Ivanova, L.; Shah, S.Q.A.; Sørum, H.; Tomova, A. Freshwater Salmon Aquaculture in Chile and Transferable Antimicrobial Resistance—Cabello—2020—Environmental Microbiology—Wiley Online Library. Environ. Microbiol. 2020, 22, 559–563. [Google Scholar] [CrossRef]
- Domingues, S.; Nielsen, K.M. Membrane Vesicles and Horizontal Gene Transfer in Prokaryotes. Curr. Opin. Microbiol. 2017, 38, 16–21. [Google Scholar] [CrossRef]
- Qureshi, Z.A.; Hittle, L.E.; O’Hara, J.A.; Rivera, J.I.; Syed, A.; Shields, R.K.; Pasculle, A.W.; Ernst, R.K.; Doi, Y. Colistin-Resistant Acinetobacter baumannii: Beyond Carbapenem Resistance. Clin. Infect. Dis. 2015, 60, 1295–1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rani, M.F.; Rahman, N.I.A.; Ismail, S.; Alattraqchi, A.G.; Cleary, D.W.; Clarke, S.C.; Yeo, C.C. Acinetobacter Spp. Infections in Malaysia: A Review of Antimicrobial Resistance Trends, Mechanisms and Epidemiology. Front. Microbiol. 2017, 8, 2479. [Google Scholar] [CrossRef] [Green Version]
- Traglia, G.M.; Place, K.; Dotto, C.; Fernandez, J.S.; Montaña, S.; Bahiense, C.d.S.; Soler-Bistue, A.; Iriarte, A.; Perez, F.; Tolmasky, M.E.; et al. Interspecies DNA Acquisition by a Naturally Competent Acinetobacter baumannii Strain. Int. J. Antimicrob. Agents 2019, 53, 483–490. [Google Scholar] [CrossRef]
- Čvančarová, M.; Moeder, M.; Filipová, A.; Cajthaml, T. Biotransformation of Fluoroquinolone Antibiotics by Ligninolytic Fungi—Metabolites, Enzymes and Residual Antibacterial Activity. Chemosphere 2015, 136, 311–320. [Google Scholar] [CrossRef]
- WHO. Critically Important Antimicrobials for Human Medicine; 6th Revision; WHO: Geneve, Switzerland, 2019; Available online: http://www.who.int/foodsafety/publications/antimicrobials-sixth/en/ (accessed on 15 July 2020).
- Quesada, S.P.; Paschoal, J.A.R.; Reyes, F.G.R. Considerations on the Aquaculture Development and on the Use of Veterinary Drugs: Special Issue for Fluoroquinolones—A Review. J. Food Sci. 2013, 78, R1321–R1333. [Google Scholar] [CrossRef]
- Wu, C.-F.; Chen, C.-H.; Wu, C.-Y.; Lin, C.-S.; Su, Y.-C.; Wu, C.-F.; Tsai, H.-P.; Fan, P.-S.; Yeh, C.-H.; Yang, W.-C.; et al. Quinolone and Organophosphorus Insecticide Residues in Bivalves and Their Associated Risks in Taiwan. Molecules 2020, 25, 3636. [Google Scholar] [CrossRef] [PubMed]
- Zarkan, A.; Matuszewska, M.; Trigg, S.B.; Zhang, M.; Belgami, D.; Croft, C.; Liu, J.; El-Ouisi, S.; Greenhalgh, J.; Duboff, J.S.; et al. Inhibition of Indole Production Increases the Activity of Quinolone Antibiotics against E. coli Persisters. Sci. Rep. 2020, 10, 11742. [Google Scholar] [CrossRef] [PubMed]
- Allen, R.C.; Popat, R.; Diggle, S.P.; Brown, S.P. Targeting Virulence: Can We Make Evolution-Proof Drugs? Nat. Rev. Microbiol. 2014, 12, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Rezzoagli, C.; Archetti, M.; Mignot, I.; Baumgartner, M.; Kümmerli, R. Combining Antibiotics with Antivirulence Compounds Can Have Synergistic Effects and Reverse Selection for Antibiotic Resistance in Pseudomonas aeruginosa. PLoS Biol. 2020, 18, e3000805. [Google Scholar] [CrossRef] [PubMed]
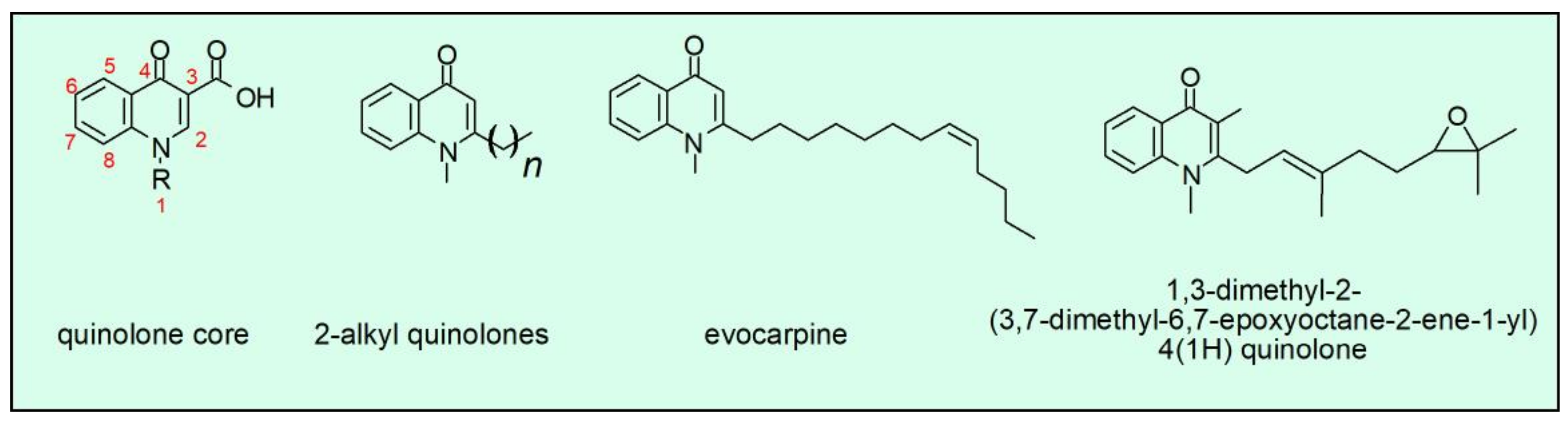
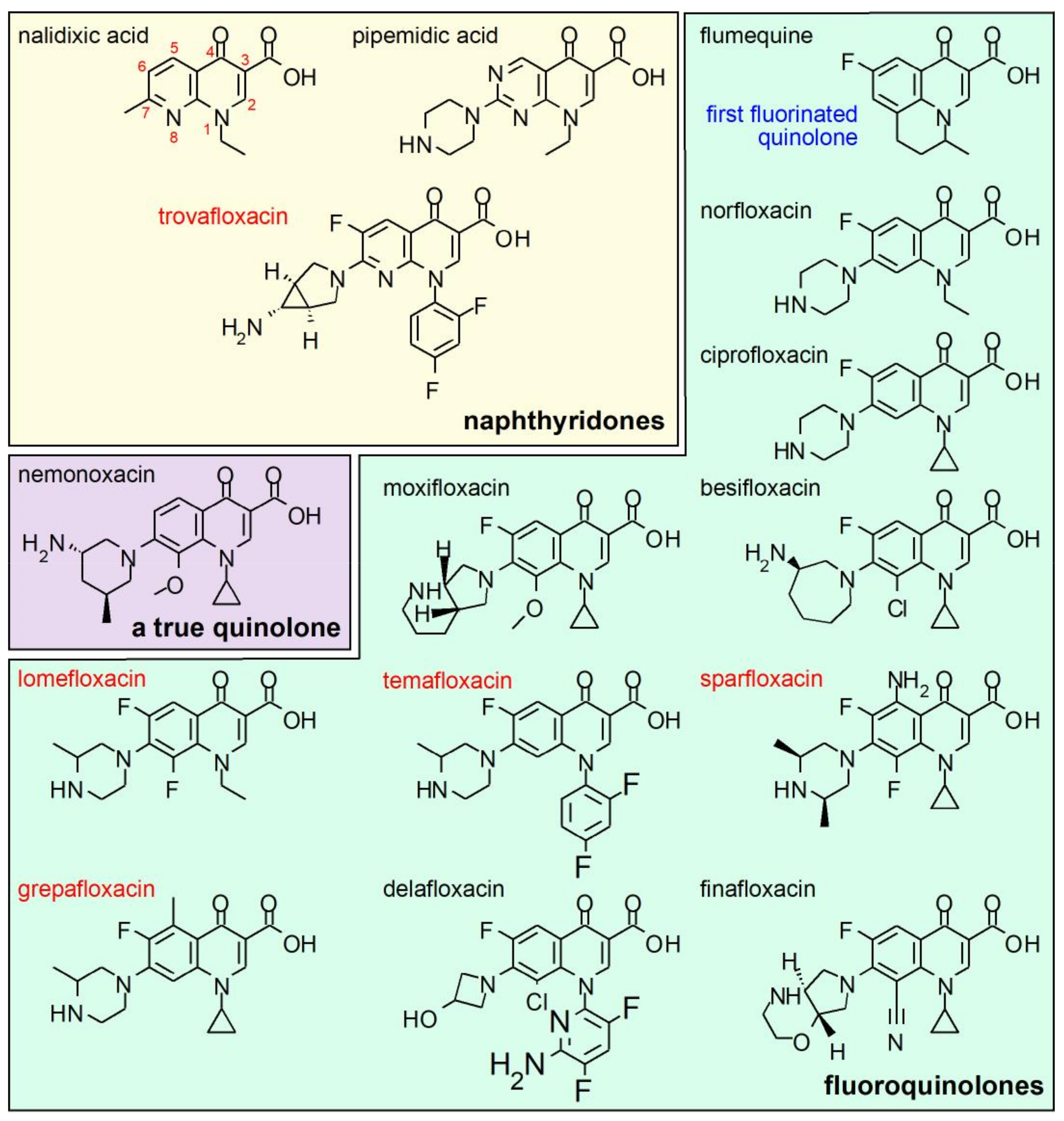
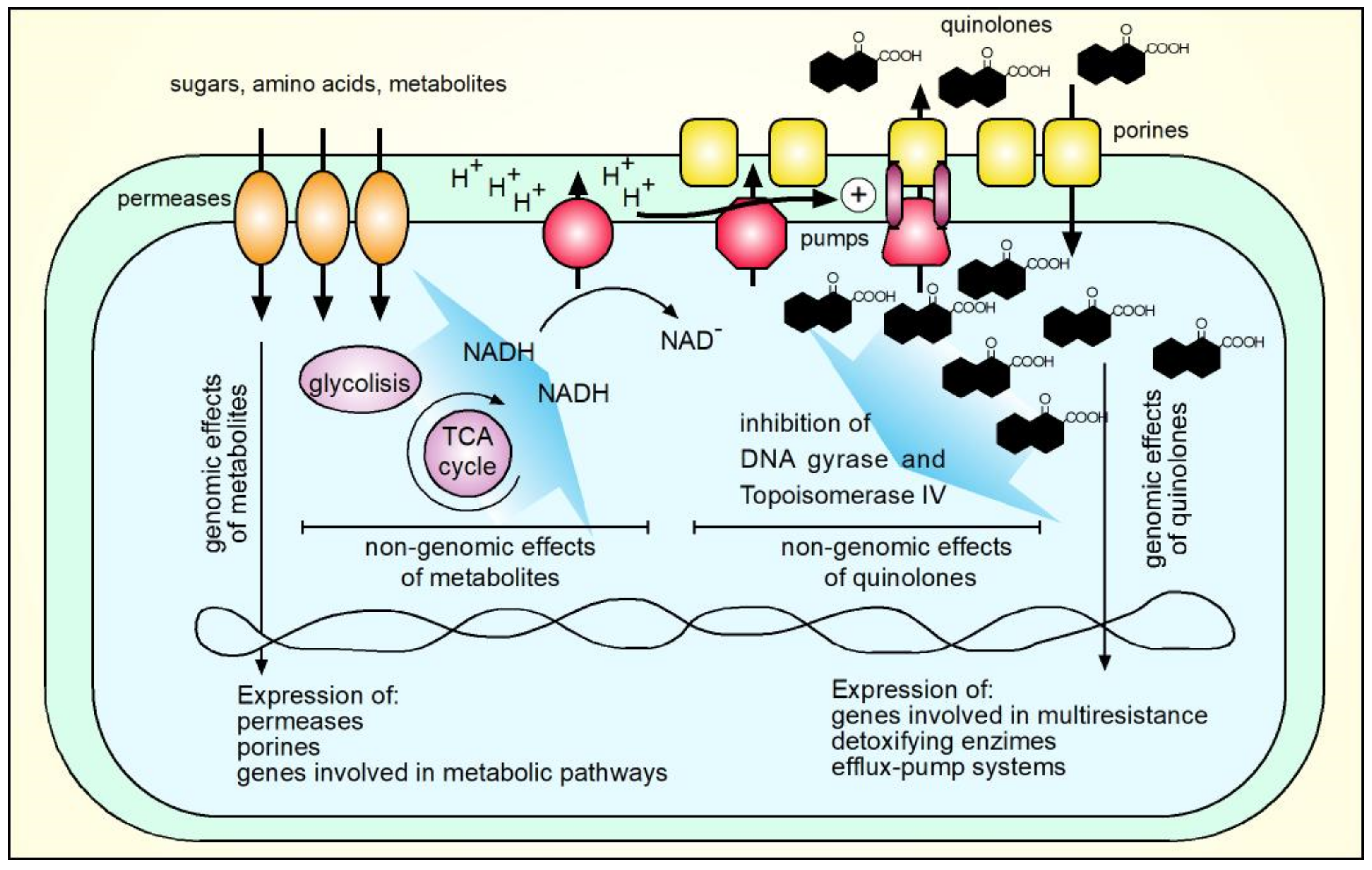
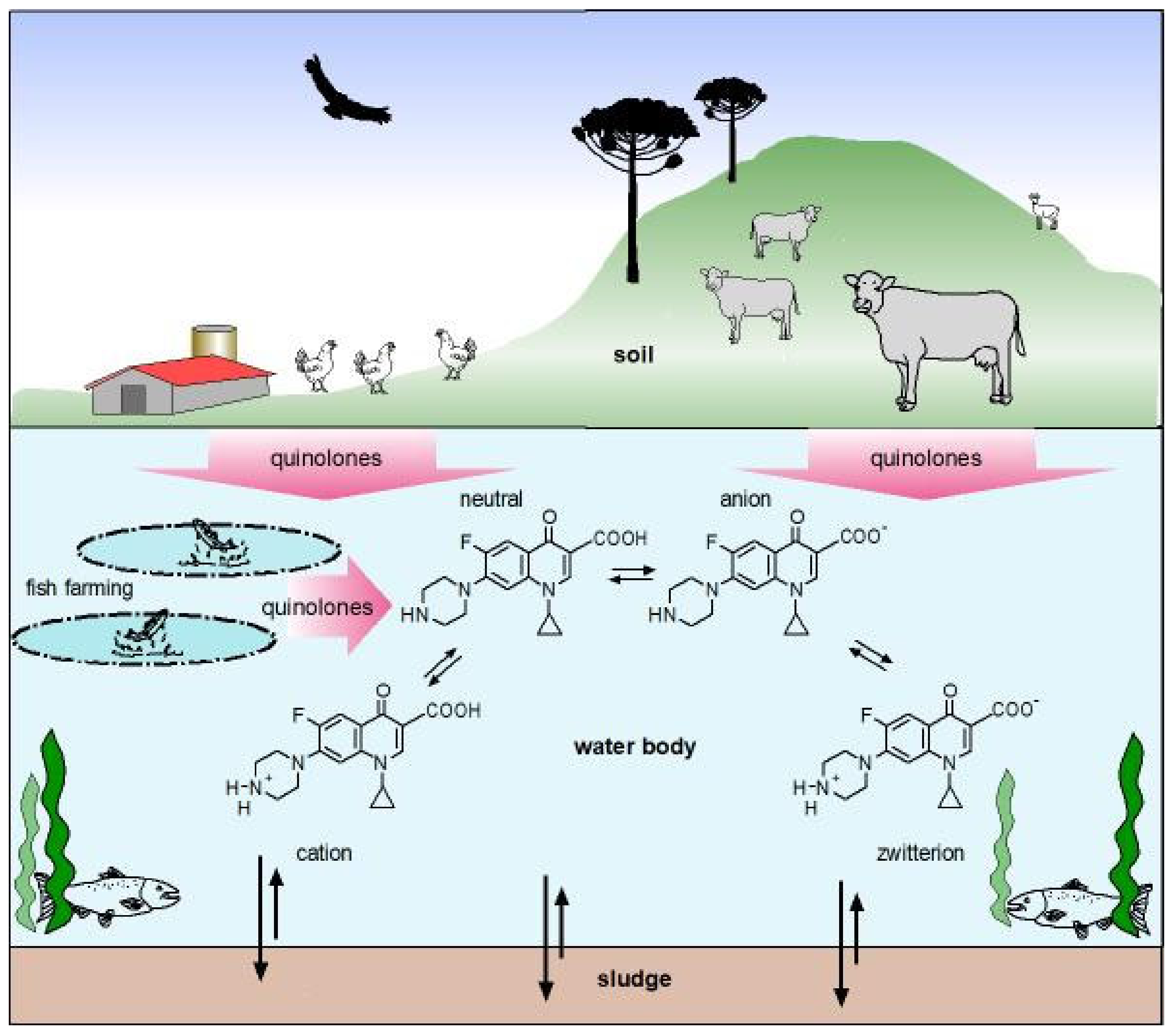

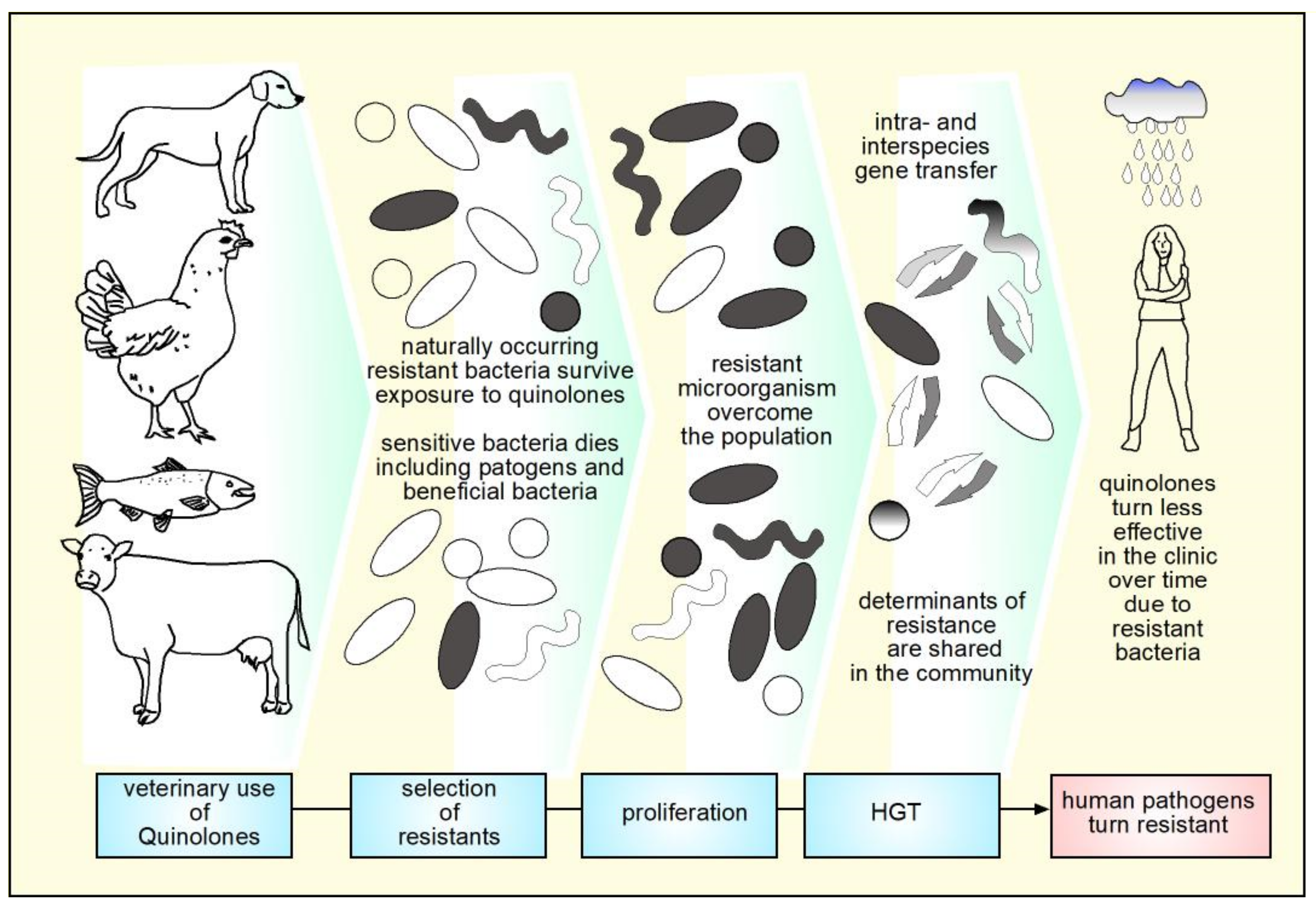
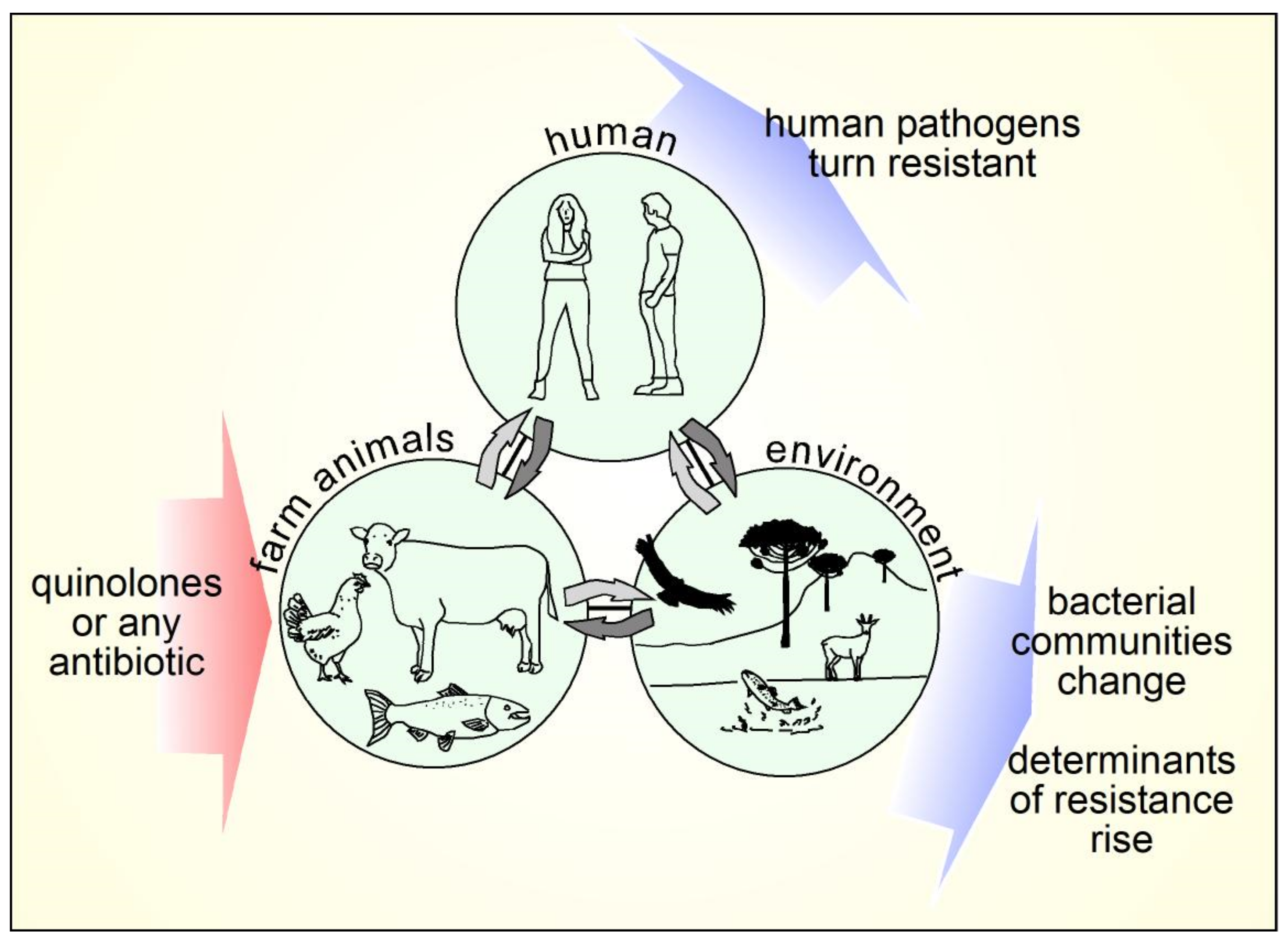
| Agents (with Antimicrobial Effect) | Relevant Effects | Reference |
|---|---|---|
| Feluric acid | Induces oxidative stress. Antimicrobial activity for Gram −/+ | [23,24] |
| CdTe-2.4 | Cadmium telluride quantum dots that after excitation produces superoxide | [25] |
| Kaempferol glycosides | Potentiate quinolones effects in MRSA and vancomycin resistant S. aureus | [30] |
| (without Antimicrobial Effect) | ||
| vitamin K3 | Inhibition and decreased transcription of efflux pump NorA | [27] |
| IMP-1700 | Inhibition of DNA-repair systems of bacteria | [28] |
| biochanin A derivatives | Inhibition of Efflux pumps non-tuberculosis Mycobacterium | [31] |
| L-serine | Enhances quinolones effects by increasing oxidative stress | [32] |
| Palmatine | Restores antimicrobial activity of ciprofloxacin in quinolones resistant E. coli | [33] |
| Ambroxol | Ambroxol combined with ciprofloxacin presented potent activity at clinical used concentrations against SARS-CoV-2 measured as qPCR of its genome, survival, and lysis plaque formation | [34] |
| Agent | Effect | Reference |
|---|---|---|
| (As Anti-Parasitic) | ||
| Fluorinated quinone amides | Antiparasitic, anti-trypanosomal | [122] |
| Metal complexes with FQs | Antiparasitic | [92] |
| Methyl quinolones-derivates | Antiparasitic | [123] |
| (As Anti-Viral) | ||
| Enrofloxacin, ofloxacin, orbifloxacin, iprofloxacin | Activity anti-HCV | [103] |
| Fluoroquinolone-isatin hybrids | Anti-HIV and anti-HCV activities | [124] |
| Quinolones derivates molecules | HIV-1 integrase inhibitors | [125] |
| (As Anti-Fungal) | ||
| Quinolone derivates molecules | Anti-fungal activity against A. flavus | [98] |
| Clinafloxacin triazole hybrids | Anti-fungal activity against C. albicans and C. mycoderma | [94] |
| (As Anti-Inflammatory) | ||
| Ciprofloxacin Levofloxacin | Anti-inflammatory. Inhibition of microglia inflammatory response | [110] |
| Moxifloxacin | Immunomodulatory activity. Reduces secretion of IL-α and TNF-α. Anti-inflammatory. Inhibited ERK1/2, p38 and p65-NFκB. Inhibits etoposide pro-inflammatory effects | [111,112,113] |
| (As Anti-Cancer/Anti-Tumor) | ||
| Moxifloxacin | Activity anti-tumor (Cooper complexes) | [119] |
| Ciprofloxacin and other FQs | Cell cycle arrest and induction of apoptosis | [115,116,117] |
| Ruthenium complex with quinolones | Anti-cancer | [126] |
| (With Other Effects) | ||
| Enrofloxacin, ofloxacin, orbifloxacin, ciprofloxacin | Inhibitory effects on CYP3A in Dogs | [127] |
| Ciprofloxacin and enoxacin | Competitive inhibitory of CYP1A2 | [78] |
| (Quinolones-Related Molecules) | ||
| Quinazolinones related compounds | Anti-convulsant | [128] |
| Quinolines | Neuroprotective, Diuretic activity | [129,130] |
| Country or Zone | Tons of FQs Used in Animal Production | Tons of Total Antimicrobials for Food-Producing Animals | Percentage | Ref. |
|---|---|---|---|---|
| China | 22,210 | 78,100 | 28.4 | [239] |
| Europe | 188 1 | 8060 | 2.3 | [267] |
| USA | 15 | 9196 | 0.2 | [258] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Millanao, A.R.; Mora, A.Y.; Villagra, N.A.; Bucarey, S.A.; Hidalgo, A.A. Biological Effects of Quinolones: A Family of Broad-Spectrum Antimicrobial Agents. Molecules 2021, 26, 7153. https://doi.org/10.3390/molecules26237153
Millanao AR, Mora AY, Villagra NA, Bucarey SA, Hidalgo AA. Biological Effects of Quinolones: A Family of Broad-Spectrum Antimicrobial Agents. Molecules. 2021; 26(23):7153. https://doi.org/10.3390/molecules26237153
Chicago/Turabian StyleMillanao, Ana R., Aracely Y. Mora, Nicolás A. Villagra, Sergio A. Bucarey, and Alejandro A. Hidalgo. 2021. "Biological Effects of Quinolones: A Family of Broad-Spectrum Antimicrobial Agents" Molecules 26, no. 23: 7153. https://doi.org/10.3390/molecules26237153
APA StyleMillanao, A. R., Mora, A. Y., Villagra, N. A., Bucarey, S. A., & Hidalgo, A. A. (2021). Biological Effects of Quinolones: A Family of Broad-Spectrum Antimicrobial Agents. Molecules, 26(23), 7153. https://doi.org/10.3390/molecules26237153






