Mass Spectrometric Identification of Antimicrobial Peptides from Medicinal Seeds
Abstract
:1. Introduction
2. Results and Discussion
2.1. AMP Predictions
2.2. Proteomic Profiling of Seed Extracts
2.3. Lipid Transfer Proteins
2.4. Defensins
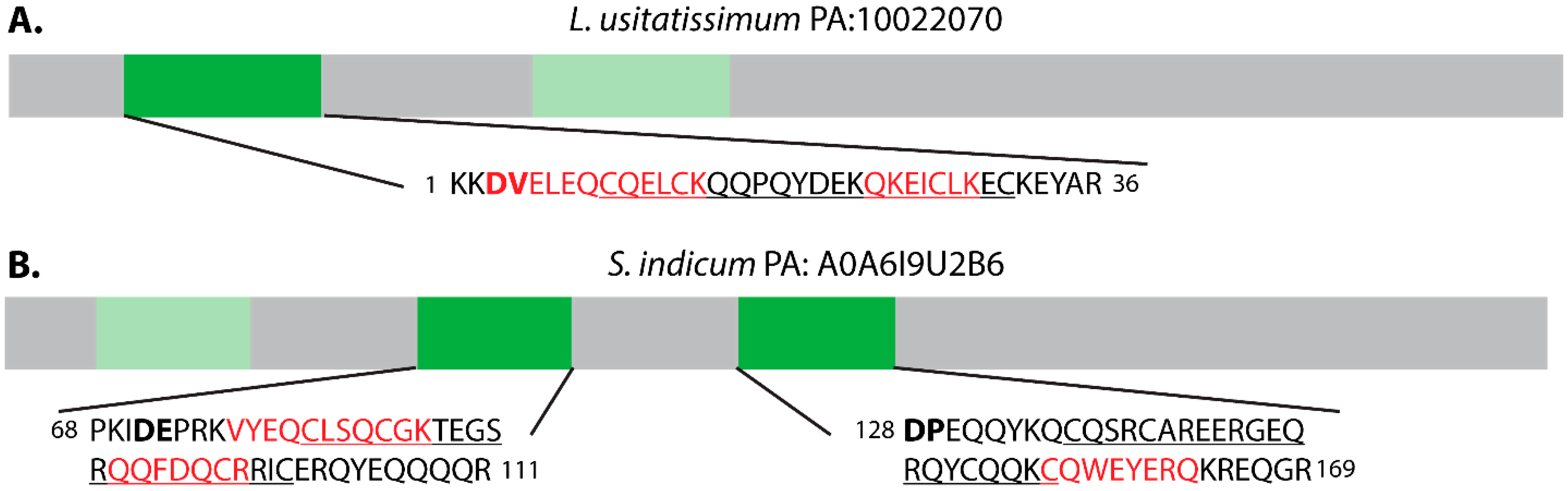
2.5. α-Hairpinins
2.6. Snakins
2.7. Plant Albumins
2.8. Bioactivity Assessment
3. Conclusions
4. Materials and Methods
4.1. AMP Prediction
4.2. Peptide Extraction and Fractionation
4.3. Reduction, Alkylation, and Trypsin Digestion
4.4. LC-MS/MS Data Acquisition
4.5. Database Searching and Peptide Identification
4.6. Sequence Alignment
4.7. Bioactivity Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Bennett, R.N.; Wallsgrove, R.M. Secondary metabolites in plant defence mechanisms. New Phytol. 1994, 127, 617–633. [Google Scholar] [CrossRef] [PubMed]
- Herbel, V.; Wink, M. Mode of action and membrane specificity of the antimicrobial peptide snakin-2. PeerJ 2016, 4, e1987. [Google Scholar] [CrossRef] [Green Version]
- Tam, J.; Wang, S.; Wong, K.; Tan, W. Antimicrobial Peptides from Plants. Pharmaceuticals 2015, 8, 711–757. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Min, K.; Cha, H.; Shin, D.H.; Hwang, K.Y.; Suh, S.W. Rice non-specific lipid transfer protein: The 1.6 Å crystal structure in the unliganded state reveals a small hydrophobic cavity. J. Mol. Biol. 1998, 276, 437–448. [Google Scholar] [CrossRef]
- Sagaram, U.S.; Pandurangi, R.; Kaur, J.; Smith, T.J.; Shah, D.M. Structure-Activity Determinants in Antifungal Plant Defensins MsDef1 and MtDef4 with Different Modes of Action against Fusarium graminearum. PLoS ONE 2011, 6, e18550. [Google Scholar] [CrossRef] [Green Version]
- Nolde, S.B.; Vassilevski, A.A.; Rogozhin, E.A.; Barinov, N.A.; Balashova, T.A.; Samsonova, O.V.; Baranov, Y.V.; Feofanov, A.V.; Egorov, T.A.; Arseniev, A.S.; et al. Disulfide-stabilized helical hairpin structure and activity of a novel antifungal peptide EcAMP1 from seeds of barnyard grass (Echinochloa crus-galli). J. Biol. Chem. 2011, 286, 25145–25153. [Google Scholar] [CrossRef] [Green Version]
- Yeung, H.; Squire, C.J.; Yosaatmadja, Y.; Panjikar, S.; López, G.; Molina, A.; Baker, E.N.; Harris, P.W.R.; Brimble, M.A. Radiation Damage and Racemic Protein Crystallography Reveal the Unique Structure of the GASA/Snakin Protein Superfamily. Angew. Chem. Int. Ed. 2016, 55, 7930–7933. [Google Scholar] [CrossRef] [PubMed]
- Porto, W.F.; Franco, O.L. Theoretical structural insights into the snakin/GASA family. Peptides 2013, 44, 163–167. [Google Scholar] [CrossRef] [Green Version]
- Jouvensal, L.; Quillien, L.; Ferrasson, E.; Rahbé, Y.; Guéguen, J.; Vovelle, F. PA1b, an insecticidal protein extracted from pea seeds (Pisum sativum): 1H-2-D NMR study and molecular modeling. Biochemistry 2003, 42, 11915–11923. [Google Scholar] [CrossRef] [PubMed]
- Shelenkov, A.A.; Slavokhotova, A.A.; Odintsova, T.I. Cysmotif Searcher Pipeline for Antimicrobial Peptide Identification in Plant Transcriptomes. Biochem. Mosc. 2018, 83, 1424–1432. [Google Scholar] [CrossRef]
- Zhou, P.; Silverstein, K.A.T.; Gao, L.; Walton, J.D.; Nallu, S.; Guhlin, J.; Young, N.D. Detecting small plant peptides using SPADA (Small Peptide Alignment Discovery Application). BMC Bioinform. 2013, 14, 335. [Google Scholar] [CrossRef] [Green Version]
- Moyer, T.B.; Allen, J.L.; Shaw, L.N.; Hicks, L.M. Multiple classes of antimicrobial peptides in Amaranthus tricolor revealed by prediction, proteomics and mass spectrometric characterization. J. Nat. Prod. 2021, 84, 444–452. [Google Scholar] [CrossRef]
- Berrocal-Lobo, M.; Segura, A.; Moreno, M.; López, G.; García-Olmedo, F.; Molina, A. Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol. 2002, 128, 951–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trabi, M.; Craik, D.J. Tissue-specific expression of head-to-tail cyclized miniproteins in violaceae and structure determination of the root cyclotide Viola hederacea root cyclotide1. Plant Cell 2004, 16, 2204–2216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meneguetti, B.T.; Machado, L.D.S.; Oshiro, K.G.; Nogueira, M.L.; Carvalho, C.M.; Franco, O.L. Antimicrobial Peptides from Fruits and Their Potential Use as Biotechnological Tools—A Review and Outlook. Front. Microbiol. 2017, 7, 2136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Xiao, Y.; Wang, X.; Pei, Y. Expression of a novel small antimicrobial protein from the seeds of motherwort (Leonurus japonicus) confers disease resistance in Tobacco. Appl. Environ. Microbiol. 2007, 73, 939–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shim, Y.Y.; Gui, B.; Arnison, P.G.; Wang, Y.; Reaney, M.J.T. Flaxseed (Linum usitatissimum L.) bioactive compounds and peptide nomenclature: A Review. Trends Food Sci. Technol. 2014, 38, 5–20. [Google Scholar] [CrossRef] [Green Version]
- Kolodziejczyk-Czepas, J. Trifolium species—The latest findings on chemical profile, ethnomedicinal use and pharmacological properties. J. Pharm. Pharmacol. 2016, 68, 845–861. [Google Scholar] [CrossRef] [Green Version]
- Esmaeili, A.K.; Taha, R.M.; Banisalam, B.; Mohajer, S.; Mahmood, N.Z. Antimicrobial Activities of Extracts Derived from in vivo and in vitro Grown Trifolium pratense (Red clover). Int. J. Environ. Sci. Dev. 2013, 4, 475–478. [Google Scholar] [CrossRef]
- Prasad MN, N.; KR, S.; Prasad, D.S.; Vijay, N.; Kothari, R.; Swamy, S.N. A Review on Nutritional and Nutraceutical Properties of Sesame. J. Nutr. Food Sci. 2012, 2, 1–6. [Google Scholar] [CrossRef]
- Kanu, P.; Bahsoon, J.; Kanu, J.; Kandeh, J. Nutraceutical Importance of Sesame Seed and Oil: A Review of the Contribution of their Lignans. Sierra Leone J. Biomed. Res. 2010, 2, 4–16. [Google Scholar] [CrossRef] [Green Version]
- Anilakumar, K.R.; Pal, A.; Khanum, F.; Bawa, A.S. Nutritional, medicinal and industrial uses of sesame (Sesamum indicum L.) seeds—An overview. Agric. Conspec. Sci. 2010, 75, 159–168. [Google Scholar]
- Shim, Y.Y.; Song, Z.; Jadhav, P.D.; Reaney, M.J.T. Orbitides from flaxseed (Linum usitatissimum L.): A comprehensive review. Trends Food Sci. Technol. 2019, 93, 197–211. [Google Scholar] [CrossRef]
- Costa, F.T.; Neto, S.M.; Bloch, C.; Franco, O.L. Susceptibility of human pathogenic bacteria to antimicrobial peptides from sesame kernels. Curr. Microbiol. 2007, 55, 162–166. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Payne, C.D.; Pouvreau, B.; Schaefer, H.; Fisher, M.F.; Taylor, N.L.; Berkowitz, O.; Whelan, J.; Rosengren, K.J.; Mylne, J.S. An Ancient Peptide Family Buried within Vicilin Precursors. ACS Chem. Biol. 2019, 14, 979–993. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Hobson, N.; Galindo, L.; Zhu, S.; Shi, D.; McDill, J.; Yang, L.; Hawkins, S.; Neutelings, G.; Datla, R.; et al. The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant J. 2012, 72, 461–473. [Google Scholar] [CrossRef] [Green Version]
- De Vega, J.J.; Ayling, S.; Hegarty, M.; Kudrna, D.; Goicoechea, J.L.; Ergon, Å.; Rognli, O.A.; Jones, C.; Swain, M.; Geurts, R.; et al. Red clover (Trifolium pratense L.) draft genome provides a platform for trait improvement. Sci. Rep. 2015, 5, 17394. [Google Scholar] [CrossRef]
- Wang, L.; Yu, S.; Tong, C.; Zhao, Y.; Liu, Y.; Song, C.; Zhang, Y.; Zhang, X.; Wang, Y.; Hua, W.; et al. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 2014, 15, R39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shelenkov, A.; Slavokhotova, A.; Odintsova, T. Predicting Antimicrobial and Other Cysteine-Rich Peptides in 1267 Plant Transcriptomes. Antibiotics 2020, 9, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kader, J.-C. Lipid-Transfer Proteins in Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol 1996, 47, 627–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, S.; You, J.; Shi, L.; Sheng, C.; Zhou, W.; Dossou, S.S.K.; Dossa, K.; Wang, L.; Zhang, X. Genome-Wide Analysis of nsLTP Gene Family and Identification of SiLTPs Contributing to High Oil Accumulation in Sesame (Sesamum indicum L.). Int. J. Mol. Sci. 2021, 22, 5291. [Google Scholar] [CrossRef] [PubMed]
- Herbert, D.B.; Gross, T.; Rupp, O.; Becker, A. Transcriptome analysis reveals major transcriptional changes during regrowth after mowing of red clover (Trifolium pratense). BMC Plant Biol. 2021, 21, 95. [Google Scholar] [CrossRef] [PubMed]
- Choi, A.M.; Lee, S.B.; Cho, S.H.; Hwang, I.; Hur, C.G.; Suh, M.C. Isolation and characterization of multiple abundant lipid transfer protein isoforms in developing sesame (Sesamum indicum L.) seeds. Plant Physiol. Biochem. 2008, 46, 127–139. [Google Scholar] [CrossRef]
- Mhaske, V.A.; Datla, R.; Qiu, S.; Harsulkar, A.M. Isolation and characterization of genes encoding lipid transfer proteins in Linum usitatissimum. Biol. Plant. 2016, 60, 285–291. [Google Scholar] [CrossRef]
- Salminen, T.A.; Blomqvist, K.; Edqvist, J. Lipid transfer proteins: Classification, nomenclature, structure, and function. Planta 2016, 244, 971–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finkina, E.I.; Melnikova, D.N.; Bogdanov, I.V.; Ovchinnikova, T.V. Lipid transfer proteins as components of the plant innate immune system: Structure, functions, and applications. Acta Nat. 2016, 8, 47–61. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, X.; Lu, C.; Zeng, X.; Li, Y.; Fu, D.; Wu, G. Non-specific lipid transfer proteins in plants: Presenting new advances and an integrated functional analysis. J. Exp. Bot. 2015, 66, 5663–5681. [Google Scholar] [CrossRef] [Green Version]
- Shafee, T.M.A.; Lay, F.T.; Hulett, M.D.; Anderson, M.A. The Defensins Consist of Two Independent, Convergent Protein Superfamilies. Mol. Biol. Evol. 2016, 33, 2345–2356. [Google Scholar] [CrossRef] [Green Version]
- Sathoff, A.E.; Velivelli, S.; Shah, D.M.; Samac, D.A. Plant defensin peptides have antifungal and antibacterial activity against human and plant pathogens. Phytopathology 2019, 109, 402–408. [Google Scholar] [CrossRef] [Green Version]
- Sathoff, A.E.; Samac, D.A. Antibacterial Activity of Plant Defensins. Mol. Plant-Microbe Interact. 2019, 32, 507–514. [Google Scholar] [CrossRef]
- de Oliveira Carvalho, A.; Moreira Gomes, V. Plant Defensins and Defensin-Like Peptides—Biological Activities and Biotechnological Applications. Curr. Pharm. Des. 2012, 17, 4270–4293. [Google Scholar] [CrossRef] [PubMed]
- Lay, F.; Anderson, M. Defensins—Components of the Innate Immune System in Plants. Curr. Protein Pept. Sci. 2005, 6, 85–101. [Google Scholar] [CrossRef] [PubMed]
- Stotz, H.U.; Thomson, J.G.; Wang, Y. Plant defensins: Defense, development and application. Plant Signal. Behav. 2009, 4, 1010–1012. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, S.; Basu, A.; Kundu, S. Biotrophy-necrotrophy switch in pathogen evoke differential response in resistant and susceptible sesame involving multiple signaling pathways at different phases. Sci. Rep. 2017, 7, 17251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vriens, K.; Cammue, B.P.A.; Thevissen, K. Antifungal plant defensins: Mechanisms of action and production. Molecules 2014, 19, 12280–12303. [Google Scholar] [CrossRef] [Green Version]
- Moyer, T.B.; Purvis, A.L.; Wommack, A.J.; Hicks, L.M. Proteomic response of Escherichia coli to a membrane lytic and iron chelating truncated Amaranthus tricolor defensin. BMC Microbiol. 2021, 21, 110. [Google Scholar] [CrossRef]
- Sathoff, A.E.; Lewenza, S.; Samac, D.A. Plant defensin antibacterial mode of action against Pseudomonas species. BMC Microbiol. 2020, 20, 173. [Google Scholar] [CrossRef]
- Rigano, M.M.; Romanelli, A.; Fulgione, A.; Nocerino, N.; D’Agostino, N.; Avitabile, C.; Frusciante, L.; Barone, A.; Capuano, F.; Capparelli, R. A novel synthetic peptide from a tomato defensin exhibits antibacterial activities against Helicobacter pylori. J. Pept. Sci. 2012, 18, 755–762. [Google Scholar] [CrossRef] [Green Version]
- Aerts, A.M.; François, I.E.J.A.; Meert, E.M.K.; Li, Q.T.; Cammue, B.P.A.; Thevissen, K. The antifungal activity of RsAFP2, a plant defensin from Raphanus sativus, involves the induction of reactive oxygen species in Candida albicans. J. Mol. Microbiol. Biotechnol. 2007, 13, 243–247. [Google Scholar] [CrossRef]
- Kaewklom, S.; Wongchai, M.; Petvises, S.; Hanpithakphong, W.; Aunpad, R. Structural and biological features of a novel plant defensin from Brugmansia × candida. PLoS ONE 2018, 13, e0201668. [Google Scholar] [CrossRef]
- Souza, G.S.; de Carvalho, L.P.; de Melo, E.J.T.; da Silva, F.C.V.; Machado, O.L.T.; Gomes, V.M.; de Oliveira Carvalho, A. A synthetic peptide derived of the β2–β3 loop of the plant defensin from Vigna unguiculata seeds induces Leishmania amazonensis apoptosis-like cell death. Amino Acids 2019, 51, 1633–1648. [Google Scholar] [CrossRef] [PubMed]
- Slavokhotova, A.A.; Rogozhin, E.A. Defense Peptides from the α-Hairpinin Family Are Components of Plant Innate Immunity. Front. Plant Sci. 2020, 11, 465. [Google Scholar] [CrossRef]
- Payne, C.D.; Vadlamani, G.; Fisher, M.F.; Zhang, J.; Clark, R.J.; Mylne, J.S.; Rosengren, K.J. Defining the Familial Fold of the Vicilin-Buried Peptide Family. J. Nat. Prod. 2020, 83, 3030–3040. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Han, M.; Cao, D.; Xu, M. Molecular and Biological Properties of Snakins: The Foremost Cysteine-Rich Plant Host Defense Peptides. J. Fungi 2020, 6, 220. [Google Scholar] [CrossRef]
- Harris, P.W.R.; Yang, S.-H.; Molina, A.; López, G.; Middleditch, M.; Brimble, M.A. Plant Antimicrobial Peptides Snakin-1 and Snakin-2: Chemical Synthesis and Insights into the Disulfide Connectivity. Chem.—Eur. J. 2014, 20, 5102–5110. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Decuadro, S.; Barraco-Vega, M.; Dans, P.D.; Pandolfi, V.; Benko-Iseppon, A.M.; Cecchetto, G. Antimicrobial and structural insights of a new snakin-like peptide isolated from Peltophorum dubium (Fabaceae). Amino Acids 2018, 50, 1245–1259. [Google Scholar] [CrossRef] [PubMed]
- Karaki, L.; Da Silva, P.; Rizk, F.; Chouabe, C.; Chantret, N.; Eyraud, V.; Gressent, F.; Sivignon, C.; Rahioui, I.; Kahn, D.; et al. Genome-wide analysis identifies gain and loss/change of function within the small multigenic insecticidal Albumin 1 family of Medicago truncatula. BMC Plant Biol. 2016, 16, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gressent, F.; Da Silva, P.; Eyraud, V.; Karaki, L.; Royer, C. Pea Albumin 1 Subunit b (PA1b), a Promising Bioinsecticide of Plant Origin. Toxins 2011, 3, 1502–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkpatrick, C.L.; Broberg, C.A.; McCool, E.N.; Lee, W.J.; Chao, A.; McConnell, E.W.; Pritchard, D.A.; Herbert, M.; Fleeman, R.; Adams, J.; et al. The “PepSAVI-MS” Pipeline for Natural Product Bioactive Peptide Discovery. Anal. Chem. 2017, 89, 1194–1201. [Google Scholar] [CrossRef]
- Al-Mohanna, T.; Nejat, N.; Iannetta, A.A.; Hicks, L.M.; Popescu, G.V.; Popescu, S.C. Arabidopsis thimet oligopeptidases are redox-sensitive enzymes active in the local and systemic plant immune response. J. Biol. Chem. 2021, 296, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Adusumilli, R.; Mallick, P. Data Conversion with ProteoWizard msConvert. Methods Mol. Biol. 2017, 1550, 339–368. [Google Scholar] [CrossRef] [PubMed]
- Brosch, M.; Yu, L.; Hubbard, T.; Choudhary, J. Accurate and sensitive peptide identification with mascot percolator. J. Proteome Res. 2009, 8, 3176–3181. [Google Scholar] [CrossRef] [Green Version]
- Vizcaíno, J.A.; Csordas, A.; del-Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez-Riverol, Y.; Reisinger, F.; Ternent, T.; et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44, D447–D456. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Plant Species | Precursor Accession | Family | Predicted Sequences | Mascot Score |
|---|---|---|---|---|
| S. indicum | A0A6I9UIH4 | Defensin | KICQRMSKTWSGVCLNSGNCDRQCRNWERAQHGACHRRGLGFACLCYFKC | 26 |
| S. indicum | A0A6I9UUK2 | Lipid transfer protein | ISCGDVQGSLAPCLAYLTGGGEPSSSCCGGVRSLAGSLQSQQDRQTACYCMKSAASSFNVRSDAASNLPGKCGVSIGMTVTPDIDCSKVS | 1373 |
| S. indicum | A0A6I9U3T5 | Lipid transfer protein | VVSCGQVQSGLSPCLGFLQGRALASQCCPGVRSVVAAARTPNDRRTACRCLQAAAKSMRNINYGNAAMLPAKCGVKIPFQISPNTDCSRVG | 591 |
| S. indicum | A0A6I9TV60 | Lipid transfer protein | AIPCGTVDMKAASCVSFATGKDAKPSAACCTGLQQLAQTVKSVDDKKAICRCLKTAVKNFSGVQDKFLSQIPTACNIKVGFPVSLSTDCEKLH | 275 |
| S. indicum | A0A6I9UAN6 | Lipid transfer protein | AIPCGTVDMKAASCVAFATGKDPKPSPTCCSGLQQLAQSVKTVDDKKAICRCLKAAVKNFAGVQDRFLSQIPAACNIKVGFPVSLSTDCEKLH | 146 |
| S. indicum | A0A6I9TZ60 | Lipid transfer protein | AIGCGTVVSYLNPCLPYVTNKGPLGSCCGGVKGLYGAAQTTQDRQSVCSCLKSLASSYKDVDLNKAAGLPGQCGVNIPYKISPSTDCSKVN | 35 |
| S. indicum | A0A6I9U3T9 | Lipid transfer protein | AISCASVMTKLSPCLSYIKSGGGLPPACCSGAKSLNDAASTTPDLQAVCGCIKILVPSLRANPAYINSIPAKCGVNLPYKYSPSLDCSKVVR | 26 |
| S. indicum | A0A6I9UAV3 | Lipid transfer protein | LTCLDIMPTVMQCASFALGMVSRPSSQCCNELSRLHGMARTTDDRRQACNCLKQIAPQYPGAMDANLLALPQLCRVALSFPIRRDTDCSKIT | 14 |
| S. indicum | A0A6I9TPG7 | Snakin* | DIETEDEVSLVARGSNRRLLPFLDCGGLCKVRCSQHSRPNVCTRACGTCCARCKCVPPGTSGNRELCGACYTDMTTHANKTKCP | 18 |
| S. indicum | A0A6I9U2B6 | α-hairpinin* | YTNPQLQEGEEESAEEGLFKCFVSCEKRRENEHELSQCEKRCVREYQERKREEREERGGRRGEETVVPKIDEPRKVYEQCLSQCGKTEGSRQQFDQCRRICERQYEQQQQREKRGGGEGTIENHHRRDPEQQYKQCQSRCAREERGEQRQYCQQKCQWEYERQKREQGREQGGGGGSTNPRKEREEEEEQEGKNPYFFESQRFDSKYRTEEGNVKVLERFSKKSELLQGVDNYRLAVLEANPNTFVLPHHFDAESVLVVAGGKGTISYVWQNRRKSYNVKLGDVMRVPAGSIVYLVNRDDNEKLYVLKLLQPVNTPGRFKEYFGVGGENPESFYRTFSNEILEAAFNVPSDRLKRLFGQQKKGVIIRASKEQIRALSQESEESSRGRREESWGPFNLLEGRPLFSNRYGQYFEASPNDYQQLKDLDVSVGFMNINKGGMVAPYYNSRSTKLVLVVGGNGRFEMACPHRSARSKQGRKERQGETTDVRYQRVSARLSIGDAFIVPAGHPIAMIASQDSNLQLVSFGIKGSYNQKYFLAGQDNIWNQVESEAKELSFKMPAREVEEIFRRQEQSYFLPGPGQGEERGKEHYVASILDFVGF | 315 |
| T. pratense | mRNA40325 | Defensin | QNKCEHLADTYKGPCFTNASCDDHCKNKEHFRSGTCHGFRCWCTHQKC | 119 |
| T. pratense | mRNA31242 | Defensin | KRCENLAGKYKGVCFGGCDHHCKTQEGAISGRCRDDFRCWCTKNC | 27 |
| T. pratense | mRNA31525 | Lipid transfer protein | FDCEATEKSLFPCGTFIIGGSVEPSTSCCSAVQNLKASTPTPDDKRNACICLKEVASHYPNIIEDLAASLPQRCGVDISFTISKNMDCDNVLNGETKGNPLTRVAPPHFR | 3106 |
| T. pratense | mRNA5174 | Lipid transfer protein | GISCGTVNGALAQCIPYLKGGPGPSPACCAGVKRLNAAAATTPDRQAACNCLKNAAGAISGLNTNNAGALPGKCGVNIPYKISTSTNCATIRA | 520 |
| T. pratense | mRNA5130 | Lipid transfer protein | AISCGAVNGALAPCIVYLRGGRGPSPACCAGVRRLKAVATTTPTRQAACNCLKSAARGISGLNNNNAGALPGRCGVSIPYKISTSTNCAIIRA | 451 |
| T. pratense | mRNA14597 | Lipid transfer protein | ALPCGQVQLTLTPCLGYLRRPGPSVPPPCCNGIRSLNNQAKTTPDRQSVCRCLKSTALSLPGLNLPAAASILAKCGVNLPYKISPSIDCNTYISLNQLSIYFHL | 128 |
| T. pratense | mRNA5120 | Lipid transfer protein | SVSCGAVTGYLVPCITYLQGGPGPSPACCDGVKKLNAAAATTPDRKAACNCLKGAAGSIARLNNNAAAALPGKCGVRIPYKFSTSTNCNSIKF | 29 |
| T. pratense | mRNA5131 | Plant albumin 1b* | ACKISCLLYKPKSCGDGCKCVPASLTYGVCVKASFEHVTNMVEEHPNLCESHDDCTKKGSGSFCARFPNPEIEYGWCFDSNSHAQASFKNAQESSNFFLKMPSAIST | 96 |
| T. pratense | mRNA38777 | Snakin* | DHEIEMEEDDELQLPDDKLLIVRDGNRRLMSDIDCGGLCGSRCSVHSRPNLCKRACGTCCVRCKCVPPGTSGNREFCGACYTDMVTHGNKTKCP | 22 |
| L. usitatissimum | 10015279 | Lipid transfer protein | AVSCGKVASALAPCVPYLRGVGAVTPACCGGVKSLNAAATTTPDRQAACRCLKSTSAGISGINYGNAGSLPGKCGVNVGYPISPTVNCNT | 205 |
| L. usitatissimum | 10001407 | Snakin | SSCFIQLSVAHVNPSSPNQATDVGRRCESKCEGRCAAAGYKERCLNYCNICCSKCRCVPSGTYGNKQECPCYRDLRDNKGRPKCP | 63 |
| L. usitatissimum | 10022070 | α-Hairpinin* | KKDVELEQCQELCKQQPQYDEKQKEICLKECKEYARKKSGRGSEETDPEKRLEECKHQCKQHKFSDEEQKKACRTKCDKQYKEGRGRIGTYYYYEEEQEESKGENPYVFTEEHFESKSQSQHGRVDVLRKFTDKSELLKGIENFRIGFLEANPQTFVPPAHFDADGVFFVAQGRGTFTMIEGNRGRMTSSSEIKRHSFNIEAGDVVRVYAGSPVYLVNKHESQKLVIIKFIRPVNLPGSFDAFHGPGGENPESFFRAFSPELLAAAFKVDKQRIQRIFQQQEGEILKATREQIRALSHGEEGGGIWPFGGESTGPFNLLHRRPTQKNTFGQLWEADPNEFEQFRDLDLLVSFANITQGAMAGPFYNSKATKIAYVVNGEGYFEMACPHVTSSSGDMGRQTRGSQSRGGQKYGKVRSQLRRGTVFIVPAGHPVVTVASANNNLEVLCFEVNAQGNFRFSLAGKDNVMSKMESEALELGFGAPAREVEQIFKNRNEEFFFPGPEWQKQQHSRGYSSA | 453 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moyer, T.B.; Brechbill, A.M.; Hicks, L.M. Mass Spectrometric Identification of Antimicrobial Peptides from Medicinal Seeds. Molecules 2021, 26, 7304. https://doi.org/10.3390/molecules26237304
Moyer TB, Brechbill AM, Hicks LM. Mass Spectrometric Identification of Antimicrobial Peptides from Medicinal Seeds. Molecules. 2021; 26(23):7304. https://doi.org/10.3390/molecules26237304
Chicago/Turabian StyleMoyer, Tessa B., Amanda M. Brechbill, and Leslie M. Hicks. 2021. "Mass Spectrometric Identification of Antimicrobial Peptides from Medicinal Seeds" Molecules 26, no. 23: 7304. https://doi.org/10.3390/molecules26237304








