Constructing Large 2D Lattices Out of DNA-Tiles
Abstract
:1. Introduction
2. Planks and Nails for DNA Lattices
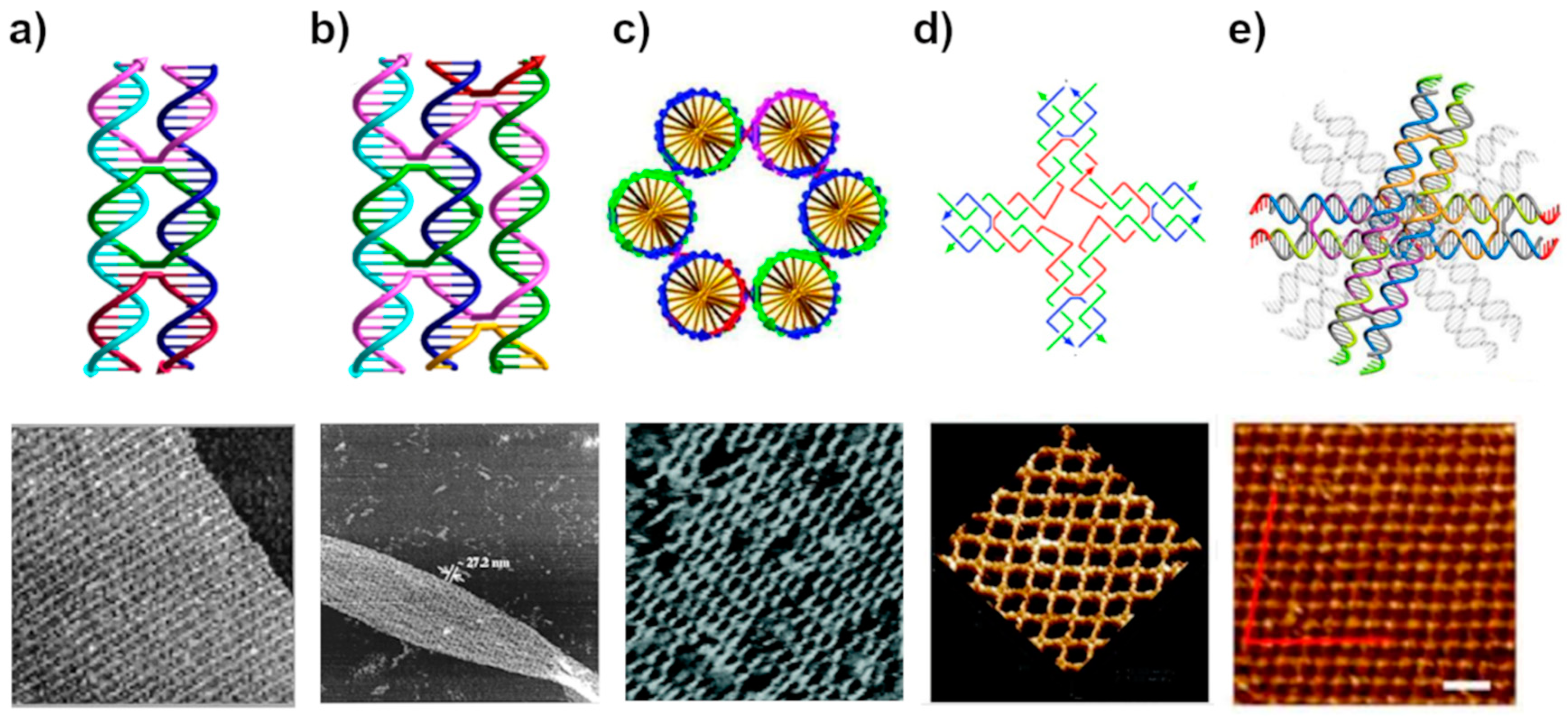
2.1. The Planks
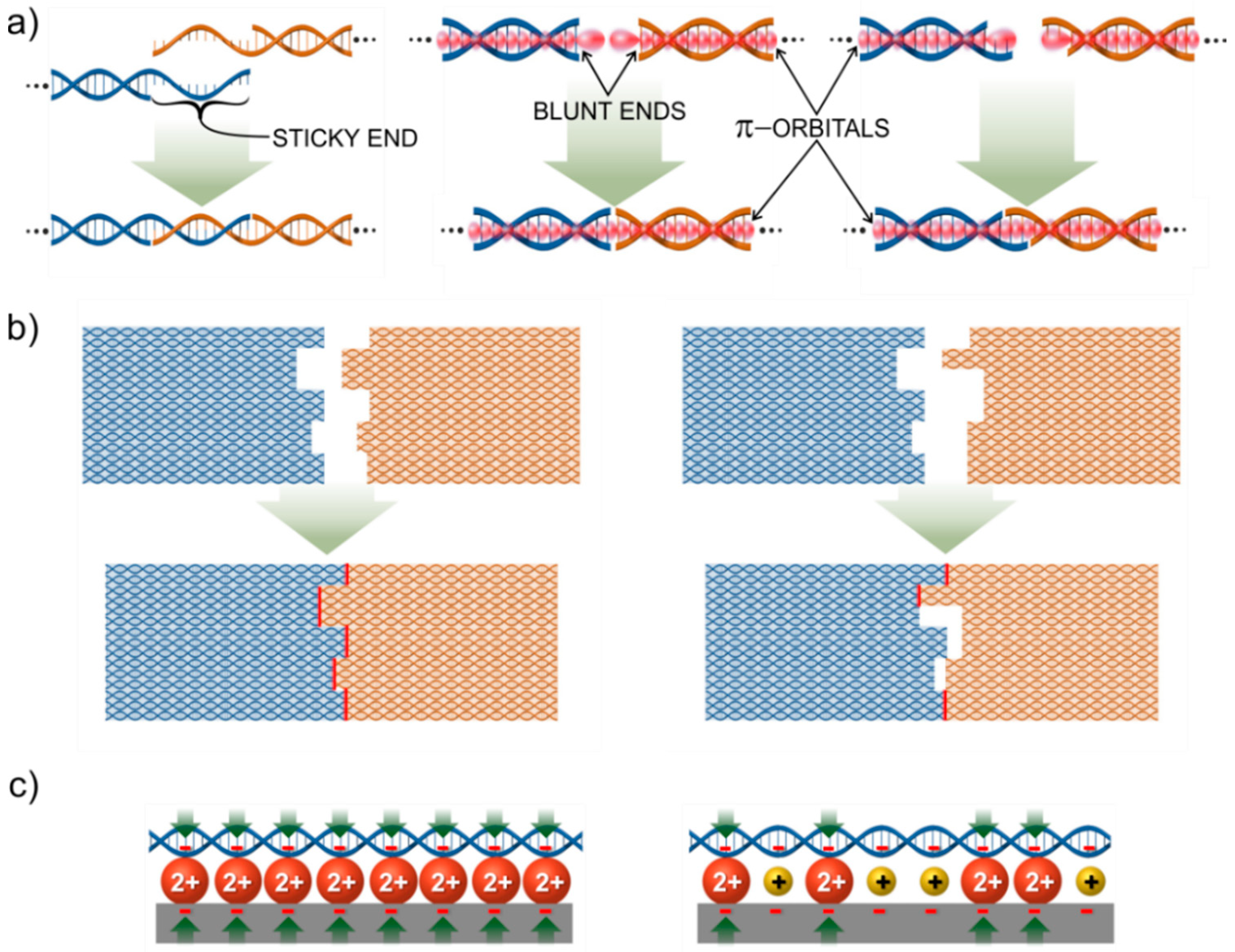
2.2. The Nails
2.3. Assembly Protocols
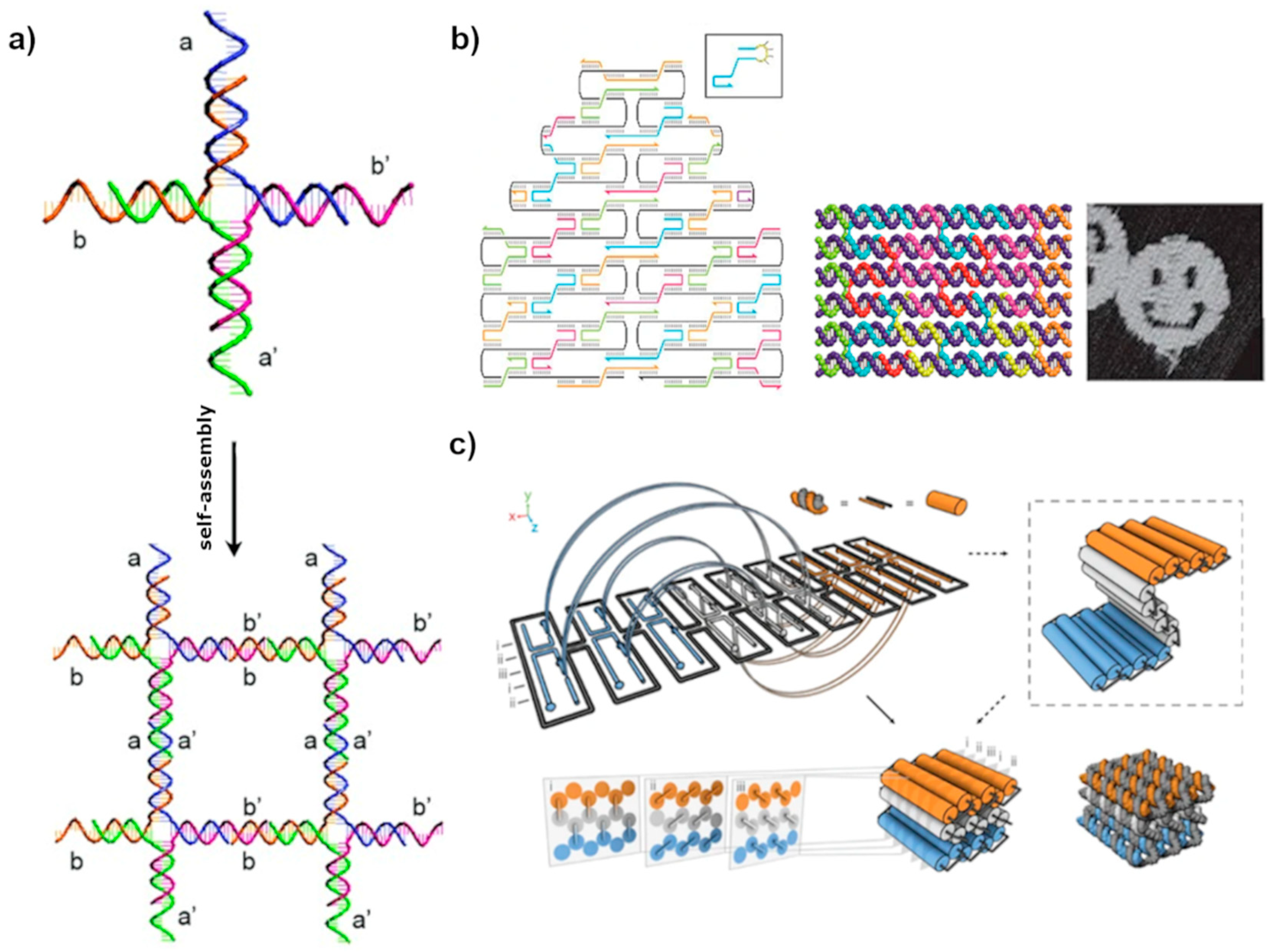
3. Assembly of DNA Lattices in Solution
3.1. Lattices by Early DNA Motif
3.2. DNA Origami-Tile Assembly
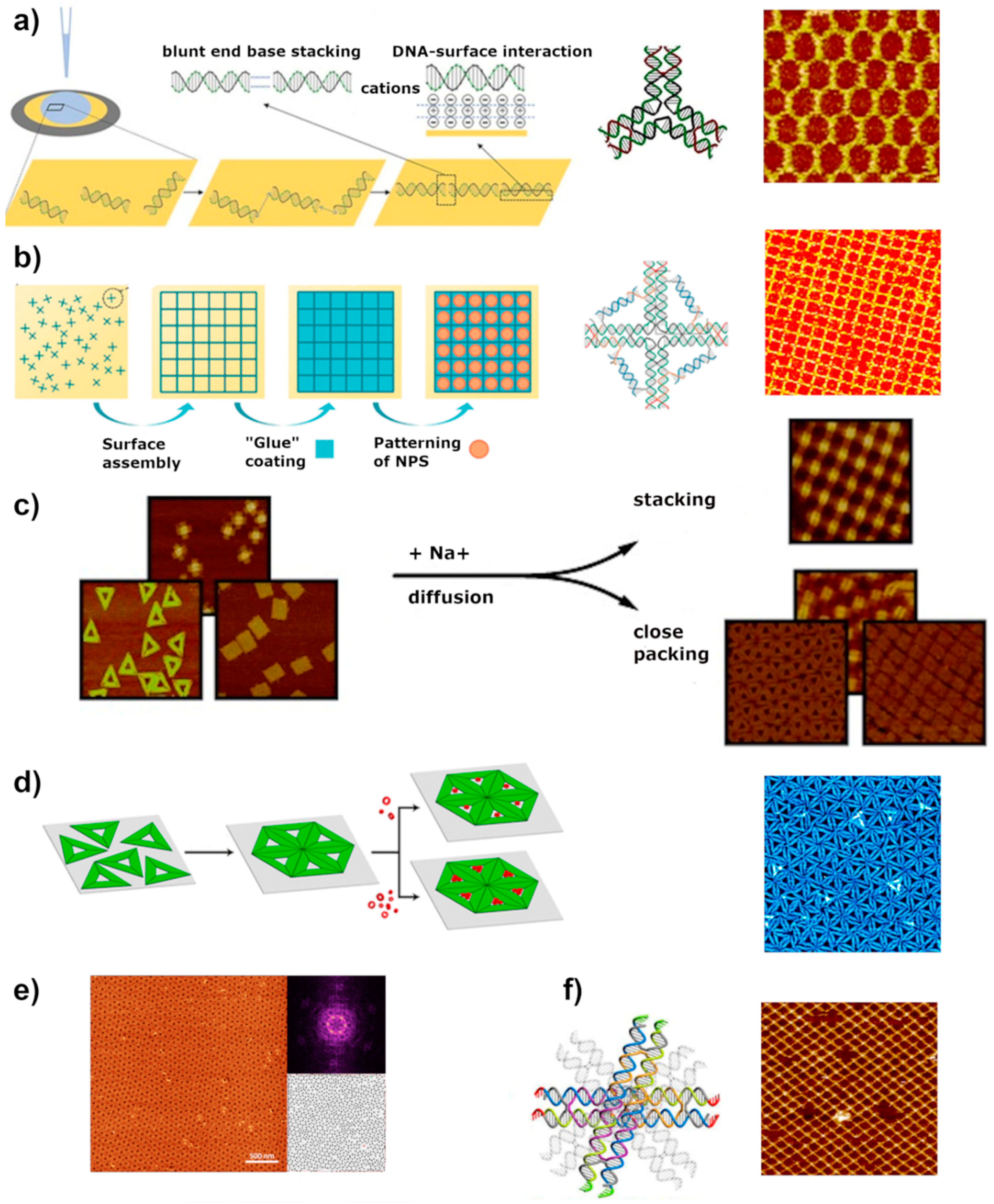
4. Surface-Assisted Assembly
4.1. Mica
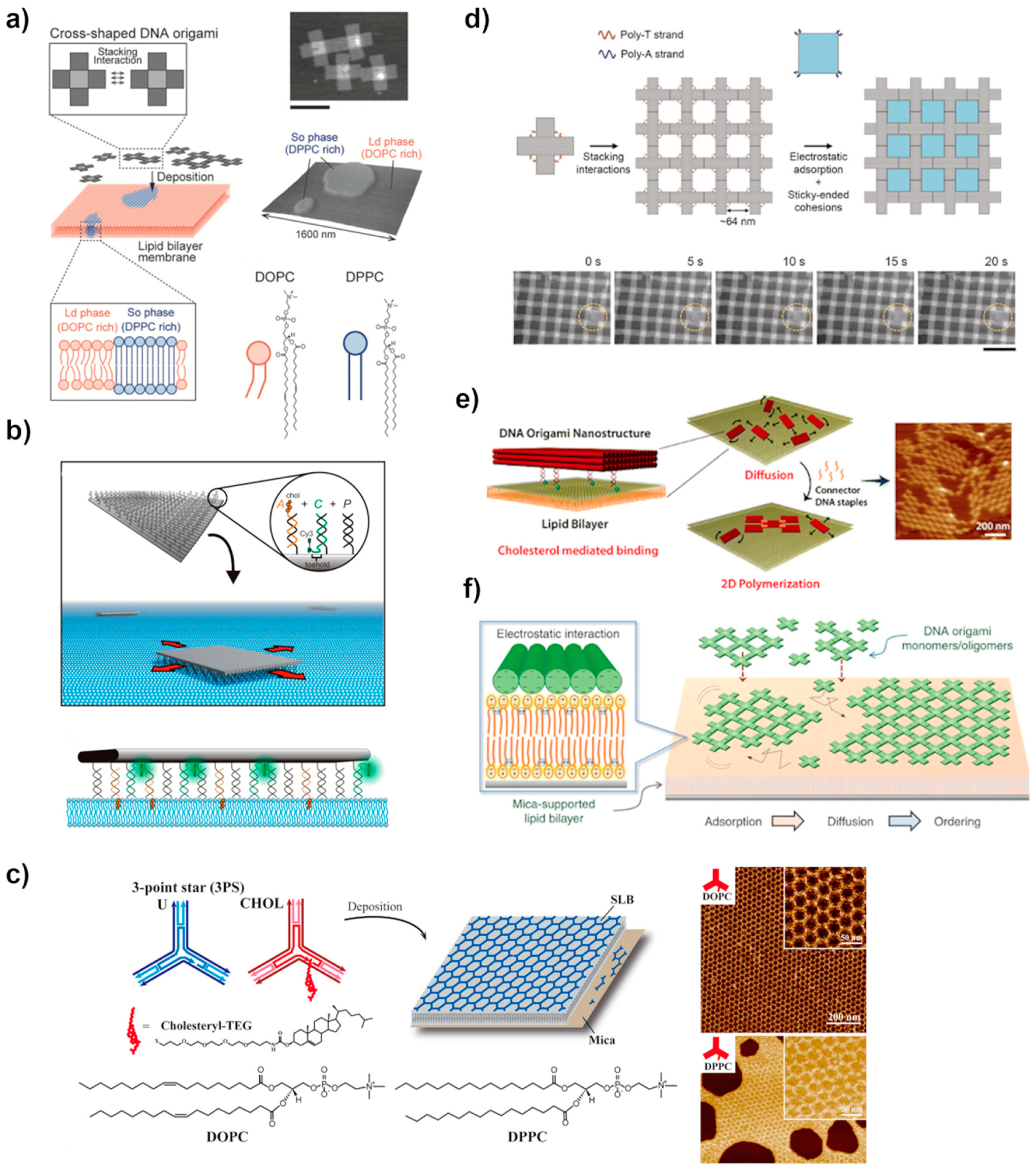
4.2. Lipid Membrane
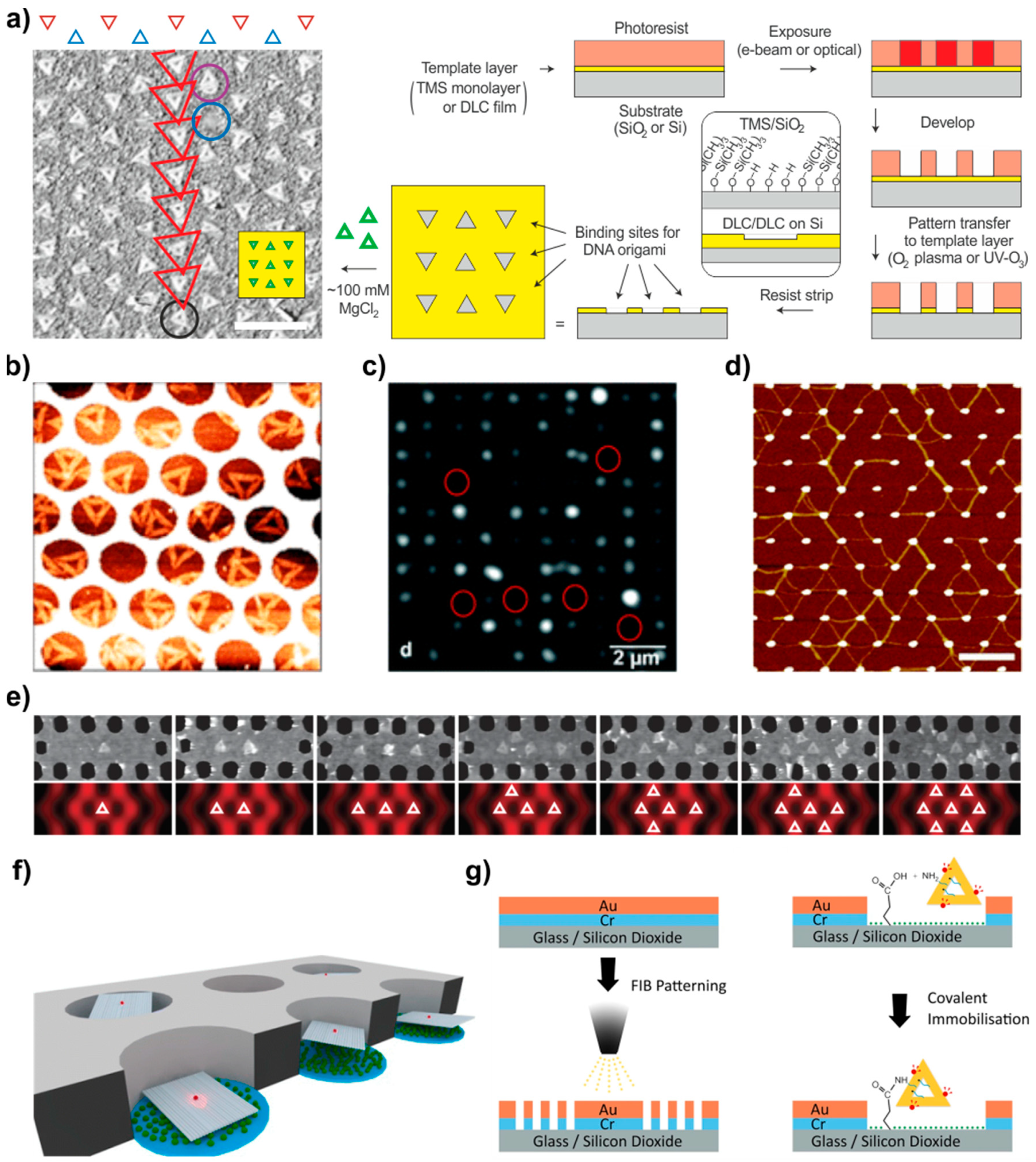
4.3. Templated Assembly (on Silicon and Other Surfaces)
5. Future Prospective
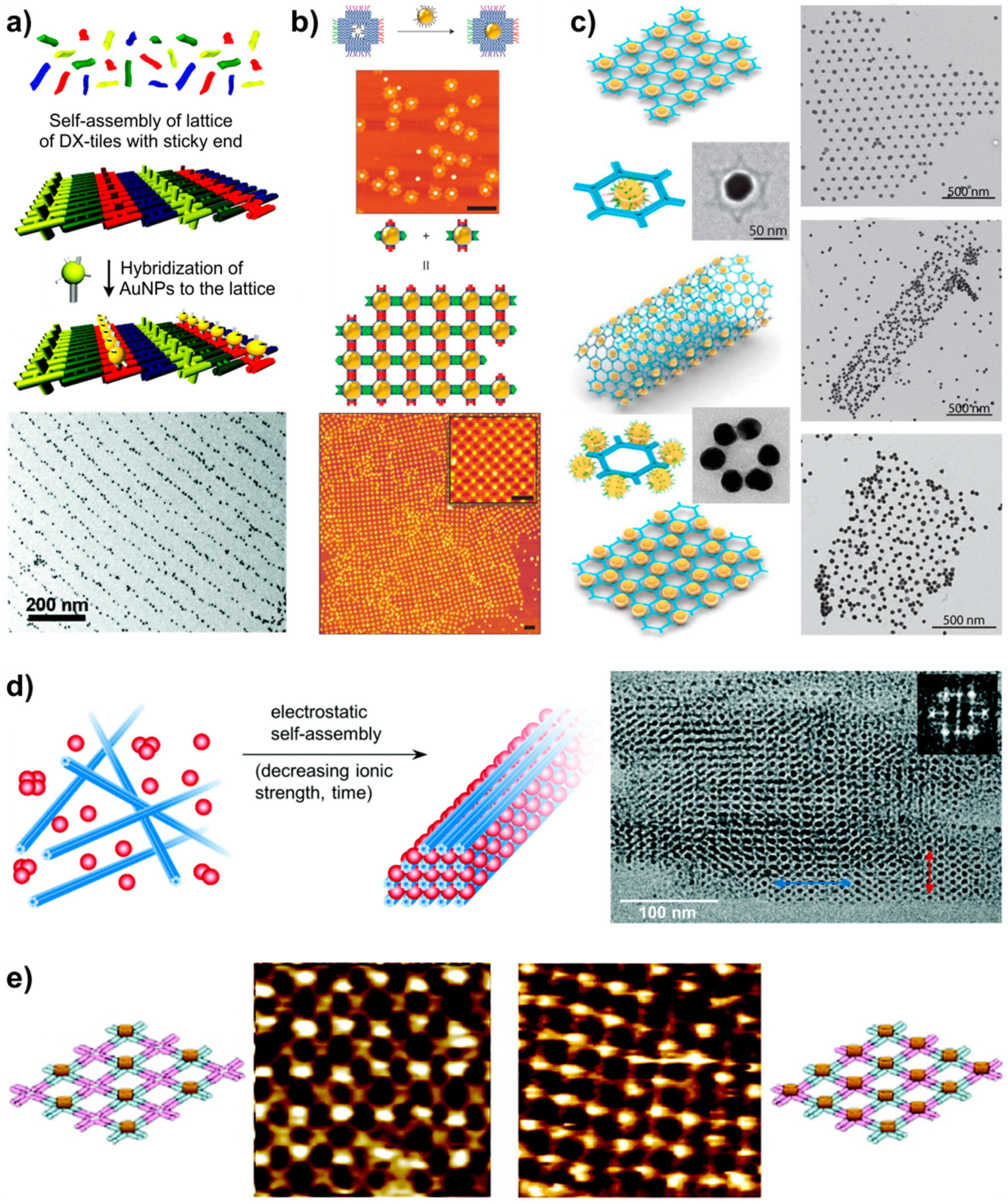
5.1. Arranging Other Components by DNA Lattices
5.2. Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Whitesides, G.M.; Grzybowski, B. Self-assembly at all scales. Science 2002, 295, 2418–2421. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S. Fabrication of novel biomaterials through molecular self-assembly. Nat. Biotechnol. 2003, 21, 1171–1178. [Google Scholar] [CrossRef]
- Grzybowski, B.A.; Huck, W.T.S. The nanotechnology of life-inspired systems. Nat. Nanotechnol. 2016, 11, 585–592. [Google Scholar] [CrossRef]
- Knowles, T.P.J.; Buehler, M.J. Nanomechanics of functional and pathological amyloid materials. Nat. Nanotechnol. 2011, 6, 469–479. [Google Scholar] [CrossRef]
- Pinheiro, A.V.; Han, D.; Shih, W.M.; Yan, H. Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotechnol. 2011, 6, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Madsen, M.; Gothelf, K.V. Chemistries for DNA nanotechnology. Chem. Rev. 2019, 119, 6384–6458. [Google Scholar] [CrossRef]
- Nickels, P.C.; Wünsch, B.; Holzmeister, P.; Bae, W.; Kneer, L.M.; Grohmann, D.; Tinnefeld, P.; Liedl, T. Molecular force spectroscopy with a DNA origami-based nanoscopic force clamp. Science 2016, 354, 305–307. [Google Scholar] [CrossRef]
- Seeman, N.C.; Sleiman, H.F. DNA nanotechnology. Nat. Rev. Mater. 2018, 3, 1–23. [Google Scholar] [CrossRef]
- Seeman, N.C. Nucleic acid junctions and lattices. J. Theor. Biol. 1982, 99, 237–247. [Google Scholar] [CrossRef]
- Seeman, N.C. Structural DNA Nanotechnology: Growing along with nano letters. Nano Lett. 2010, 10, 1971–1978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nat. Cell Biol. 2006, 440, 297–302. [Google Scholar] [CrossRef] [Green Version]
- Douglas, S.M.; Dietz, H.; Liedl, T.; Högberg, B.; Graf, F.; Shih, W.M. Self-assembly of DNA into nanoscale three-dimensional shapes. Nat. Cell Biol. 2009, 459, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Tapio, K.; Bald, I. The potential of DNA origami to build multifunctional materials. Multifunct. Mater. 2020, 3, 032001. [Google Scholar] [CrossRef]
- Wang, P.; Meyer, T.A.; Pan, V.; Dutta, P.K.; Ke, Y. The beauty and utility of DNA origami. Chem 2017, 2, 359–382. [Google Scholar] [CrossRef] [Green Version]
- Sacca, B.; Niemeyer, C.M. DNA origami: The art of folding DNA. Angew. Chem. Int. Ed. 2011, 51, 58–66. [Google Scholar] [CrossRef]
- Acuna, G.P.; Moller, F.M.; Holzmeister, P.; Beater, S.; Lalkens, B.; Tinnefeld, P. Fluorescence enhancement at docking sites of DNA-directed self-assembled nanoantennas. Science 2012, 338, 506–510. [Google Scholar] [CrossRef]
- Roller, E.-M.; Argyropoulos, C.; Högele, A.; Liedl, T.; Pilo-Pais, M. Plasmon–exciton coupling using DNA templates. Nano Lett. 2016, 16, 5962–5966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuzyk, A.; Schreiber, R.; Fan, Z.; Pardatscher, G.; Roller, E.-M.; Högele, A.; Simmel, F.C.; Govorov, A.O.; Liedl, T. DNA-based self-assembly of chiral plasmonic nanostructures with tailored optical response. Nature 2012, 483, 311–314. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Duan, X.; Liu, N. DNA-nanotechnology-enabled chiral plasmonics: From static to dynamic. Acc. Chem. Res. 2017, 50, 2906–2914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuzyk, A.; Schreiber, R.; Zhang, H.; Govorov, A.O.; Liedl, T.; Liu, N. Reconfigurable 3D plasmonic metamolecules. Nat. Mater. 2014, 13, 862–866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A Logic-Gated Nanorobot for targeted transport of molecular payloads. Science 2012, 335, 831–834. [Google Scholar] [CrossRef] [PubMed]
- Linko, V.; Ora, A.; Kostiainen, M.A. DNA Nanostructures as smart drug-delivery vehicles and molecular devices. Trends Biotechnol. 2015, 33, 586–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, Y.; Kotthoff, L.; Olejko, L.; Resch-Genger, U.; Bald, I. DNA origami-based förster resonance energy-transfer nanoarrays and their application as ratiometric sensors. ACS Appl. Mater. Interfaces 2018, 10, 23295–23302. [Google Scholar] [CrossRef] [PubMed]
- Bartnik, K.; Barth, A.; Pilo-Pais, M.; Crevenna, A.H.; Liedl, T.; Lamb, D.C. A DNA origami platform for single-pair förster resonance energy transfer investigation of DNA–DNA interactions and ligation. J. Am. Chem. Soc. 2019, 142, 815–825. [Google Scholar] [CrossRef]
- Langecker, M.; Arnaut, V.; Martin, T.G.; List, J.; Renner, S.; Mayer, M.; Dietz, H.; Simmel, F.C. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science 2012, 338, 932–936. [Google Scholar] [CrossRef] [Green Version]
- Bujold, K.E.; Lacroix, A.; Sleiman, H.F. DNA nanostructures at the interface with biology. Chem 2018, 4, 495–521. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Chao, J.; Pan, D.; Liu, H.; Huang, Q.; Fan, C. Folding super-sized DNA origami with scaffold strands from long-range PCR. Chem. Commun. 2012, 48, 6405–6407. [Google Scholar] [CrossRef] [PubMed]
- Marchi, A.N.; Saaem, I.; Vogen, B.N.; Brown, S.; LaBean, T.H. Toward larger DNA origami. Nano Lett. 2014, 14, 5740–5747. [Google Scholar] [CrossRef]
- Castro, C.E.; Kilchherr, F.; Kim, D.-N.; Shiao, E.L.; Wauer, T.; Wortmann, P.; Bathe, M.; Dietz, H. A primer to scaffolded DNA origami. Nat. Methods 2011, 8, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Wagenbauer, K.F.; Sigl, C.; Dietz, H. Gigadalton-scale shape-programmable DNA assemblies. Nat. Cell Biol. 2017, 552, 78–83. [Google Scholar] [CrossRef]
- Pfeifer, W.; Saccà, B. From nano to macro through hierarchical self-assembly: The DNA paradigm. ChemBioChem 2016, 17, 1063–1080. [Google Scholar] [CrossRef]
- Li, Z.; Liu, M.; Wang, L.; Nangreave, J.; Yan, H.; Liu, Y. Molecular behavior of DNA origami in higher-order self-assembly. J. Am. Chem. Soc. 2010, 132, 13545–13552. [Google Scholar] [CrossRef] [Green Version]
- Tigges, T.; Heuser, T.; Tiwari, R.; Walther, A. 3D DNA origami cuboids as monodisperse patchy nanoparticles for switchable hierarchical self-assembly. Nano Lett. 2016, 16, 7870–7874. [Google Scholar] [CrossRef]
- Liu, W.; Zhong, H.; Wang, R.; Seeman, N.C. Crystalline two-dimensional DNA-origami arrays. Angew. Chem. Int. Ed. 2010, 50, 264–267. [Google Scholar] [CrossRef] [Green Version]
- Jungmann, R.; Scheible, M.; Kuzyk, A.; Pardatscher, G.; Castro, C.E.; Simmel, F.C. DNA origami-based nanoribbons: Assembly, length distribution, and twist. Nanotechnology 2011, 22, 275301. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, W.; Lill, P.; Gatsogiannis, C.; Saccà, B. Hierarchical assembly of DNA filaments with designer elastic properties. ACS Nano 2018, 12, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Han, N.; Nangreave, J.; Liu, Y.; Yan, H. DNA Origami with double-stranded DNA as a unified scaffold. ACS Nano 2012, 6, 8209–8215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C.; Liu, Y.; Rinker, S.; Yan, H. DNA tile based self-assembly: Building complex nanoarchitectures. ChemPhysChem 2006, 7, 1641–1647. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Zhang, F.; Liu, Y.; Yan, H. DNA origami: Scaffolds for creating higher order structures. Chem. Rev. 2017, 117, 12584–12640. [Google Scholar] [CrossRef]
- Xin, Y.; Rivadeneira, S.M.; Grundmeier, G.; Castro, M.; Keller, A. Self-assembly of highly ordered DNA origami lattices at solid-liquid interfaces by controlling cation binding and exchange. Nano Res. 2020, 13, 3142–3150. [Google Scholar] [CrossRef]
- Julin, S.; Nummelin, S.; Kostiainen, M.A.; Linko, V. DNA nanostructure-directed assembly of metal nanoparticle superlat-tices. J. Nanopart. Res. 2018, 20, 119. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, Y.; Endo, M.; Sugiyama, H. Lipid-bilayer-assisted two-dimensional self-assembly of dna origami nanostructures. Nat. Commun. 2015, 6, 8052. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Mao, Y. DNA Origami as scaffolds for self-assembly of lipids and proteins. ChemBioChem 2019, 20, 2422–2431. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Linko, V.; Tapio, K.; Pikker, S.; Lemma, T.; Gopinath, A.; Gothelf, K.V.; Kostiainen, M.A.; Toppari, J.J. Plasmonic Nanostructures through DNA-assisted lithography. Sci. Adv. 2018, 4, eaap8978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, C.G.; Winfree, E. DNA sticky end design and assignment for robust algorithmic self-assembly. Comput. Vis. 2013, 8141, 61–75. [Google Scholar] [CrossRef] [Green Version]
- Evans, C.G.; Winfree, E. Physical principles for DNA tile self-assembly. Chem. Soc. Rev. 2017, 46, 3808–3829. [Google Scholar] [CrossRef] [PubMed]
- Seeman, N.C. Structural DNA nanotechnology: An overview. Breast Cancer 2005, 303, 143–166. [Google Scholar]
- Tsu-Ju, F.; Seeman, N.C. DNA double-crossover molecules. Biochemistry 1993, 32, 3211–3220. [Google Scholar]
- LaBean, T.H.; Yan, H.; Kopatsch, J.; Liu, F.; Winfree, E.; Reif, J.H.; Seeman, N.C. Construction, analysis, ligation, and self-assembly of DNA triple crossover complexes. J. Am. Chem. Soc. 2000, 122, 1848–1860. [Google Scholar] [CrossRef]
- Dietz, H.; Douglas, S.M.; Shih, W.M. Folding DNA into twisted and curved nanoscale shapes. Science 2009, 325, 725–730. [Google Scholar] [CrossRef] [Green Version]
- Yin, P.; Hariadi, R.F.; Sahu, S.; Choi, H.M.T.; Park, S.H.; LaBean, T.H.; Reif, J.H. Programming DNA tube circumferences. Science 2008, 321, 824–826. [Google Scholar] [CrossRef] [Green Version]
- Wei, B.; Dai, M.; Yin, P. Complex shapes self-assembled from single-stranded DNA tiles. Nat. Cell Biol. 2012, 485, 623–626. [Google Scholar] [CrossRef] [Green Version]
- Han, D.; Pal, S.; Yang, Y.; Jiang, S.; Nangreave, J.; Liu, Y.; Yan, H. DNA gridiron nanostructures based on four-arm junctions. Science 2013, 339, 1412–1415. [Google Scholar] [CrossRef] [Green Version]
- Benson, E.; Mohammed, A.; Gardell, J.; Masich, S.; Czeizler, E.; Orponen, P.; Högberg, B. DNA rendering of polyhedral meshes at the nanoscale. Nat. Cell Biol. 2015, 523, 441–444. [Google Scholar] [CrossRef] [Green Version]
- Veneziano, R.; Ratanalert, S.; Zhang, K.; Zhang, F.; Yan, H.; Chiu, W.; Bathe, M. Designer nanoscale DNA assemblies programmed from the top down. Science 2016, 352, 1534. [Google Scholar] [CrossRef] [Green Version]
- Ban, E.; Picu, C.R. Strength of DNA sticky end links. Biomacromolecules 2013, 15, 143–149. [Google Scholar] [CrossRef]
- Iinuma, R.; Ke, Y.; Jungmann, R.; Schlichthaerle, T.; Woehrstein, J.B.; Yin, P. Polyhedra self-assembled from DNA tripods and characterized with 3D DNA-PAINT. Science 2014, 344, 65–69. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Wang, M.; Zhang, F.; Zhu, J. DNA polygonal cavities with tunable shapes and sizes. Chem. Commun. 2015, 51, 16247–16250. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Gorday, K.; Nuckolls, C.; Wind, S.J. Control of DNA origami inter-tile connection with vertical linkers. Chem. Commun. 2016, 52, 1610–1613. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Kuzuya, A.; Liu, W.; Seeman, N.C. Blunt-ended DNA stacking interactions in a 3-helix motif. Chem. Commun. 2010, 46, 4905–4907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Gaitanaros, S.; Lee, S.; Bathe, M.; Shih, W.M.; Ke, Y. Programming self-assembly of DNA origami honeycomb two-dimensional lattices and plasmonic metamaterials. J. Am. Chem. Soc. 2016, 138, 7733–7740. [Google Scholar] [CrossRef]
- Suzuki, Y.; Sugiyama, H.; Endo, M. Complexing DNA origami frameworks through sequential self-assembly based on directed docking. Angew. Chem. Int. Ed. 2018, 57, 7061–7065. [Google Scholar] [CrossRef]
- Marras, A.E.; Zhou, L.; Su, H.-J.; Castro, C.E. Programmable motion of DNA origami mechanisms. Proc. Natl. Acad. Sci. USA 2015, 112, 713–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rafat, A.A.; Pirzer, T.; Scheible, M.B.; Kostina, A.; Simmel, F.C. Surface-assisted large-scale ordering of DNA origami tiles. Angew. Chem. Int. Ed. 2014, 53, 7665–7668. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.; Rothemund, P.W.K. Self-assembly of two-dimensional DNA origami lattices using cation-controlled surface diffusion. Nat. Commun. 2014, 5, 1–11. [Google Scholar]
- Liu, L.; Li, Y.; Wang, Y.; Zheng, J.; Mao, C. Regulating DNA self-assembly by DNA-surface interactions. ChemBioChem 2017, 18, 2404–2407. [Google Scholar] [CrossRef]
- Winfree, E.; Liu, F.; Wenzler, L.A.; Seeman, N.C. Design and self-assembly of two-dimensional DNA crystals. Nat. Cell Biol. 1998, 394, 539–544. [Google Scholar] [CrossRef]
- Liu, F.; Sha, R.; Seeman, N.C. Modifying the surface features of two-dimensional DNA crystals. J. Am. Chem. Soc. 1999, 121, 917–922. [Google Scholar] [CrossRef]
- Liu, Y.; Ke, Y.; Yan, H. Self-assembly of symmetric finite-size DNA nanoarrays. J. Am. Chem. Soc. 2005, 127, 17140–17141. [Google Scholar] [CrossRef]
- Park, S.H.; Pistol, C.; Ahn, S.J.; Reif, J.H.; Lebeck, A.R.; Dwyer, C.; LaBean, T.H. Finite-size, fully addressable DNA tile lattices formed by hierarchical assembly procedures. Angew. Chem. Int. Ed. 2006, 45, 735–739. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Ko, S.H.; Zhang, C.; Ribbe, A.E.; Mao, C. Surface-mediated DNA self-assembly. J. Am. Chem. Soc. 2009, 131, 13248–13249. [Google Scholar] [CrossRef]
- Lund, K.; Liu, Y.; Lindsay, S.; Yan, H. Self-assembling a molecular pegboard. J. Am. Chem. Soc. 2005, 127, 17606–17607. [Google Scholar] [CrossRef] [PubMed]
- Patitz, M.J.; Summers, S.M. Self-assembly of discrete self-similar fractals. Nat. Comput. 2009, 9, 135–172. [Google Scholar] [CrossRef] [Green Version]
- Rothemund, P.W.K.; Papadakis, N.; Winfree, E. Algorithmic self-assembly of DNA sierpinski triangles. PLoS Biol. 2004, 2, e424. [Google Scholar] [CrossRef] [PubMed]
- Schulman, R.; Yurke, B.; Winfree, E. Robust self-replication of combinatorial information via crystal growth and scission. Proc. Natl. Acad. Sci. USA 2012, 109, 6405–6410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barish, R.D.; Schulman, R.; Rothemund, P.W.K.; Winfree, E. An information-bearing seed for nucleating algorithmic self-assembly. Proc. Natl. Acad. Sci. USA 2009, 106, 6054–6059. [Google Scholar] [CrossRef] [Green Version]
- Tikhomirov, G.; Petersen, P.; Qian, L. Programmable disorder in random DNA tilings. Nat. Nanotechnol. 2016, 12, 251–259. [Google Scholar] [CrossRef]
- Zhao, Z.; Liu, Y.; Yan, H. Organizing DNA origami tiles into larger structures using preformed scaffold frames. Nano Lett. 2011, 11, 2997–3002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, H.; Park, S.H.; Finkelstein, G.; Reif, J.H.; LaBean, T.H. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science 2003, 301, 1882–1884. [Google Scholar] [CrossRef]
- Malo, J.; Mitchell, J.C.; Vénien-Bryan, C.; Harris, J.R.; Wille, H.; Sherratt, D.J.; Turberfield, A.J. Engineering a 2D protein-DNA crystal. Angew. Chem. Int. Ed. 2005, 44, 3057–3061. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Constantinou, P.E.; Micheel, C.; Alivisatos, A.P.; Kiehl, R.A.; Seeman, N.C. Two-dimensional nanoparticle arrays show the organizational power of robust DNA motifs. Nano Lett. 2006, 6, 1502–1504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.; Birktoft, J.J.; Chen, Y.; Wang, T.; Sha, R.; Constantinou, P.E.; Ginell, S.L.; Mao, C.; Seeman, N.C. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nat. Cell Biol. 2009, 461, 74–77. [Google Scholar] [CrossRef] [PubMed]
- Kallenbach, N.R.; Ma, R.-I.; Seeman, N.C. An immobile nucleic acid junction constructed from oligonucleotides. Nat. Cell Biol. 1983, 305, 829–831. [Google Scholar] [CrossRef]
- Wang, Y.; Mueller, J.E.; Kemper, B.; Seeman, N.C. Assembly and characterization of five-arm and six-arm DNA branched junctions. Biochemistry 1991, 30, 5667–5674. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.-I.; Kallenbach, N.R.; Sheardy, R.D.; Petrillo, M.L.; Seeman, N.C. Three-arm nucleic acid junctions are flexible. Nucleic Acids Res. 1986, 14, 9745–9753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Seeman, N.C. Assembly and characterization of 8-arm and 12-arm DNA branched junctions. J. Am. Chem. Soc. 2007, 129, 8169–8176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Lin, T.; Zhang, S.; Bai, T.; Mi, Y.; Wei, B. Self-assembly of fully addressable DNA nanostructures from double crossover tiles. Nucleic Acids Res. 2016, 44, 7989–7996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Lin, C.; Li, H.; Yan, H. Aptamer-directed self-assembly of protein arrays on a DNA nanostructure. Angew. Chem. Int. Ed. 2005, 44, 4333–4338. [Google Scholar] [CrossRef]
- Liu, D.; Park, S.H.; Reif, J.H.; LaBean, T.H. DNA Nanotubes Self-Assembled from Triple-Crossover Tiles as Templates for conductive nanowires. Proc. Natl. Acad. Sci. USA 2004, 101, 717–722. [Google Scholar] [CrossRef] [Green Version]
- Park, S.H.; Barish, R.; Li, H.; Reif, J.H.; Finkelstein, G.; Yan, H.; LaBean, T.H. Three-helix bundle DNA tiles self-assemble into 2D lattice or 1D templates for silver nanowires. Nano Lett. 2005, 5, 693–696. [Google Scholar] [CrossRef]
- Mathieu, F.; Liao, S.; Kopatsch, J.; Wang, T.; Mao, C.; Seeman, N.C. Six-helix bundles designed from DNA. Nano Lett. 2005, 5, 661–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, L.; Chao, T.C.; Camacho-Alanis, F.; Ros, A. Six-helix bundle and triangle DNA origami insulator-based dielectro-phoresis. Anal. Chem. 2013, 85, 11427–11434. [Google Scholar] [CrossRef] [PubMed]
- Julin, S.; Korpi, A.; Nonappa; Shen, B.; Liljeström, V.; Ikkala, O.; Keller, A.; Linko, V.; Kostiainen, M.A. DNA origami directed 3D nanoparticle superlattice: Via electrostatic assembly. Nanoscale 2019, 11, 4546–4551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teschome, B.; Facsko, S.; Schönherr, T.; Kerbusch, J.; Keller, A.; Erbe, A. Temperature-dependent charge transport through individually contacted DNA origami-based au nanowires. Langmuir 2016, 32, 10159–10165. [Google Scholar] [CrossRef]
- Ding, B.; Wu, H.; Xu, W.; Zhao, Z.; Liu, Y.; Yu, H.; Yan, H. Interconnecting gold islands with DNA origami nanotubes. Nano Lett. 2010, 10, 5065–5069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zadegan, R.M.; Norton, M.L. Structural DNA nanotechnology: From design to applications. Int. J. Mol. Sci. 2012, 13, 7149–7162. [Google Scholar] [CrossRef]
- Hong, F.; Jiang, S.; Lan, X.; Narayanan, R.P.; Šulc, P.; Zhang, F.; Liu, Y.; Yan, H. Layered-crossover tiles with precisely tunable angles for 2D and 3D DNA crystal engineering. J. Am. Chem. Soc. 2018, 140, 14670–14676. [Google Scholar] [CrossRef]
- He, Y.; Tian, Y.; Ribbe, A.A.E.; Mao, C. Highly connected two-dimensional crystals of DNA six-point-stars. J. Am. Chem. Soc. 2006, 128, 15978–15979. [Google Scholar] [CrossRef]
- He, Y.; Chen, Y.; Liu, H.; Ribbe, A.A.E.; Mao, C. Self-assembly of hexagonal DNA two-dimensional (2D) arrays. J. Am. Chem. Soc. 2005, 127, 12202–12203. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.; Ke, A.Y.; Yan, H. Periodic square-like gold nanoparticle arrays templated by self-assembled 2D DNA nanogrids on a surface. Nano Lett. 2006, 6, 248–251. [Google Scholar] [CrossRef]
- Tikhomirov, G.; Petersen, P.; Qian, L. Triangular DNA origami tilings. J. Am. Chem. Soc. 2018, 140, 17361–17364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shih, W.M.; Quispe, J.D.; Joyce, G.F. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nat. Cell Biol. 2004, 427, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Mi, Y. A new triple crossover triangle (TXT) motif for DNA self-assembly. Biomacromolecules 2005, 6, 2528–2532. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Sun, A.W.; Seeman, N.C. Designed two-dimensional DNA holliday junction arrays visualized by atomic force microscopy. J. Am. Chem. Soc. 1999, 121, 5437–5443. [Google Scholar] [CrossRef]
- Endo, M.; Sugita, T.; Katsuda, Y.; Hidaka, K.; Sugiyama, H. Programmed-assembly system using DNA jigsaw pieces. Chem. A Eur. J. 2010, 16, 5362–5368. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.A.B.; Tuckwell, A.J.; Berengut, J.F.; Bath, J.; Benn, F.; Duff, A.P.; Whitten, A.E.; Dunn, K.E.; Hynson, R.M.; Turberfield, A.J.; et al. Dimensions and global twist of single-layer DNA origami measured by small-angle X-ray scattering. ACS Nano 2018, 12, 5791–5799. [Google Scholar] [CrossRef] [Green Version]
- Woo, S.; Rothemund, P.W.K. Programmable molecular recognition based on the geometry of DNA nanostructures. Nat. Chem. 2011, 3, 620–627. [Google Scholar] [CrossRef]
- Endo, M.; Sugita, T.; Rajendran, A.; Katsuda, Y.; Emura, T.; Hidaka, K.; Sugiyama, H. Two-dimensional DNA origami assemblies using a four-way connector. Chem. Commun. 2011, 47, 3213–3215. [Google Scholar] [CrossRef]
- Rajendran, A.; Endo, M.; Katsuda, Y.; Hidaka, K.; Sugiyama, H. Programmed two-dimensional self-assembly of multiple DNA origami jigsaw pieces. ACS Nano 2010, 5, 665–671. [Google Scholar] [CrossRef]
- Tikhomirov, G.; Petersen, P.; Qian, L. Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns. Nat. Cell Biol. 2017, 552, 67–71. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Halverson, J.; Tian, Y.; Tkachenko, A.V.; Gang, O. Self-organized architectures from assorted DNA-framed nanoparticles. Nat. Chem. 2016, 8, 867–873. [Google Scholar] [CrossRef]
- Lu, L.; Dai, Y.; Du, H.; Liu, M.; Wu, J.; Zhang, Y.; Liang, Z.; Raza, S.; Wang, D.; Jia, C. Atomic scale understanding of the epitaxy of perovskite oxides on flexible mica substrate. Adv. Mater. Interfaces 2020, 7, 1–8. [Google Scholar] [CrossRef]
- Endo, M.; Sugiyama, H. Single-molecule imaging of dynamic motions of biomolecules in DNA origami nanostructures using high-speed atomic force microscopy. Acc. Chem. Res. 2014, 47, 1645–1653. [Google Scholar] [CrossRef] [PubMed]
- Kielar, C.; Ramakrishnan, S.; Fricke, S.; Grundmeier, G.; Keller, A. Dynamics of DNA origami lattice formation at solid-liquid interfaces. ACS Appl. Mater. Interfaces 2018, 10, 44844–44853. [Google Scholar] [CrossRef]
- Liu, L.; Zheng, M.; Li, Z.; Li, Q.; Mao, C. Patterning nanoparticles with DNA molds. ACS Appl. Mater. Interfaces 2019, 11, 13853–13858. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Subramaniam, S.; Stewart, A.F.; Grundmeier, G.; Keller, A. Regular nanoscale protein patterns via directed adsorption through self-assembled DNA origami masks. ACS Appl. Mater. Interfaces 2016, 8, 31239–31247. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Ji, X.; Grundmeier, G.; Keller, A. Dynamics of lattice defects in mixed DNA origami monolayers. Nanoscale 2020, 12, 9733–9743. [Google Scholar] [CrossRef]
- Liu, W.-J.; Zhang, P.; Sun, T.; Li, L.; Wei, Y.-H.; Wang, K.-Z.; Li, B.; Liu, L. Effect of concentration and adsorption time on the formation of a large-scale origami pattern. Nucl. Sci. Tech. 2019, 30, 111. [Google Scholar] [CrossRef]
- Zhou, W.; Saran, R.; Liu, J. Metal sensing by DNA. Chem. Rev. 2017, 117, 8272–8325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duguid, J.; Bloomfield, V.A.; Benevides, J.; Thomas, G.J. Raman spectroscopy of DNA-metal complexes. I. Interactions and conformational effects of the divalent cations: Mg, Ca, Sr, Ba, Mn, Co, Ni, Cu, Pd, and Cd. Biophys. J. 1993, 65, 1916–1928. [Google Scholar] [CrossRef] [Green Version]
- Pastré, D.; Piétrement, O.; Fusil, S.; Landousy, F.; Jeusset, J.; David, M.-O.; Hamon, L.; Le Cam, E.; Zozime, A. Adsorption of DNA to mica mediated by divalent counterions: A theoretical and experimental study. Biophys. J. 2003, 85, 2507–2518. [Google Scholar] [CrossRef] [Green Version]
- Kan, Y.; Tan, Q.; Wu, G.; Si, W.; Chen, Y. Study of DNA adsorption on mica surfaces using a surface force apparatus. Sci. Rep. 2015, 5, srep08442. [Google Scholar] [CrossRef] [Green Version]
- Piétrement, O.; Pastré, D.; Fusil, S.; Jeusset, J.; David, M.-O.; Landousy, F.; Hamon, L.; Zozime, A.A.; Le Cam, E. Reversible binding of DNA on NiCl2-treated mica by varying the ionic strength. Langmuir 2003, 19, 2536–2539. [Google Scholar] [CrossRef]
- Suzuki, Y.; Endo, M.; Yang, Y.; Sugiyama, H. Dynamic assembly/disassembly processes of photoresponsive DNA origami nanostructures directly visualized on a lipid membrane surface. J. Am. Chem. Soc. 2014, 136, 1714–1717. [Google Scholar] [CrossRef]
- Sato, Y.; Endo, M.; Morita, M.; Takinoue, M.; Sugiyama, H.; Murata, S.; Nomura, S.; Shin-ichiro, M.; Suzuki, Y. Environment-dependent self-assembly of DNA origami lattices on phase-separated lipid membranes. Adv. Mater. Interfaces 2018, 5, 1–6. [Google Scholar] [CrossRef]
- Johnson-Buck, A.; Jiang, S.; Yan, H.; Walter, N.G. DNA–Cholesterol barges as programmable membrane-exploring agents. ACS Nano 2014, 8, 5641–5649. [Google Scholar] [CrossRef]
- Kempter, S.; Khmelinskaia, A.; Strauss, M.T.; Schwille, P.; Jungmann, R.; Liedl, T.; Bae, W. Single particle tracking and super-resolution imaging of membrane-assisted stop-and-go diffusion and lattice assembly of DNA origami. ACS Nano 2019, 13, 996–1002. [Google Scholar] [CrossRef]
- Avakyan, N.; Conway, J.W.; Sleiman, H.F. Long-range ordering of blunt-ended DNA tiles on supported lipid bilayers. J. Am. Chem. Soc. 2017, 139, 12027–12034. [Google Scholar] [CrossRef]
- Kocabey, S.; Kempter, S.; List, J.; Xing, Y.; Bae, W.; Schiffels, D.; Shih, W.M.; Simmel, F.C.; Liedl, T. Membrane-assisted growth of DNA origami nanostructure arrays. ACS Nano 2015, 9, 3530–3539. [Google Scholar] [CrossRef]
- Burns, J.R.; Stulz, E.; Howorka, S. Self-assembled DNA nanopores that span lipid bilayers. Nano Lett. 2013, 13, 2351–2356. [Google Scholar] [CrossRef] [Green Version]
- Burns, J.R.; Göpfrich, K.; Wood, J.W.; Thacker, V.V.; Stulz, E.; Keyser, U.F.; Howorka, S. Lipid-bilayer-spanning DNA nanopores with a bifunctional porphyrin anchor. Angew. Chem. Int. Ed. 2013, 52, 12069–12072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kershner, R.J.; Bozano, L.D.; Micheel, C.M.; Hung, A.M.; Fornof, A.R.; Cha, J.N.; Rettner, C.T.; Bersani, M.; Frommer, J.; Rothemund, P.W.K.; et al. Placement and orientation of individual DNA shapes on lithographically patterned surfaces. Nat. Nanotechnol. 2009, 4, 557–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, A.M.; Micheel, C.M.; Bozano, L.D.; Osterbur, L.W.; Wallraff, G.M.; Cha, J.N. Large-area spatially ordered arrays of gold nanoparticles directed by lithographically confined DNA origami. Nat. Nanotechnol. 2009, 5, 121–126. [Google Scholar] [CrossRef]
- Penzo, E.; Wang, R.; Palma, M.; Wind, S.J. Selective placement of DNA origami on substrates patterned by nanoimprint Lithography. J. Vac. Sci. Technol. B 2011, 29, 6. [Google Scholar] [CrossRef]
- Gopinath, A.; Rothemund, P.W.K. Optimized assembly and covalent coupling of single-molecule DNA origami nanoarrays. ACS Nano 2014, 8, 12030–12040. [Google Scholar] [CrossRef] [Green Version]
- Gao, B.; Sarveswaran, K.; Bernstein, G.H.; Lieberman, M. Guided deposition of individual DNA nanostructures on silicon substrates. Langmuir 2010, 26, 12680–12683. [Google Scholar] [CrossRef]
- Gerdon, A.E.; Oh, S.S.; Hsieh, K.; Ke, Y.; Yan, H.; Soh, H.T. Controlled delivery of DNA origami on patterned surfaces. Small 2009, 5, 1942–1946. [Google Scholar] [CrossRef] [PubMed]
- Brassat, K.; Ramakrishnan, S.; Bürger, J.; Hanke, M.; Doostdar, M.; Lindner, J.K.N.; Grundmeier, G.; Keller, A. On the Adsorption of DNA origami nanostructures in nanohole arrays. Langmuir 2018, 34, 14757–14765. [Google Scholar] [CrossRef]
- Yun, J.M.; Kim, K.N.; Kim, J.Y.; Shin, D.O.; Lee, W.J.; Lee, S.H.; Lieberman, M.; Kim, S.O. DNA origami nanopatterning on chemically modified graphene. Angew. Chem. Int. Ed. 2011, 51, 912–915. [Google Scholar] [CrossRef]
- Shaali, M.; Woller, J.G.; Johansson, P.G.; Hannestad, J.K.; De Battice, L.; Aissaoui, N.; Brown, T.; El-Sagheer, A.H.; Ku-batkin, S.; Lara-Avila, S.; et al. Site-selective immobilization of functionalized DNA origami on nano-patterned teflon AF. J. Mater. Chem. C 2017, 5, 7637–7643. [Google Scholar] [CrossRef]
- Hawkes, W.; Huang, D.; Reynolds, P.; Hammond, L.; Ward, M.; Gadegaard, N.; Marshall, J.F.; Iskratsch, T.; Palma, M. Probing the nanoscale organisation and multivalency of cell surface receptors: DNA origami nanoarrays for cellular studies with single-molecule control. Faraday Discuss. 2019, 219, 203–219. [Google Scholar] [CrossRef] [Green Version]
- Gopinath, A.; Miyazono, E.; Faraon, A.; Rothemund, P.W.K. Engineering and mapping nanocavity emission via precision placement of DNA origami. Nature 2016, 535, 401–405. [Google Scholar] [CrossRef]
- Pibiri, E.; Holzmeister, P.; Lalkens, B.; Acuna, G.P.; Tinnefeld, P. Single-molecule positioning in zeromode waveguides by DNA origami nanoadapters. Nano Lett. 2014, 14, 3499–3503. [Google Scholar] [CrossRef]
- Huang, D.; Freeley, M.; Palma, M. DNA-mediated patterning of single quantum dot nanoarrays: A reusable platform for single-molecule control. Sci. Rep. 2017, 7, srep45591. [Google Scholar] [CrossRef] [PubMed]
- Weiss, P.S. A conversation with Prof. Ned Seeman: Founder of DNA nanotechnology. ACS Nano 2008, 2, 1089–1096. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Lin, Y.; Liu, H.; Wang, L.; Fan, C. DNA-based plasmonic nanostructures. Mater. Today 2015, 18, 326–335. [Google Scholar] [CrossRef]
- Mirkin, C.A.; Letsinger, R.L.; Mucic, R.C.; Storhoff, J.J. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nat. Cell Biol. 1996, 382, 607–609. [Google Scholar] [CrossRef] [PubMed]
- Alivisatos, A.P.; Johnsson, K.P.; Peng, X.; Wilson, T.E.; Loweth, C.J.; Bruchez, M.P.; Schultz, P.G. Organization of ‘nanocrystal molecules’ using DNA. Nat. Cell Biol. 1996, 382, 609–611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, M.R.; Seeman, N.C.; Mirkin, C.A. Programmable materials and the nature of the DNA bond. Science 2015, 347, 1260901. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Kostiainen, M.A.; Linko, V. DNA origami nanophotonics and plasmonics at interfaces. Langmuir 2018, 34, 14911–14920. [Google Scholar] [CrossRef] [Green Version]
- Xiao, S.; Liu, F.; Rosen, A.E.; Hainfeld, J.F.; Seeman, N.C.; Musier-Forsyth, K.; Kiehl, R.A. Selfassembly of metallic NA-noparticle arrays by DNA scaffolding. J. Nanopart. Res. 2002, 4, 313–317. [Google Scholar] [CrossRef]
- Le, J.D.; Pinto, Y.; Seeman, N.C.; Musier-Forsyth, K.; Taton, A.T.A.; Kiehl, R.A. DNA-Templated self-assembly of metallic nanocomponent arrays on a surface. Nano Lett. 2004, 4, 2343–2347. [Google Scholar] [CrossRef]
- Pinto, Y.Y.; Le, J.D.; Seeman, N.C.; Musier-Forsyth, K.; Taton, T.A.; Kiehl, R.A. Sequence-encoded self-assembly of mul-tiple-nanocomponent arrays by 2D DNA scaffolding. Nano Lett. 2005, 5, 2399–2402. [Google Scholar] [CrossRef]
- Park, S.H.; Yin, P.; Liu, Y.; Reif, J.H.; Labean, T.H.; Yan, H. Programmable DMA self-assemblies for nanoscale organization of ligands and proteins. Nano Lett. 2005, 5, 729–733. [Google Scholar] [CrossRef]
- Sharma, J.; Chhabra, R.; Cheng, A.; Brownell, J.; Liu, Y.; Yan, H. Control of self-assembly of DNA tubules through integration of gold nanoparticles. Science 2009, 323, 112–116. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Zhang, Y.; Wang, T.; Xin, H.L.; Li, H.; Gang, O. Lattice engineering through nanoparticle–DNA frameworks. Nat. Mater. 2016, 15, 654–661. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Tagawa, M.; Xin, H.L.; Wang, T.; Emamy, H.; Li, H.; Yager, K.G.; Starr, F.W.; Tkachenko, A.V.; Gang, O. Diamond family of nanoparticle superlattices. Science 2016, 351, 582–586. [Google Scholar] [CrossRef] [Green Version]
- Williams, B.A.R.; Lund, K.; Liu, Y.; Yan, H.; Chaput, J.C. Self-assembled peptide nanoarrays: An approach to studying protein-protein interactions. Angew. Chem. Int. Ed. 2007, 46, 3051–3054. [Google Scholar] [CrossRef] [PubMed]
- Cigler, P.; Lytton-Jean, A.K.R.; Anderson, D.G.; Finn, M.G.; Park, S.Y. DNA-controlled assembly of a NaTl lattice structure from gold nanoparticles and protein nanoparticles. Nat. Mater. 2010, 9, 918–922. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Niemeyer, C.M. From DNA nanotechnology to material systems engineering. Adv. Mater. 2019, 31, 1806294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, D.; Hartman, M.R.; Derrien, T.L.; Hamada, S.; An, D.; Yancey, K.G.; Cheng, R.; Ma, M.; Luo, D. DNA materials: Bridging nanotechnology and biotechnology. Acc. Chem. Res. 2014, 47, 1902–1911. [Google Scholar] [CrossRef]
- Di Felice, R.; Porath, D. DNA-based nanoelectronics. NanoBioTechnology 2008, 141–185. [Google Scholar] [CrossRef]
- Song, X.; Reif, J. Nucleic acid databases and molecular-scale computing. ACS Nano 2019, 13, 6256–6268. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Li, H.; Wang, L.; Gu, H.; Fan, C. DNA nanotechnology-enabled drug delivery systems. Chem. Rev. 2018, 119, 6459–6506. [Google Scholar] [CrossRef] [PubMed]
- Aryal, B.R.; Ranasinghe, D.R.; Westover, T.R.; Calvopiña, D.G.; Davis, R.C.; Harb, J.N.; Woolley, A.T. DNA origami mediated electrically connected metal-semiconductor junctions. Nano Res. 2020, 13, 1419–1426. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parikka, J.M.; Sokołowska, K.; Markešević, N.; Toppari, J.J. Constructing Large 2D Lattices Out of DNA-Tiles. Molecules 2021, 26, 1502. https://doi.org/10.3390/molecules26061502
Parikka JM, Sokołowska K, Markešević N, Toppari JJ. Constructing Large 2D Lattices Out of DNA-Tiles. Molecules. 2021; 26(6):1502. https://doi.org/10.3390/molecules26061502
Chicago/Turabian StyleParikka, Johannes M., Karolina Sokołowska, Nemanja Markešević, and J. Jussi Toppari. 2021. "Constructing Large 2D Lattices Out of DNA-Tiles" Molecules 26, no. 6: 1502. https://doi.org/10.3390/molecules26061502
APA StyleParikka, J. M., Sokołowska, K., Markešević, N., & Toppari, J. J. (2021). Constructing Large 2D Lattices Out of DNA-Tiles. Molecules, 26(6), 1502. https://doi.org/10.3390/molecules26061502






