PepFun: Open Source Protocols for Peptide-Related Computational Analysis
Abstract
1. Introduction
2. Results and Discussion
2.1. The PepFun Code and Functionalities
2.2. Installing and Running PepFun
2.3. PepFun Tutorial and Examples
2.3.1. Analysis of Sequence Properties
| python pepfun.py - m sequence - s [SEQUENCE] |
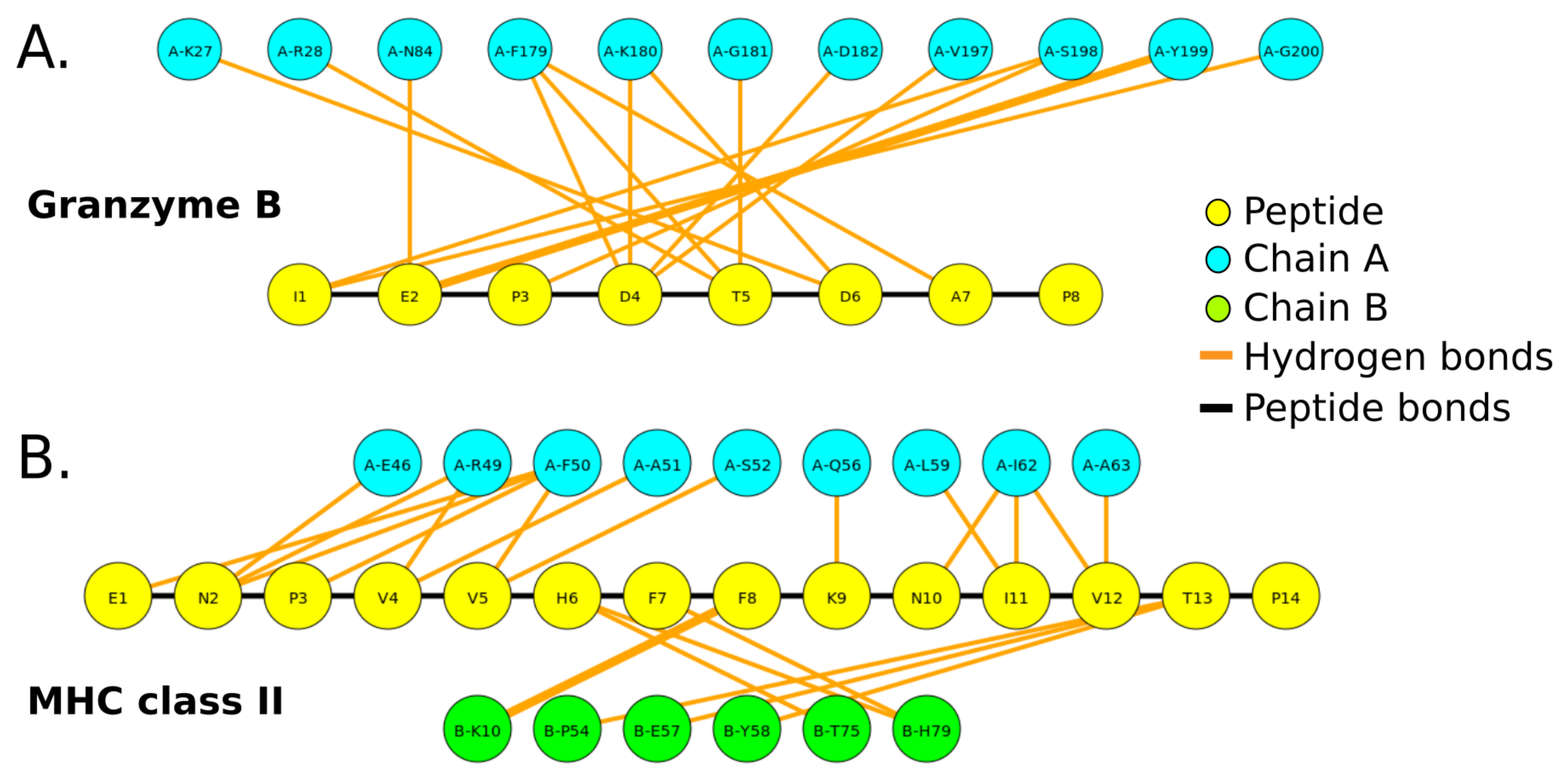
2.3.2. Structural Analysis of Peptides in Complex with Protein Targets
| python pepfun.py - m structure - p [STRUCTURE_FILE] - c [CHAIN] - t [THRESHOLD] |
2.3.3. Peptide Libraries
3. Materials and Methods
3.1. PepFun Technical Considerations
3.2. PepFun Functionalities
3.2.1. Sequence-Based Functionalities
3.2.2. Structure-Based Functionalities
3.2.3. Functions for Customizing Peptide Libraries
3.3. Test of PepFun with Sets of Known Peptide Binders
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Uhlig, T.; Kyprianou, T.; Martinelli, F.G.; Oppici, C.A.; Heiligers, D.; Hills, D.; Calvo, X.R.; Verhaert, P. The emergence of peptides in the pharmaceutical business: From exploration to exploitation. EuPA Open Proteom. 2014, 4, 58–69. [Google Scholar] [CrossRef]
- Fosgerau, K.; Hoffmann, T. Peptide therapeutics: Current status and future directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Vlieghe, P.; Lisowski, V.; Martinez, J.; Khrestchatisky, M. Synthetic therapeutic peptides: Science and market. Drug Discov. Today 2010, 15, 40–56. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Park, J.P.; Dooley, K.; Cropek, D.M.; West, A.C.; Banta, S. Rapid Development of New Protein Biosensors Utilizing Peptides Obtained via Phage Display. PLoS ONE 2011, 6, e24948. [Google Scholar] [CrossRef] [PubMed]
- Skwarczynski, M.; Toth, I. Peptide-based synthetic vaccines. Chem. Sci. 2016, 7, 842–854. [Google Scholar] [CrossRef]
- Vanhee, P.; van der Sloot, A.M.; Verschueren, E.; Serrano, L.; Rousseau, F.; Schymkowitz, J. Computational design of peptide ligands. Trends Biotechnol. 2011, 29, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Zaidman, D.; Wolfson, H.J. PinaColada: Peptide-inhibitor ant colony ad-hoc design algorithm. Bioinformatics 2016, 32, 2289–2296. [Google Scholar] [CrossRef]
- Jenson, J.M.; Xue, V.; Stretz, L.; Mandal, T.; Reich, L.L.; Keating, A.E. Peptide design by optimization on a data-parameterized protein interaction landscape. Proc. Natl. Acad. Sci. USA 2018, 115, E10342–E10351. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; De Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; McGinnis, S.; Madden, T.L. BLAST: Improvements for better sequence analysis. Nucleic Acids Res. 2006, 34, W6–W9. [Google Scholar] [CrossRef] [PubMed]
- Cheol Jeong, J.; Lin, X.; Chen, X.W. On position-specific scoring matrix for protein function prediction. IEEE/ACM Trans. Comput. Biol. Bioinform. 2010, 8, 308–315. [Google Scholar] [CrossRef]
- Barley, M.H.; Turner, N.J.; Goodacre, R. Improved Descriptors for the Quantitative Structure–Activity Relationship Modeling of Peptides and Proteins. J. Chem. Inf. Model. 2018, 58, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.W.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2011, 2, 31–45. [Google Scholar]
- Raveh, B.; London, N.; Schueler-Furman, O. Sub-angstrom modeling of complexes between flexible peptides and globular proteins. Proteins Struct. Funct. Bioinform. 2010, 78, 2029–2040. [Google Scholar] [CrossRef] [PubMed]
- London, N.; Raveh, B.; Schueler-Furman, O. Peptide docking and structure-based characterization of peptide binding: From knowledge to know-how. Curr. Opin. Struct. Biol. 2013, 23, 894–902. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Antes, I. DynaDock: A now molecular dynamics-based algorithm for protein-peptide docking including receptor flexibility. Proteins Struct. Funct. Bioinform. 2010, 78, 1084–1104. [Google Scholar] [CrossRef]
- Kamenik, A.S.; Lessel, U.; Fuchs, J.E.; Fox, T.; Liedl, K.R. Peptidic Macrocycles - Conformational Sampling and Thermodynamic Characterization. J. Chem. Inf. Model. 2018, 58, 982–992. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Zhang, D.; Huang, S.Y. Efficient conformational ensemble generation of protein-bound peptides. J. Cheminform. 2017, 9, 59. [Google Scholar] [CrossRef]
- Tu, M.; Cheng, S.; Lu, W.; Du, M. Advancement and prospects of bioinformatics analysis for studying bioactive peptides from food-derived protein: Sequence, structure, and functions. TrAC Trends Anal. Chem. 2018, 105, 7–17. [Google Scholar] [CrossRef]
- Shen, Y.; Maupetit, J.; Derreumaux, P.; Tufféry, P. Improved PEP-FOLD Approach for Peptide and Miniprotein Structure Prediction. J. Chem. Theory Comput. 2014, 10, 4745–4758. [Google Scholar] [CrossRef]
- Cock, P.J.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, D.; Weiss, R.M.; Terwilliger, T.C. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc. Natl. Acad. Sci. USA 1984, 81, 140–144. [Google Scholar] [CrossRef]
- Mannhold, R.; Van de Waterbeemd, H. Substructure and whole molecule approaches for calculating logP. J. Comput.-Aided Mol. Des. 2001, 15, 337–354. [Google Scholar] [CrossRef]
- Wieczorek, M.; Abualrous, E.T.; Sticht, J.; Álvaro-Benito, M.; Stolzenberg, S.; Noé, F.; Freund, C. Major histocompatibility complex (MHC) class I and MHC class II proteins: Conformational plasticity in antigen presentation. Front. Immunol. 2017, 8, 1–16. [Google Scholar] [CrossRef]
- Santos, G.B.; Ganesan, A.; Emery, F.S. Oral Administration of Peptide-Based Drugs: Beyond Lipinski’s Rule. ChemMedChem 2016, 11, 2245–2251. [Google Scholar] [CrossRef] [PubMed]
- Ochoa, R.; Magnitov, M.; Laskowski, R.A.; Cossio, P.; Thornton, J.M. An automated protocol for modelling peptide substrates to proteases. BMC Bioinform. 2020, 21, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, S.; Kanehisa, M. AAindex: Amino acid index database. Nucleic Acids Res. 2000, 28, 374. [Google Scholar] [CrossRef]
- McGinnis, S.; Madden, T.L. BLAST: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004, 32, W20–W25. [Google Scholar] [CrossRef] [PubMed]
- Guerin, M.E.; Stirnemann, G.; Giganti, D. Conformational entropy of a single peptide controlled under force governs protease recognition and catalysis. Proc. Natl. Acad. Sci. USA 2018, 115, 11525–11530. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, J.E.; von Grafenstein, S.; Huber, R.G.; Margreiter, M.A.; Spitzer, G.M.; Wallnoefer, H.G.; Liedl, K.R. Cleavage Entropy as Quantitative Measure of Protease Specificity. PLoS Comput. Biol. 2013, 9, e1003007. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple Ligand–Protein Interaction Diagrams for Drug Discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Gajdoš, P.; Ježowicz, T.; Uher, V.; Dohnálek, P. A parallel Fruchterman–Reingold algorithm optimized for fast visualization of large graphs and swarms of data. Swarm Evol. Comput. 2016, 26, 56–63. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Zhang, T.; Li, H.; Xi, H.; Stanton, R.V.; Rotstein, S.H. HELM: A hierarchical notation language for complex biomolecule structure representation. J. Chem. Inf. Model. 2012, 52, 2796–2806. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Ebejer, J.P.; Morris, G.M.; Deane, C.M. Freely available conformer generation methods: How good are they? J. Chem. Inf. Model. 2012, 52, 1146–1158. [Google Scholar] [CrossRef] [PubMed]
- Spellmeyer, D.C.; Wong, A.K.; Bower, M.J.; Blaney, J.M. Conformational analysis using distance geometry methods. J. Mol. Graph. Model. 1997, 15, 18–36. [Google Scholar] [CrossRef]
- Halgren, T.A. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 1996, 17, 490–519. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. Inter J. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Ochoa, R.; Laio, A.; Cossio, P. Predicting the Affinity of Peptides to Major Histocompatibility Complex Class II by Scoring Molecular Dynamics Simulations. J. Chem. Inf. Model. 2019, 59, 3464–3473. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Sidney, J.; Kim, Y.; Sette, A.; Lund, O.; Nielsen, M.; Peters, B. Peptide Binding Predictions for HLA DR, DP and DQ Molecules. BMC Bioinform. 2010, 11, 568. [Google Scholar] [CrossRef]
- Loffler, P.; Schmitz, S.; Hupfeld, E.; Sterner, R.; Merkl, R.; Hughes, M. Rosetta:MSF: A modular framework for multi-state computational protein design. PLoS Comput. Biol. 2017, 13, e1005600. [Google Scholar] [CrossRef]
- Ochoa, R.; Soler, M.A.; Laio, A.; Cossio, P. Assessing the capability of in silico mutation protocols for predicting the finite temperature conformation of amino acids. Phys. Chem. Chem. Phys. 2018, 20, 25901–25909. [Google Scholar] [CrossRef] [PubMed]
- Hedstrom, L. Serine protease mechanism and specificity. Chem. Rev. 2002, 102, 4501–4523. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef] [PubMed]




| Property | Average Values MHC Set | Average Values Protease Set |
|---|---|---|
| Net charge | 0.457 | −1.872 |
| Molecular weight (g/mol) | 1696.111 | 884.133 |
| LogP | −5.810 | −4.235 |
| Hydrophobicity | 1.735 | 0.002 |
| Aromaticity | 0.099 | 0.043 |
| Instability index | 33.883 | 37.095 |
| Isoelectric point | 7.290 | 4.375 |
| Number hydrogen donors | 24.183 | 13.337 |
| Number hydrogen acceptors | 23.670 | 13.578 |
| Number of solubility rules failed | 2 | 1.5 |
| Number of synthesis rules failed | 4.5 | 0.5 |
| Peptide Position | Average Number of Contacts |
|---|---|
| P4 | 13.64 ± 6.57 |
| P3 | 12.63 ± 1.66 |
| P2 | 18.70 ± 12.37 |
| P1 | 45.59 ± 4.90 |
| P1’ | 15.25 ± 8.68 |
| P2’ | 7.89 ± 6.71 |
| P3’ | 2.41 ± 1.85 |
| P4’ | 0.02 ± 0.19 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ochoa, R.; Cossio, P. PepFun: Open Source Protocols for Peptide-Related Computational Analysis. Molecules 2021, 26, 1664. https://doi.org/10.3390/molecules26061664
Ochoa R, Cossio P. PepFun: Open Source Protocols for Peptide-Related Computational Analysis. Molecules. 2021; 26(6):1664. https://doi.org/10.3390/molecules26061664
Chicago/Turabian StyleOchoa, Rodrigo, and Pilar Cossio. 2021. "PepFun: Open Source Protocols for Peptide-Related Computational Analysis" Molecules 26, no. 6: 1664. https://doi.org/10.3390/molecules26061664
APA StyleOchoa, R., & Cossio, P. (2021). PepFun: Open Source Protocols for Peptide-Related Computational Analysis. Molecules, 26(6), 1664. https://doi.org/10.3390/molecules26061664







