l-Aspartate: An Essential Metabolite for Plant Growth and Stress Acclimation
Abstract
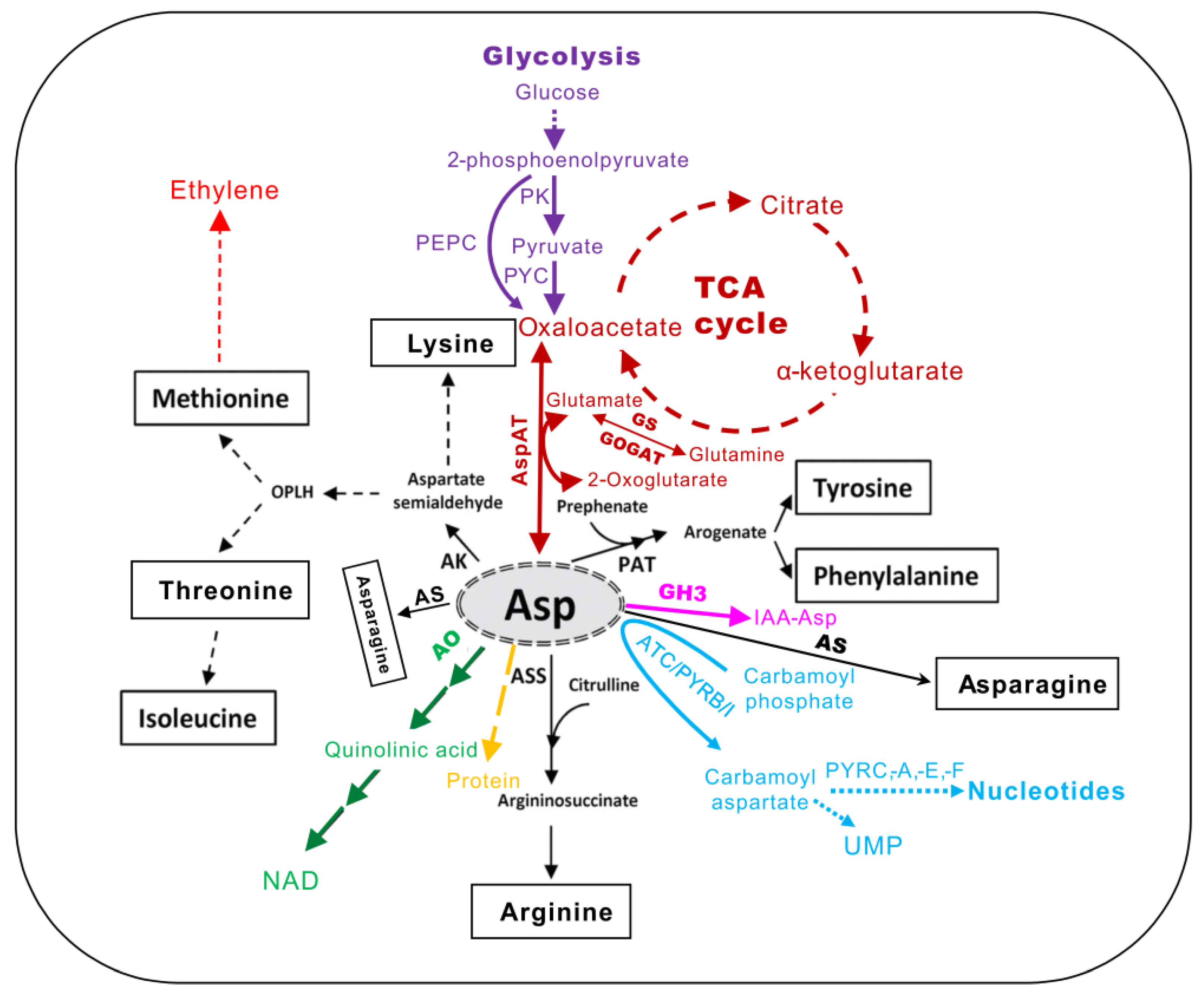
:1. Introduction
2. The Biosynthesis and Transformation of Asp in Plants
2.1. Key Enzymes Involved in Asp Anabolism and Catabolism
2.1.1. Aspartate Aminotransferase
2.1.2. Asparate Kinase (AK)
2.1.3. Aspartate Oxidase (AO)
2.1.4. Argininosuccinate Synthase (ASS)
2.1.5. Aspartate Transcarbamylase (ATC)
2.1.6. Malate–Aspartate Shuttle
2.2. Aspartate Transporters
2.3. The Effect of Asp/Asn Homeostasis on Plants
3. Role of Asp in Growth and Stresses
3.1. Asp is an Endogenous Metabolic Limitation for Cell Proliferation
3.2. Asp in Plants Coordinates Nitrogen Assimilation into Amino Acids
3.3. Asp is a Drought Stress-Specific Responsive Metabolite
3.4. The Variation of Asp Level Is Closely Linked to Stress Acclimation
3.5. Asp Acts as a Biomarker of Biotic Stress and Environment-Induced Exposure
4. Asp Signaling and Its Association with Phytohormones
5. Conclusions and Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Caspi, R.; Billington, R.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Midford, P.E.; Ong, W.K.; Paley, S.; Subhraveti, P.; Karp, P.D. The MetaCyc database of metabolic pathways and enzymes–A 2019 update. Nucleic Acids Res. 2020, 48, D445–D453. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wei, H.; Wang, T.; Xu, Q.; Zhang, C.; Fan, X.; Ma, Q.; Chen, N.; Xie, X. Current status on metabolic engineering for the production of l-aspartate family amino acids and derivatives. Bioresour. Technol. 2017, 245, 1588–1602. [Google Scholar] [CrossRef]
- Lam, H.M.; Peng, S.; Coruzzi, G.M. Metabolic Regulation of the Gene Encoding Glutamine-Dependent Asparagine Synthetase in Arabidopsis thaliana. Plant Physiol. 1994, 106, 1347–1357. [Google Scholar] [CrossRef] [Green Version]
- Gaufichon, L.; Rothstein, S.J.; Suzuki, A. Asparagine Metabolic Pathways in Arabidopsis. Plant Cell Physiol. 2016, 57, 675–689. [Google Scholar] [CrossRef] [Green Version]
- de la Torre, F.; Canas, R.A.; Pascual, M.B.; Avila, C.; Canovas, F.M. Plastidic aspartate aminotransferases and the biosynthesis of essential amino acids in plants. J. Exp. Bot. 2014, 65, 5527–5534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birsoy, K.; Wang, T.; Chen, W.W.; Freinkman, E.; Abu-Remaileh, M.; Sabatini, D.M. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell 2015, 162, 540–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chai, M.-F.; Chen, Q.-J.; An, R.; Chen, Y.-M.; Chen, J.; Wang, X.-C. NADK2, an Arabidopsis Chloroplastic NAD Kinase, Plays a Vital Role in Both Chlorophyll Synthesis and Chloroplast Protection. Plant Mol. Biol. 2005, 59, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Vanderauwera, S.; De Block, M.; Van de Steene, N.; van de Cotte, B.; Metzlaff, M.; Van Breusegem, F. Silencing of poly(ADP-ribose) polymerase in plants alters abiotic stress signal transduction. Proc. Natl. Acad. Sci. USA 2007, 104, 15150–15155. [Google Scholar] [CrossRef] [Green Version]
- Hashida, S.; Takahashi, H.; Uchimiya, H. The role of NAD biosynthesis in plant development and stress responses. Ann. Bot. 2009, 103, 819–824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dornfeld, K.; Madden, M.; Skildum, A.; Wallace, K.B. Aspartate facilitates mitochondrial function, growth arrest and survival during doxorubicin exposure. Cell Cycle 2015, 14, 3282–3291. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, L.B.; Gui, D.Y.; Hosios, A.M.; Bush, L.N.; Freinkman, E.; Vander Heiden, M.G. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell 2015, 162, 552–563. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Bermudez, J.; Baudrier, L.; La, K.; Zhu, X.G.; Fidelin, J.; Sviderskiy, V.O.; Papagiannakopoulos, T.; Molina, H.; Snuderl, M.; Lewis, C.A.; et al. Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat. Cell Biol. 2018, 20, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Alkan, H.F.; Walter, K.E.; Luengo, A.; Madreiter-Sokolowski, C.T.; Stryeck, S.; Lau, A.N.; Al-Zoughbi, W.; Lewis, C.A.; Thomas, C.J.; Hoefler, G.; et al. Cytosolic Aspartate Availability Determines Cell Survival When Glutamine Is Limiting. Cell Metab. 2018, 28, 706–720.e6. [Google Scholar] [CrossRef] [Green Version]
- Meléndez-Rodríguez, F.; Urrutia, A.A.; Lorendeau, D.; Rinaldi, G.; Roche, O.; Böğürcü-Seidel, N.; Ortega Muelas, M.; Mesa-Ciller, C.; Turiel, G.; Bouthelier, A.; et al. HIF1α Suppresses Tumor Cell Proliferation through Inhibition of Aspartate Biosynthesis. Cell Rep. 2019, 26, 2257–2265.e4. [Google Scholar] [CrossRef] [Green Version]
- Alkan, H.F.; Bogner-Strauss, J.G. Maintaining cytosolic aspartate levels is a major function of the TCA cycle in proliferating cells. Mol. Cell. Oncol. 2019, 6, e1536843. [Google Scholar] [CrossRef] [Green Version]
- Ritterhoff, J.; Young, S.; Villet, O.; Shao, D.; Neto, F.C.; Bettcher, L.F.; Hsu, Y.-W.A.; Kolwicz, S.C.; Raftery, D.; Tian, R. Metabolic Remodeling Promotes Cardiac Hypertrophy by Directing Glucose to Aspartate Biosynthesis. Circ. Res. 2020, 126, 182–196. [Google Scholar] [CrossRef]
- Fu, H.; Zhang, J.; Tepper, P.G.; Bunch, L.; Jensen, A.A.; Poelarends, G.J. Chemoenzymatic Synthesis and Pharmacological Characterization of Functionalized Aspartate Analogues As Novel Excitatory Amino Acid Transporter Inhibitors. J. Med. Chem. 2018, 61, 7741–7753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, J.L.; Forcat, S.; Beckmann, M.; Bennett, M.; Miller, S.J.; Baker, J.M.; Hawkins, N.D.; Vermeer, C.P.; Lu, C.; Lin, W.; et al. The metabolic transition during disease following infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Plant J. 2010, 63, 443–457. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Mancuso, A.; Daikhin, E.; Nissim, I.; Yudkoff, M.; Wehrli, S.; Thompson, C.B. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 19345–19350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toney, M.D. Aspartate aminotransferase: An old dog teaches new tricks. Arch. Biochem. Biophys. 2014, 544, 119–127. [Google Scholar] [CrossRef] [Green Version]
- Hasegawa, D.; Kito, K.; Maeda, T.; Rai, V.; Cha-um, S.; Tanaka, Y.; Fukaya, M.; Takabe, T. Functional characterization of aminotransferase involved in serine and aspartate metabolism in a halotolerant cyanobacterium, Aphanothece halophytica. Protoplasma 2019, 256, 1727–1736. [Google Scholar] [CrossRef] [PubMed]
- Wrenger, C.; Muller, I.B.; Silber, A.M.; Jordanova, R.; Lamzin, V.S.; Groves, M.R. Aspartate Aminotransferase - Bridging Carbohydrate and Energy Metabolism in Plasmodium Falciparum. Curr. Drug Metab. 2012, 13, 332–336. [Google Scholar] [CrossRef]
- Wadsworth, G.J. The plant aspartate aminotransferase gene family. Physiol. Plant. 1997, 100, 998–1006. [Google Scholar] [CrossRef]
- de la Torre, F.; De Santis, L.; Suárez, M.F.; Crespillo, R.; Cánovas, F.M. Identification and functional analysis of a prokaryotic-type aspartate aminotransferase: Implications for plant amino acid metabolism. Plant J. 2006, 46, 414–425. [Google Scholar] [CrossRef]
- Graindorge, M.; Giustini, C.; Jacomin, A.C.; Kraut, A.; Curien, G.; Matringe, M. Identification of a plant gene encoding glutamate/aspartate-prephenate aminotransferase: The last homeless enzyme of aromatic amino acids biosynthesis. FEBS Lett. 2010, 584, 4357–4360. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Dudareva, N. The Shikimate Pathway and Aromatic Amino Acid Biosynthesis in Plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef]
- Schultz, C.J.; Coruzzi, G.M. The aspartate aminotransferase gene family of Arabidopsis encodes isozymes localized to three distinct subcellular compartments. Plant J. 1995, 7, 61–75. [Google Scholar] [CrossRef]
- Schultz, C.J.C.J.; Hsu, M.; Miesak, B.; Coruzzi, G.M. Arabidopsis mutants define an in vivo role for isoenzymes of aspartate aminotransferase in plant nitrogen assimilation. Genetics 1998, 149, 491–499. [Google Scholar]
- de la Torre, F.; Suarez, M.F.; De Santis, L.; Canovas, F.M. The aspartate aminotransferase family in conifers: Biochemical analysis of a prokaryotic-type enzyme from maritime pine. Tree Physiol. 2007, 27, 1283–1291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angelovici, R.; Fait, A.; Fernie, A.R.; Galili, G. A seed high-lysine trait is negatively associated with the TCA cycle and slows down Arabidopsis seed germination. New Phytol. 2011, 189, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Martin, M.N.; Hudson, A.O.; Lee, J.; Muhitch, M.J.; Leustek, T. Methionine and threonine synthesis are limited by homoserine availability and not the activity of homoserine kinase in Arabidopsis thaliana. Plant J. 2005, 41, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, M.; Wang, G.; Galili, G. New insights into the metabolism of aspartate-family amino acids in plant seeds. Plant Reprod. 2018, 31, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Nasu, S.; Wicks, F.D.; Gholson, R.K. L-Aspartate Oxidase, a Newly Discovered Enzyme of Escherichia coli, Is the B Protein of Quinolinate Synthetase. J. Biol. Chem. 1982, 257, 626–632. [Google Scholar] [CrossRef]
- Katoh, A.; Uenohara, K.; Akita, M.; Hashimoto, T. Early Steps in the Biosynthesis of NAD in Arabidopsis Start with Aspartate and Occur in the Plastid. Plant Physiol. 2006, 141, 851–857. [Google Scholar] [CrossRef] [Green Version]
- Hao, J.; Pétriacq, P.; de Bont, L.; Hodges, M.; Gakière, B. Characterization of l -aspartate oxidase from Arabidopsis thaliana. Plant Sci. 2018, 271, 133–142. [Google Scholar] [CrossRef]
- Gakière, B.; Hao, J.; de Bont, L.; Pétriacq, P.; Nunes-Nesi, A.; Fernie, A.R. NAD + Biosynthesis and Signaling in Plants. CRC. Crit. Rev. Plant Sci. 2018, 37, 259–307. [Google Scholar] [CrossRef]
- Slocum, R.D. Genes, enzymes and regulation of arginine biosynthesis in plants. Plant Physiol. Biochem. 2005, 43, 729–745. [Google Scholar] [CrossRef]
- de la Torre, F.; El-Azaz, J.; Avila, C.; Canovas, F.M. Deciphering the Role of Aspartate and Prephenate Aminotransferase Activities in Plastid Nitrogen Metabolism. PLANT Physiol. 2014, 164, 92–104. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.-T.; Qi, Y.; Wang, Y.-C.; Chi, K.K.; Chung, Y.; Ouyang, C.; Chen, Y.-R.; Oh, M.E.; Sheng, X.; Tang, Y.; et al. Arginine starvation kills tumor cells through aspartate exhaustion and mitochondrial dysfunction. Commun. Biol. 2018, 1, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, C.; Vaishnav, A.; Edwards, B.F.P.; Evans, D.R. Characterization and assembly of the Pseudomonas aeruginosa aspartate transcarbamoylase-pseudo dihydroorotase complex. PLoS ONE 2020, 15, e0229494. [Google Scholar] [CrossRef] [Green Version]
- Kantrowitz, E.R. Allostery and cooperativity in Escherichia coli aspartate transcarbamoylase. Arch. Biochem. Biophys. 2012, 519, 81–90. [Google Scholar] [CrossRef] [Green Version]
- Kafer, C.; Zhou, L.; Santoso, D.; Guirgis, A.; Weers, B.; Park, S.; Thornburg, R. Regulation of pyrimidine metabolism in plants. Front. Biosci. 2004, 9, 1611–1625. [Google Scholar] [CrossRef] [PubMed]
- Witz, S.; Jung, B.; Fürst, S.; Möhlmann, T. De Novo Pyrimidine Nucleotide Synthesis Mainly Occurs outside of Plastids, but a Previously Undiscovered Nucleobase Importer Provides Substrates for the Essential Salvage Pathway in Arabidopsis. Plant Cell 2012, 24, 1549–1559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borst, P. The malate–aspartate shuttle (Borst cycle): How it started and developed into a major metabolic pathway. IUBMB Life 2020, 72, 2241–2259. [Google Scholar] [CrossRef]
- Easlon, E.; Tsang, F.; Skinner, C.; Wang, C.; Lin, S.-J. The malate-aspartate NADH shuttle components are novel metabolic longevity regulators required for calorie restriction-mediated life span extension in yeast. Genes Dev. 2008, 22, 931–944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Contreras, L.; Satrústegui, J. Calcium Signaling in Brain Mitochondria. J. Biol. Chem. 2009, 284, 7091–7099. [Google Scholar] [CrossRef] [Green Version]
- Monné, M.; Miniero, D.V.; Bisaccia, F.; Fiermonte, G. The mitochondrial oxoglutarate carrier: From identification to mechanism. J. Bioenerg. Biomembr. 2013, 45, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Winefield, C.S.; Farnden, K.J.F.; Reynolds, P.H.S.; Marshall, C.J. Evolutionary analysis of aspartate aminotransferases. J. Mol. Evol. 1995, 40, 455–463. [Google Scholar] [CrossRef]
- Amoedo, N.D.; Punzi, G.; Obre, E.; Lacombe, D.; De Grassi, A.; Pierri, C.L.; Rossignol, R. AGC1/2, the mitochondrial aspartate-glutamate carriers. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 2394–2412. [Google Scholar] [CrossRef]
- Bröer, S.; Palacín, M. The role of amino acid transporters in inherited and acquired diseases. Biochem. J. 2011, 436, 193–211. [Google Scholar] [CrossRef] [Green Version]
- Monné, M.; Daddabbo, L.; Gagneul, D.; Obata, T.; Hielscher, B.; Palmieri, L.; Miniero, D.V.; Fernie, A.R.; Weber, A.P.M.; Palmieri, F. Uncoupling proteins 1 and 2 (UCP1 and UCP2) from Arabidopsis thaliana are mitochondrial transporters of aspartate, glutamate, and dicarboxylates. J. Biol. Chem. 2018, 293, 4213–4227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorgoglione, R.; Porcelli, V.; Santoro, A.; Daddabbo, L.; Vozza, A.; Monné, M.; Di Noia, M.A.; Palmieri, L.; Fiermonte, G.; Palmieri, F. The human uncoupling proteins 5 and 6 (UCP5/SLC25A14 and UCP6/SLC25A30) transport sulfur oxyanions, phosphate and dicarboxylates. Biochim. Biophys. Acta Bioenerg. 2019, 1860, 724–733. [Google Scholar] [CrossRef]
- Noguchi, K.; Yoshida, K. Interaction between photosynthesis and respiration in illuminated leaves. Mitochondrion 2008, 8, 87–99. [Google Scholar] [CrossRef]
- Monné, M.; Vozza, A.; Lasorsa, F.M.; Porcelli, V.; Palmieri, F. Mitochondrial carriers for aspartate, glutamate and other amino acids: A review. Int. J. Mol. Sci. 2019, 20, 4456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, N.; Gu, M.; Hu, J.; Qu, H.; Xu, G. Rice OsLHT1 Functions in Leaf-to-Panicle Nitrogen Allocation for Grain Yield and Quality. Front. Plant Sci. 2020, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Camargos, L.S.; Aguiar, L.F.; Azevedo, R.A. Variation in the Amino Acid Concentration During Development of Canavalia ensiformes. Biol. Plant. 2004, 48, 309–312. [Google Scholar] [CrossRef]
- Pratelli, R.; Pilot, G. Regulation of amino acid metabolic enzymes and transporters in plants. J. Exp. Bot. 2014, 65, 5535–5556. [Google Scholar] [CrossRef] [PubMed]
- Yabuki, Y.; Ohashi, M.; Imagawa, F.; Ishiyama, K.; Beier, M.P.; Konishi, N.; Umetsu-Ohashi, T.; Hayakawa, T.; Yamaya, T.; Kojima, S. A temporal and spatial contribution of asparaginase to asparagine catabolism during development of rice grains. Rice 2017, 10. [Google Scholar] [CrossRef] [Green Version]
- Deng, L.; Yao, P.; Li, L.; Ji, F.; Zhao, S.; Xu, C.; Lan, X.; Jiang, P. p53-mediated control of aspartate-asparagine homeostasis dictates LKB1 activity and modulates cell survival. Nat. Commun. 2020, 11, 1755. [Google Scholar] [CrossRef] [PubMed]
- Wan, T.-F.; Shao, G.-H.; Shan, X.-C.; Zeng, N.-Y.; Lam, H.-M. Correlation between AS1 Gene Expression and Seed Protein Contents in Different Soybean (Glycine Max [L.] Merr.) Cultivars. Plant Biol. 2006, 8, 271–276. [Google Scholar] [CrossRef]
- Antunes, F.; Aguilar, M.; Pineda, M.; Sodek, L. Nitrogen stress and the expression of asparagine synthetase in roots and nodules of soybean ( Glycine max ). Physiol. Plant. 2008, 133, 736–743. [Google Scholar] [CrossRef]
- Lam, H.-M.; Wong, P.; Chan, H.-K.; Yam, K.-M.; Chen, L.; Chow, C.-M.; Coruzzi, G.M. Overexpression of the ASN1 Gene Enhances Nitrogen Status in Seeds of Arabidopsis. Plant Physiol. 2003, 132, 926–935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seiffert, B.; Zhou, Z.; Wallbraun, M.; Lohaus, G.; Mollers, C. Expression of a bacterial asparagine synthetase gene in oilseed rape (Brassica napus) and its effect on traits related to nitrogen efficiency. Physiol. Plant. 2004, 121, 656–665. [Google Scholar] [CrossRef]
- Galili, G. The aspartate-family pathway of plants: Linking production of essential amino acids with energy and stress regulation. Plant Signal. Behav. 2011, 6, 192–195. [Google Scholar] [CrossRef] [Green Version]
- Galili, G.; Höfgen, R. Metabolic Engineering of Amino Acids and Storage Proteins in Plants. Metab. Eng. 2002, 4, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Galili, G. Increased lysine synthesis coupled with a knockout of its catabolism synergistically boosts lysine content and also transregulates the metabolism of other amino acids in Arabidopsis seeds. Plant Cell 2003, 15, 845–853. [Google Scholar] [CrossRef] [Green Version]
- Yagi, T.; Sako, M.; Moriuti, S.; Shounaka, M.; Masaki, K.; Yamamoto, S.; Yagi, T.; Sako, M.; Moriuti, S.; Shounaka, M. Purification and Characterization of Aspartate Aminotransferase Isoenzymes from Rice Bran Purification and Characterization of Aspartate Aminotransferase Isoenzymes from Rice. Biosci. Biotechnol. Biochem. 2014, 57, 2074–2080. [Google Scholar] [CrossRef] [Green Version]
- Kirma, M.; Araujo, W.L.; Fernie, A.R.; Galili, G. The multifaceted role of aspartate-family amino acids in plant metabolism. J. Exp. Bot. 2012, 63, 4995–5001. [Google Scholar] [CrossRef] [Green Version]
- Lea, P.J.; Sodek, L.; Parry, M.A.J.; Shewry, P.R.; Halford, N.G. Asparagine in plants. Ann. Appl. Biol. 2007, 150, 1–26. [Google Scholar] [CrossRef]
- Okumoto, S.; Pilot, G. Amino Acid Export in Plants: A Missing Link in Nitrogen Cycling. Mol. Plant 2011, 4, 453–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lima, J.D.; Sodek, L. N-stress alters aspartate and asparagine levels of xylem sap in soybean. Plant Sci. 2003, 165, 649–656. [Google Scholar] [CrossRef]
- Schlüter, U.; Colmsee, C.; Scholz, U.; Bräutigam, A.; Weber, A.P.M.; Zellerhoff, N.; Bucher, M.; Fahnenstich, H.; Sonnewald, U. Adaptation of maize source leaf metabolism to stress related disturbances in carbon, nitrogen and phosphorus balance. BMC Genom. 2013, 14, 442. [Google Scholar] [CrossRef] [Green Version]
- Zerche, S.; Haensch, K.T.; Druege, U.; Hajirezaei, M.R. Nitrogen remobilisation facilitates adventitious root formation on reversible dark-induced carbohydrate depletion in Petunia hybrida. BMC Plant Biol. 2016, 16, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vitor, S.C.; do Amarante, L.; Sodek, L. Are phloem-derived amino acids the origin of the elevated malate concentration in the xylem sap following mineral N starvation in soybean? Planta 2018, 248, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, J.K.; Devi, S.; Buckseth, T.; Ali, N.; Singh, R.K.; Zinta, R.; Dua, V.K.; Chakrabarti, S.K. Precision phenotyping of contrasting potato (Solanum tuberosum L.) varieties in a novel aeroponics system for improving nitrogen use efficiency: In search of key traits and genes. J. Integr. Agric. 2020, 19, 51–61. [Google Scholar] [CrossRef]
- Silvente, S. Molecular cloning of the cDNA encoding aspartate aminotransferase from bean root nodules and determination of its role in nodule nitrogen metabolism. J. Exp. Bot. 2003, 54, 1545–1551. [Google Scholar] [CrossRef]
- Good, A.G.; Zaplachinski, S.T. The effects of drought stress on free amino acid accumulation and protein synthesis in Brassica napus. Physiol. Plant. 1994, 90, 9–14. [Google Scholar] [CrossRef]
- Jia, X.; Sun, C.; Zuo, Y.; Li, G.; Li, G.; Ren, L.; Chen, G. Integrating transcriptomics and metabolomics to characterise the response of Astragalus membranaceus Bge. var. mongolicus (Bge.) to progressive drought stress. BMC Genom. 2016, 17, 188. [Google Scholar] [CrossRef] [Green Version]
- Ullah, N.; Yüce, M.; Neslihan Öztürk Gökçe, Z.; Budak, H. Comparative metabolite profiling of drought stress in roots and leaves of seven Triticeae species. BMC Genom. 2017, 18, 1–12. [Google Scholar] [CrossRef]
- Gargallo-Garriga, A.; Preece, C.; Sardans, J.; Oravec, M.; Urban, O.; Peñuelas, J. Root exudate metabolomes change under drought and show limited capacity for recovery. Sci. Rep. 2018, 8, 12696. [Google Scholar] [CrossRef] [Green Version]
- Khan, N.; Bano, A.; Rahman, M.A.; Guo, J.; Kang, Z.; Babar, M.A. Comparative Physiological and Metabolic Analysis Reveals a Complex Mechanism Involved in Drought Tolerance in Chickpea (Cicer arietinum L.) Induced by PGPR and PGRs. Sci. Rep. 2019, 9, 2097. [Google Scholar] [CrossRef]
- Barickman, T.C.; Ku, K.-M.; Sams, C.E. Differing precision irrigation thresholds for kale (Brassica oleracea L. var. acephala) induces changes in physiological performance, metabolites, and yield. Environ. Exp. Bot. 2020, 180, 104253. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, G.; Zhao, P.; Zhou, Q.; Zhao, X. The abundance of certain metabolites responds to drought stress in the highly drought tolerant plant Caragana korshinskii. Acta Physiol. Plant. 2017, 39. [Google Scholar] [CrossRef]
- Ali, Q.; Athar, H.-R.; Haider, M.Z.; Shahid, S.; Aslam, N.; Shehzad, F.; Naseem, J.; Ashraf, R.; Ali, A.; Hussain, S.M. Role of Amino Acids in Improving Abiotic Stress Tolerance to Plants. In Plant Tolerance to Environmental Stress; CRC Press: Boca Raton, FL, USA, 2019; pp. 175–204. ISBN 9780203705315. [Google Scholar]
- Li, M.; Guo, R.; Jiao, Y.; Jin, X.; Zhang, H.; Shi, L. Comparison of salt tolerance in Soja based on metabolomics of seedling roots. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Paidi, M.K.; Agarwal, P.; More, P.; Agarwal, P.K. Chemical Derivatization of Metabolite Mass Profiling of the Recretohalophyte Aeluropus lagopoides Revealing Salt Stress Tolerance Mechanism. Mar. Biotechnol. 2017, 19, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, R.; Khan, A.L.; Bilal, S.; Waqas, M.; Kang, S.M.; Lee, I.J. Inoculation of abscisic acid-producing endophytic bacteria enhances salinity stress tolerance in Oryza sativa. Environ. Exp. Bot. 2017, 136, 68–77. [Google Scholar] [CrossRef]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments: A review. Plant Signal. Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef] [Green Version]
- El-Shintinawy, F.; El-Shourbagy, M.N. Alleviation of changes in protein metabolism in NaCl-stressed wheat seedlings by thiamine. Biol. Plantarum 2001, 44, 541–545. [Google Scholar] [CrossRef]
- Koehler, G.; Rohloff, J.; Wilson, R.C.; Kopka, J.; Erban, A.; Winge, P.; Bones, A.M.; Davik, J.; Alsheikh, M.K.; Randall, S.K. Integrative “omic” analysis reveals distinctive cold responses in leaves and roots of strawberry, fragaria × ananassa ‘Korona’. Front. Plant Sci. 2015, 6, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byeon, S.-E.; Lee, J. Differential responses of fruit quality and major targeted metabolites in three different cultivars of cold-stored figs (Ficus carica L.). Sci. Hortic. 2020, 260, 108877. [Google Scholar] [CrossRef]
- Janmohammadi, M.; Sabaghnia, N.; Mahfoozi, S. Frost tolerance and metabolite changes of rye (Secale cereale) during the cold hardening and overwintering. Acta Physiol. Plant. 2018, 40, 42. [Google Scholar] [CrossRef]
- Nguyen, V.L.; Palmer, L.; Roessner, U.; Stangoulis, J. Genotypic Variation in the Root and Shoot Metabolite Profiles of Wheat (Triticum aestivum L.) Indicate Sustained, Preferential Carbon Allocation as a Potential Mechanism in Phosphorus Efficiency. Front. Plant Sci. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schützendübel, A.; Nikolova, P.; Rudolf, C.; Polle, A. Cadmium and H2O2-induced oxidative stress in Populus x canescens roots. Plant Physiol. Biochem. 2002, 40, 577–584. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehmann, M.; Laxa, M.; Sweetlove, L.J.; Fernie, A.R.; Obata, T. Metabolic recovery of Arabidopsis thaliana roots following cessation of oxidative stress. Metabolomics 2012, 8, 143–153. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, M.; Schwarzländer, M.; Obata, T.; Sirikantaramas, S.; Burow, M.; Olsen, C.E.; Tohge, T.; Fricker, M.D.; Møller, B.L.; Fernie, A.R.; et al. The metabolic response of Arabidopsis roots to oxidative stress is distinct from that of heterotrophic cells in culture and highlights a complex relationship between the levels of transcripts, metabolites, and flux. Mol. Plant 2009, 2, 390–406. [Google Scholar] [CrossRef] [Green Version]
- Okunev, R.V. Free Amino Acid Accumulation in Soil and Tomato Plants (Solanum lycopersicum L.) Associated with Arsenic Stress. Water Air Soil Pollut. 2019, 230, 1–10. [Google Scholar] [CrossRef]
- Yan, L.; Riaz, M.; Liu, Y.; Zeng, Y.; Jiang, C. Aluminum toxicity could be mitigated with boron by altering the metabolic patterns of amino acids and carbohydrates rather than organic acids in trifoliate orange. Tree Physiol. 2019, 39, 1572–1582. [Google Scholar] [CrossRef]
- Gaude, N.; Bortfeld, S.; Erban, A.; Kopka, J.; Krajinski, F. Symbiosis dependent accumulation of primary metabolites in arbuscule-containing cells. BMC Plant Biol. 2015, 15, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Kasote, D.M.; Jayaprakasha, G.K.; Singh, J.; Ong, K.; Crosby, K.M.; Patil, B.S. Metabolomics-based biomarkers of Fusarium wilt disease in watermelon plants. J. Plant Dis. Prot. 2020, 127, 591–596. [Google Scholar] [CrossRef]
- Bali, S.; Kaur, P.; Jamwal, V.L.; Gandhi, S.G.; Sharma, A.; Ohri, P.; Bhardwaj, R.; Ali, M.A.; Ahmad, P. Seed priming with jasmonic acid counteracts root knot nematode infection in tomato by modulating the activity and expression of antioxidative enzymes. Biomolecules 2020, 10, 98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsubara, Y.; Okada, T.; Liu, J. Suppression of Fusarium Crown Rot and Increase in Several Free Amino Acids in Mycorrhizal Asparagus. Am. J. Plant Sci. 2014, 05, 235–240. [Google Scholar] [CrossRef] [Green Version]
- Fritz, C.; Mueller, C.; Matt, P.; Feil, R.; Stitt, M. Impact of the C-N status on the amino acid profile in tobacco source leaves. Plant Cell Environ. 2006, 29, 2055–2076. [Google Scholar] [CrossRef] [PubMed]
- Ueda, S.; Ikeda, M.; Yamakawa, T. Provision of carbon skeletons for amide synthesis in non-nodulated soybean and pea roots in response to the source of nitrogen supply. Soil Sci. Plant Nutr. 2008, 54, 732–737. [Google Scholar] [CrossRef] [Green Version]
- Yoneyama, T.; Suzuki, A. Light-independent nitrogen assimilation in plant leaves: Nitrate incorporation into glutamine, glutamate, aspartate, and asparagine traced by 15N. Plants 2020, 9, 1303. [Google Scholar] [CrossRef]
- Abbes, Z.; Kharrat, M.; Delavault, P.; Chaïbi, W.; Simier, P. Nitrogen and carbon relationships between the parasitic weed Orobanche foetida and susceptible and tolerant faba bean lines. Plant Physiol. Biochem. 2009, 47, 153–159. [Google Scholar] [CrossRef]
- Reggiani, R.; Cantu, C.A.; Brambilla, I.; Bertani, A. Accumulation and interconversion of amino acids in rice roots under anoxia. Plant Cell Physiol. 1988, 29, 981–987. [Google Scholar] [CrossRef]
- Gao, H.; Jia, Y.; Guo, S.; Lv, G.; Wang, T.; Juan, L. Exogenous calcium affects nitrogen metabolism in root-zone hypoxia-stressed muskmelon roots and enhances short-term hypoxia tolerance. J. Plant Physiol. 2011, 168, 1217–1225. [Google Scholar] [CrossRef]
- Milburn, M.V.; Privé, G.G.; Milligan, D.L.; Scott, W.G.; Yeh, J.; Jancarik, J.; Koshland, D.E.; Kim, S.H. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science 1991, 254, 1342–1347. [Google Scholar] [CrossRef] [Green Version]
- Scott, W.G.; Stoddard, B.L. Transmembrane signalling and the aspartate receptor. Structure 1994, 2, 877–887. [Google Scholar] [CrossRef] [Green Version]
- Ottemann, K.M.; Xiao, W.; Shin, Y.K.; Koshland, D.E. A piston model for transmembrane signaling of the aspartate receptor. Science 1999, 285, 1751–1754. [Google Scholar] [CrossRef]
- Korolik, V. Aspartate chemosensory receptor signalling in Campylobacter jejuni. Virulence 2010, 1, 414–417. [Google Scholar] [CrossRef] [Green Version]
- Mise, T.; Matsunami, H.; Samatey, F.A.; Maruyama, I.N. Crystallization and preliminary X-ray diffraction analysis of the periplasmic domain of the Escherichia coli aspartate receptor Tar and its complex with aspartate. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2014, 70, 1219–1223. [Google Scholar] [CrossRef] [Green Version]
- Falke, J.; Koshland, D. Global flexibility in a sensory receptor: A site-directed cross-linking approach. Science 1987, 237, 1596–1600. [Google Scholar] [CrossRef]
- Lemaire, L.; Deleu, C.; Le Deunff, E. Modulation of ethylene biosynthesis by ACC and AIB reveals a structural and functional relationship between the K15NO3 uptake rate and root absorbing surfaces. J. Exp. Bot. 2013, 64, 2725–2737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Z.; Zhong, S.; Grierson, D. Recent advances in ethylene research. J. Exp. Bot. 2009, 60, 3311–3336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehta, P.K.; Hale, T.I.; Christen, P. Aminotransferases: Demonstration of homology and division into evolutionary subgroups. Eur. J. Biochem. 1993, 214, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Christen, P.; Mehta, P.K. From cofactor to enzymes. The molecular evolution of pyridoxal-5′-phosphate-dependent enzymes. Chem. Rec. 2001, 1, 436–447. [Google Scholar] [CrossRef] [PubMed]
- Capitani, G.; Hohenester, E.; Feng, L.; Storici, P.; Kirsch, J.F.; Jansonius, J.N. Structure of 1-aminocyclopropane-1-carboxylate synthase, a key enzyme in the biosynthesis of the plant hormone ethylene. J. Mol. Biol. 1999, 294, 745–756. [Google Scholar] [CrossRef]
- Yamagami, T.; Tsuchisaka, A.; Yamada, K.; Haddon, W.F.; Harden, L.A.; Theologis, A. Biochemical Diversity among the 1-Amino-cyclopropane-1-Carboxylate Synthase Isozymes Encoded by the Arabidopsis Gene Family. J. Biol. Chem. 2003, 278, 49102–49112. [Google Scholar] [CrossRef] [Green Version]
- Tsuchisaka, A.; Theologis, A. Heterodimeric interactions among the 1-amino-cyclopropane-1-carboxylate synthase polypeptides encoded by the Arabidopsis gene family. Proc. Natl. Acad. Sci. USA 2004, 101, 2275–2280. [Google Scholar] [CrossRef] [Green Version]
- Le Deunff, E. From Aspartate to Ethylene: Central Role of N, C, and S Shuttles by Aminotransferases during Biosynthesis of a Major Plant Growth Hormone; Springer: Cham, Switzerland, 2018; pp. 253–293. [Google Scholar] [CrossRef]
- Brunoni, F.; Collani, S.; Casanova-Sáez, R.; Šimura, J.; Karady, M.; Schmid, M.; Ljung, K.; Bellini, C. Conifers exhibit a characteristic inactivation of auxin to maintain tissue homeostasis. New Phytol. 2020, 226, 1753–1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Östin, A.; Kowalyczk, M.; Bhalerao, R.P.; Sandberg, G. Metabolism of Indole-3-Acetic Acid in Arabidopsis. Plant Physiol. 1998, 118, 285–296. [Google Scholar] [CrossRef] [Green Version]
- Žižková, E.; Kubeš, M.; Dobrev, P.I.; Přibyl, P.; Šimura, J.; Zahajská, L.; Záveská Drábková, L.; Novák, O.; Motyka, V. Control of cytokinin and auxin homeostasis in cyanobacteria and algae. Ann. Bot. 2017, 119, 151–166. [Google Scholar] [CrossRef] [Green Version]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis Enzyme Family That Conjugates Amino Acids to Indole-3-Acetic Acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Böttcher, C.; Keyzers, R.A.; Boss, P.K.; Davies, C. Sequestration of auxin by the indole-3-acetic acid-amido synthetase GH3-1 in grape berry (Vitis vinifera L.) and the proposed role of auxin conjugation during ripening. J. Exp. Bot. 2010, 61, 3615–3625. [Google Scholar] [CrossRef] [Green Version]
- Park, J.-E.; Park, J.-Y.; Kim, Y.-S.; Staswick, P.E.; Jeon, J.; Yun, J.; Kim, S.-Y.; Kim, J.; Lee, Y.-H.; Park, C.-M. GH3-mediated Auxin Homeostasis Links Growth Regulation with Stress Adaptation Response in Arabidopsis. J. Biol. Chem. 2007, 282, 10036–10046. [Google Scholar] [CrossRef] [Green Version]
- González-Lamothe, R.; El Oirdi, M.; Brisson, N.; Bouarab, K. The Conjugated Auxin Indole-3-Acetic Acid–Aspartic Acid Promotes Plant Disease Development. Plant Cell 2012, 24, 762–777. [Google Scholar] [CrossRef]
- Campanella, J.J.; Smith, S.M.; Leibu, D.; Wexler, S.; Ludwig-Müller, J. The Auxin Conjugate Hydrolase Family of Medicago truncatula and Their Expression During the Interaction with Two Symbionts. J. Plant Growth Regul. 2008, 27, 26–38. [Google Scholar] [CrossRef]
- Porco, S.; Pěnčík, A.; Rashed, A.; Voß, U.; Casanova-Sáez, R.; Bishopp, A.; Golebiowska, A.; Bhosale, R.; Swarup, R.; Swarup, K.; et al. Dioxygenase-encoding AtDAO1 gene controls IAA oxidation and homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 11016–11021. [Google Scholar] [CrossRef] [Green Version]
- Ostrowski, M.; Ciarkowska, A.; Jakubowska, A. The auxin conjugate indole-3-acetyl-aspartate affects responses to cadmium and salt stress in Pisum sativum L. J. Plant Physiol. 2016, 191, 63–72. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Pavlović, I.; Pěnčík, A.; Novák, O.; Vujčić, V.; Radić Brkanac, S.; Lepeduš, H.; Strnad, M.; Salopek-Sondi, B. Short-term salt stress in Brassica rapa seedlings causes alterations in auxin metabolism. Plant Physiol. Biochem. 2018, 125, 74–84. [Google Scholar] [CrossRef] [PubMed]
| Stress | Species | Tissues (Stress Period) | Asp Fold Change | Change of Asp-Associated Metabolites | Physiological Role | Ref. |
|---|---|---|---|---|---|---|
| Drought | Astragalus membranaceus | Roots (10 days) | 2.3 | ↑Asp family metabolism, ↑glutamate, ↑GABA,↑TCA cycle, ↑sucrose | Sensing water status | [78] |
| Cicer arietinum L. (chickpea) | Leaves | −2.5~−6.1 | ↑Thr, ↑Met, ↓Asn, ↑citrulline | Osmoregulation | [81] | |
| Caragana korshinskii | Leaves and roots | −0.32~−0.63 | ↑Asn, ↑sugars/glycosides, ↓Glu,↓isocitric acid | Drought-responsive metabolites | [83] | |
| Triticeae | Roots and leaves | >2 | ↑Succinate, ↑Trehalose, ↑Glu, ↑Asn, ↑Met, ↑Phe | Drought stress-specific responsive metabolites | [79] | |
| Brassica oleracea L. var. acephala (kale) | Leaves | −1.3 | ↓Glu, ↓Thr, ↓Ala, ↑Pro | Biomarker for drought tolerance | [82] | |
| Salinity | Aeluropus lagopoides | Shoots and roots | 6.2~11 | ↑Asn, ↑Lys, ↓malate | Stomatal opening, inhibited Ca2+ uptake | [86] |
| Wheat | Seedlings (17 days) | 15.75 | ↑Ile, ↑Lys, ↑Phe, ↑Pro, ↓Glu, ↓Arg, ↓Met | Protein metabolism, osmoprotection | [89] | |
| N starvation or low N | Non- nodulated soybean | Phloem sap (4 days) | −3.7 | ↓Asn, ↓Glu, ↑malate, ↑GABA | Transform to malate to deliver the amino acids | [74] |
| Maize | Leaves | ≈2 | ↓Asn, ↓Glu | Regulation of N mobilization | [72] | |
| Solanum tuberosum L. (potato) | Shoots and tubers of potato cv. Kufri Jyoti | >5 | ↑Thr, ↑Asn, ↑Glu, | NUE efficiency | [75] | |
| Tobacco | Leaves | >−2 | ↑Glu, ↑Lys, ↑Ile, ↓Gln, ↓Arg, ↓Phe | Represents a significant proportion of the total amino acid pool | [104] | |
| Soybean | Xylem sap | ≈8 | ↓Asn, ↓Gln, ↑Glu, ↑Ala, ↑GABA | N recycling, source of N in alanine formation | [71] | |
| Supplementation of nitrate | Soybean | Roots | ≈3 | ↑Asn, ↑Glu, ↑Gln | Provide C skeleton for the synthesis of Asn | [105] |
| Low C | Tobacco | Leaves | >−2 | ↑Glu, ↑Asn, ↓Phe | Represents a significant proportion of the total amino acid pool | [104] |
| Light | Sunflower | Leaf discus | ≈2 | ↑Glu, ↑Gln | Convert to Asn for N storage and transport in the dark | [106] |
| Tobacco | Leaves | 2.6 | ↑Phe | Light-responsive marker metabolites | [104] | |
| Cold | Fragaria × ananassa (strawberry) | Leaves and roots of Duch. “Korona” | 3–5 | ↑Ile, ↑hexoses, ↑pentoses | Protective metabolites | [90] |
| Secale cereale (rye) | Plant crown | 3 | ↑Glu, ↑Pro | Frost tolerance improvement | [92] | |
| Ficus carica L. (fig) | Fruits | >2 | ↑Glu, ↑Glucose, ↑fructose, ↓Arg, ↓GABA, ↓Phe, ↓Ile, ↓Pro | Cold-responsive marker metabolites | [91] | |
| Low P | Triticum aestivum L. (Wheat) | Leaves | 1.2 | ↑Gln, ↑β-alanine, ↑raffinose, ↑1-kestose | Enhanced PUE | [93] |
| Fusarium wilt | Citrullus vulgaris (watermelon) | Leaves, stems, and roots | 33–43 | ↑Lys, ↑Arg, ↑citrulline | Biomarker of Fusarium wilt disease | [101] |
| Fusarium crown rot | Asparagus officinalis L., cv. “Welcome” | Mycorrhizal asparagus shoots | ≈1.7 | ↑Glu, ↑Arg, ↑citrulline, ↑GABA | Disease tolerance | [103] |
| Parasitic weed | Faba bean | Tubercles of tolerant line | ≈−0.4 | ↓Asn, ↓Glu, ↓Gln, ↓GABA,↓sucrose | N metabolism of the parasite | [107] |
| Arbuscule | Medicago truncatula | Mycorrhizal roots | >10 | ↑Glu, ↑Asn, ↑Gln, ↑sucrose, ↑trehalose | Associated with higher N availability | [100] |
| JA (100 nM) | Tomato | Seedlings | 1.6 | ↑Asn, ↑Glu, ↓Gln,↓Lys, ↓Met,↓Arg | Osmoregulation | [102] |
| Oxidative stress | Arabidopsis thaliana | Roots (6 h) | ≈2 | ↓Glu, ↓malate, ↓succinate, ↓fumarate, ↓hexose phosphates, ↑2-OG, ↑pyruvate, ↑citrate | Oxidative stress-responsive metabolites | [96] |
| Hypoxia | Muskmelon | Roots (6 days) | 1.23 | ↑Thr, ↑Glu, ↑Lys, ↑GABA | Hypoxia-responsive metabolites | [109] |
| Anoxia | Rice | Excised roots | ≈−2 | ↑GABA, ↑Pro, ↑pyruvate,↓Glu,↓Gln, ↓Asn, ↓2-OG | Corresponds to a weak fall in cytoplasmic pH | [108] |
| Arsenate (As(V)) | Tomato | Aboveground tissues and roots | 2.4–3.1 | ↑Asn, ↑Gln, ↑Glu, ↑Arg, ↑Lys, ↑Ile | Marker for As(V) stress | [98] |
| Aluminum (Al) | Trifoliate orange | Roots | −2 | ↓Ile, ↓Glu, ↓malate,↓sugars, ↑Asn, ↑Lys, ↑Gln | Marker for Al stress | [99] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, M.; Zhang, C.; Suglo, P.; Sun, S.; Wang, M.; Su, T. l-Aspartate: An Essential Metabolite for Plant Growth and Stress Acclimation. Molecules 2021, 26, 1887. https://doi.org/10.3390/molecules26071887
Han M, Zhang C, Suglo P, Sun S, Wang M, Su T. l-Aspartate: An Essential Metabolite for Plant Growth and Stress Acclimation. Molecules. 2021; 26(7):1887. https://doi.org/10.3390/molecules26071887
Chicago/Turabian StyleHan, Mei, Can Zhang, Peter Suglo, Shuyue Sun, Mingyao Wang, and Tao Su. 2021. "l-Aspartate: An Essential Metabolite for Plant Growth and Stress Acclimation" Molecules 26, no. 7: 1887. https://doi.org/10.3390/molecules26071887
APA StyleHan, M., Zhang, C., Suglo, P., Sun, S., Wang, M., & Su, T. (2021). l-Aspartate: An Essential Metabolite for Plant Growth and Stress Acclimation. Molecules, 26(7), 1887. https://doi.org/10.3390/molecules26071887







