The Bioactive Peptide SL-13R Expands Human Umbilical Cord Blood Hematopoietic Stem and Progenitor Cells In Vitro
Abstract
:1. Introduction
2. Results
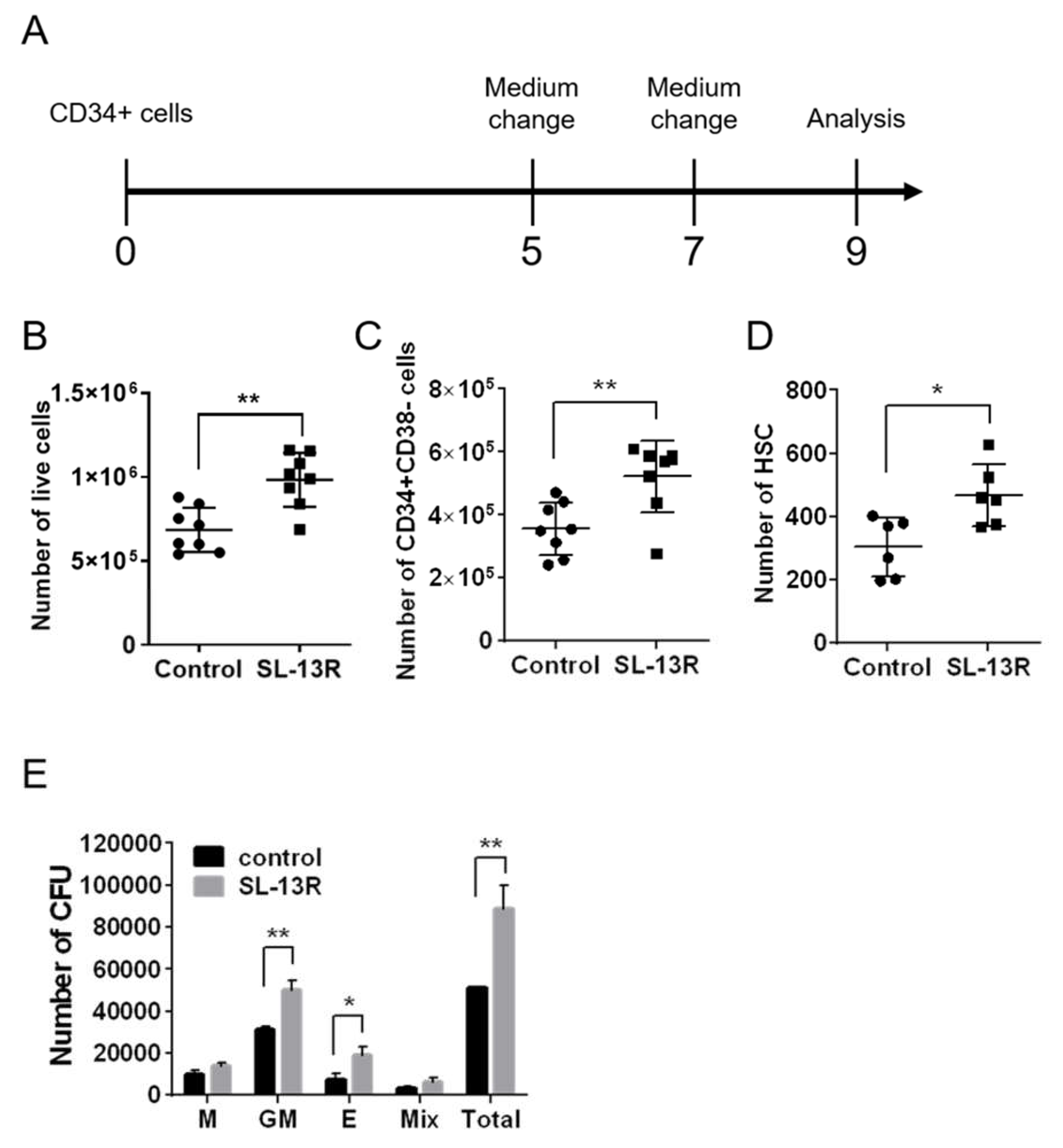
2.1. SL-13R Enhances Expansion of UCB-Derived Hematopoietic Cells
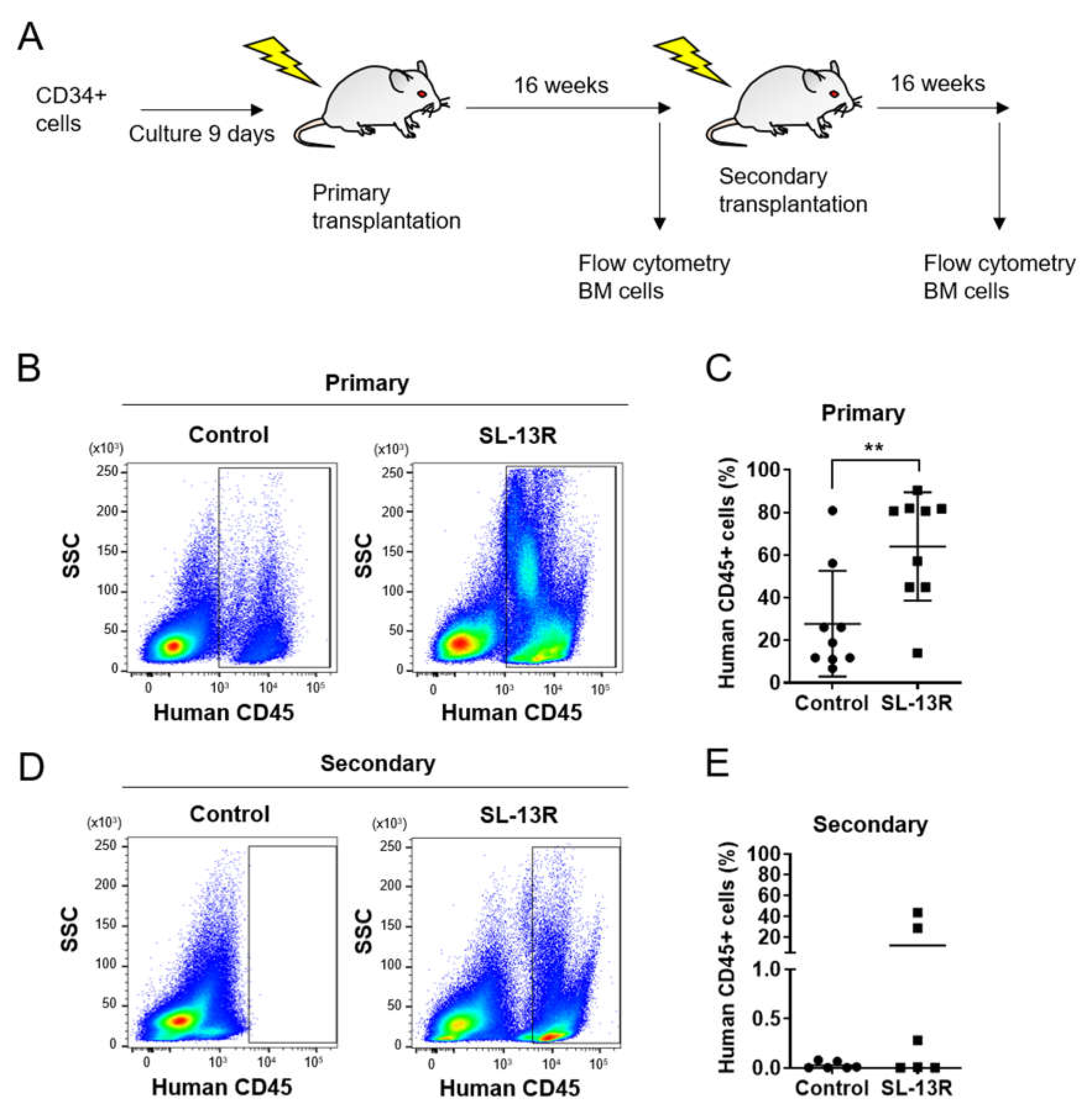
2.2. Ex Vivo Expanded CD34+ Cells Treated with SL-13R Possess Long-Term Reconstitution and Self-Renewal Ability
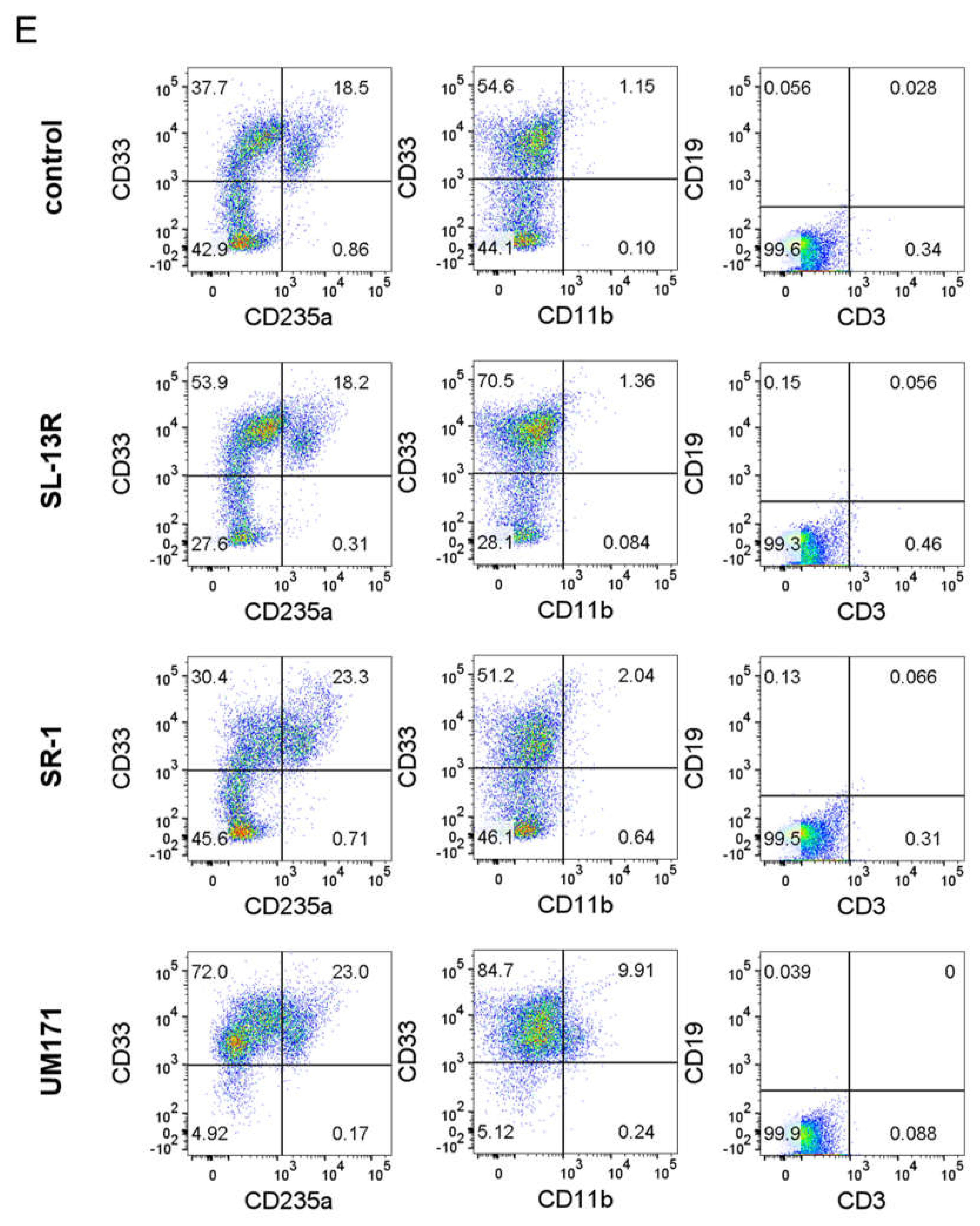
2.3. SL-13R Induced Subset of CD34+ Cells Expansion as Potently as SR-1 and UM171
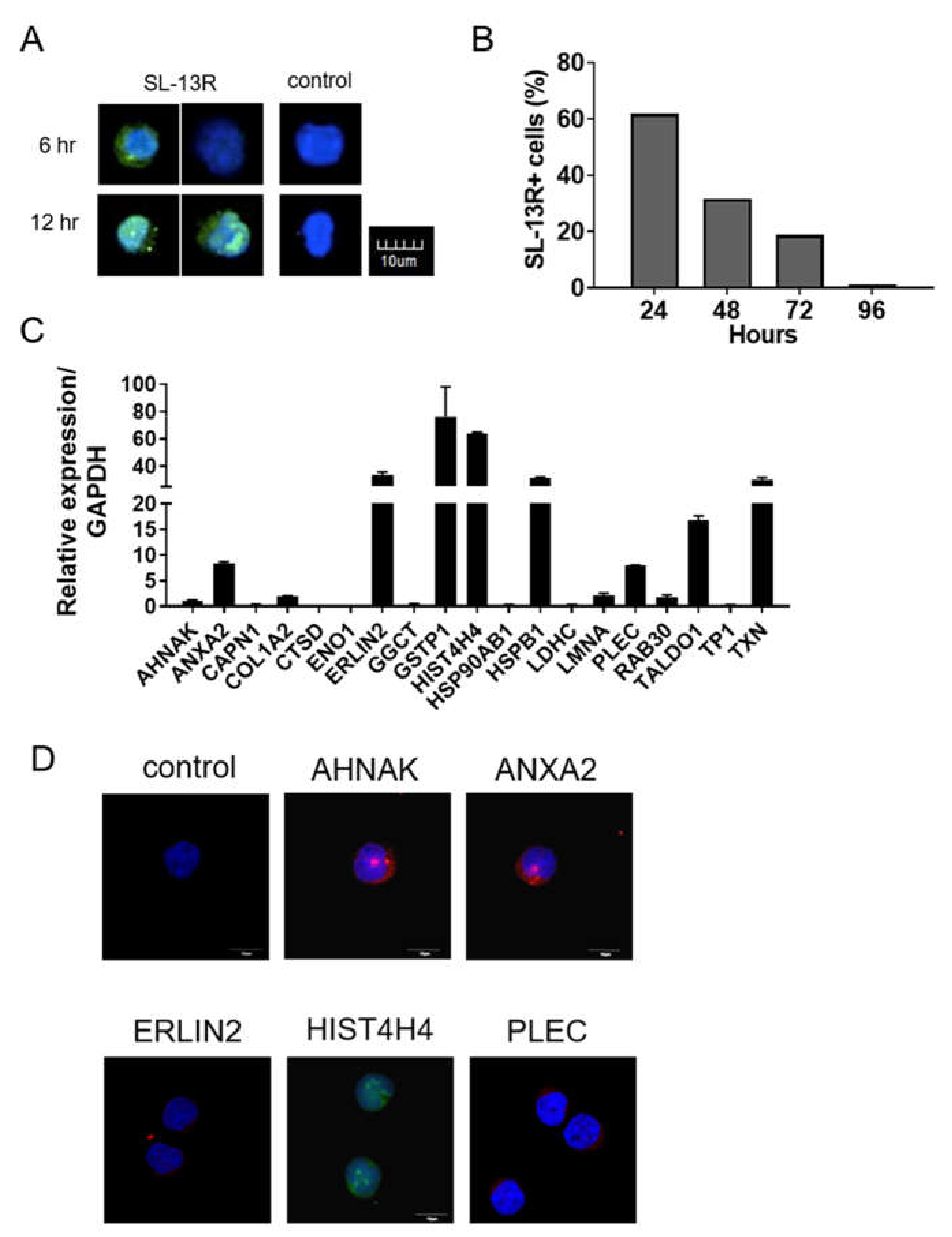
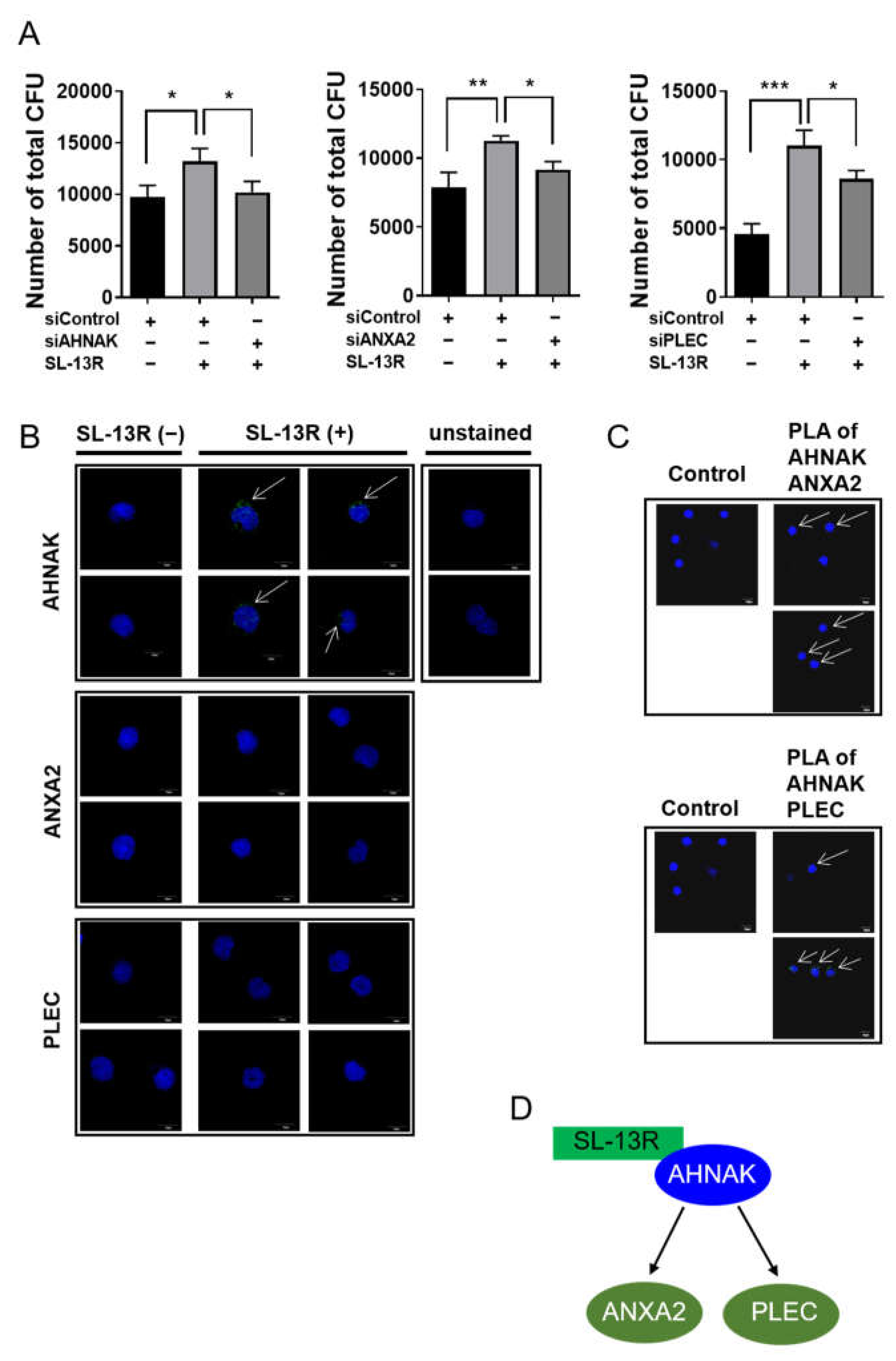
2.4. Identification of Proteins Binding to SL-13R Peptide
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Flow Cytometry
4.3. Colony Formation Unit (CFU) Assay
4.4. Reconstitution Analysis
4.5. siRNA Transfection
4.6. RNA Extraction and Quantitative Reverse Transcription Polymerase Chain Reaction, RT-PCR (qRT-PCR)
4.7. Gene Expression Microarrays
4.8. Cell Proliferation Assay
4.9. Intracellular Detection of SL-13R
4.10. Immunoprecipitation and LC-MS/MS Analysis
4.11. Immunocytochemistry
4.12. Proximity Ligation Assay
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Juric, M.K.; Ghimire, S.; Ogonek, J.; Weissinger, E.M.; Holler, E.; van Rood, J.J.; Oudshoorn, M.; Dickinson, A.; Greinix, H.T. Milestones of Hematopoietic Stem Cell Transplantation—From First Human Studies to Current Developments. Front. Immunol. 2016, 7, 470. [Google Scholar] [CrossRef] [Green Version]
- Cutler, C.; Antin, J.H. Peripheral blood stem cells for allogeneic transplantation: A review. Stem Cells 2001, 19, 108–117. [Google Scholar] [CrossRef]
- Gluckman, E.; Rocha, V. Cord blood transplantation: State of the art. Haematologica 2009, 94, 451–454. [Google Scholar] [CrossRef] [Green Version]
- Rocha, V.; Labopin, M.; Sanz, G.; Arcese, W.; Schwerdtfeger, R.; Bosi, A.; Jacobsen, N.; Ruutu, T.; de Lima, M.; Finke, J.; et al. Transplants of umbilical-cord blood or bone marrow from unrelated donors in adults with acute leukemia. N. Engl. J. Med. 2004, 351, 2276–2285. [Google Scholar] [CrossRef]
- Miller, P.H.; Knapp, D.J.; Eaves, C.J. Heterogeneity in hematopoietic stem cell populations: Implications for transplantation. Curr. Opin. Hematol. 2013, 20, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Heike, T.; Nakahata, T. Ex vivo expansion of hematopoietic stem cells by cytokines. Biochim. Biophys. Acta 2002, 1592, 313–321. [Google Scholar] [CrossRef] [Green Version]
- Ueda, T.; Tsuji, K.; Yoshino, H.; Ebihara, Y.; Yagasaki, H.; Hisakawa, H.; Mitsui, T.; Manabe, A.; Tanaka, R.; Kobayashi, K.; et al. Expansion of human NOD/SCID-repopulating cells by stem cell factor, Flk2/Flt3 ligand, thrombopoietin, IL-6, and soluble IL-6 receptor. J. Clin. Invest. 2000, 105, 1013–1021. [Google Scholar] [CrossRef] [PubMed]
- Fares, I.; Chagraoui, J.; Gareau, Y.; Gingras, S.; Ruel, R.; Mayotte, N.; Csaszar, E.; Knapp, D.J.; Miller, P.; Ngom, M.; et al. Cord blood expansion. Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self-renewal. Science 2014, 345, 1509–1512. [Google Scholar] [CrossRef] [Green Version]
- Boitano, A.E.; Wang, J.; Romeo, R.; Bouchez, L.C.; Parker, A.E.; Sutton, S.E.; Walker, J.R.; Flaveny, C.A.; Perdew, G.H.; Denison, M.S.; et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science 2010, 329, 1345–1348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horwitz, M.E.; Chao, N.J.; Rizzieri, D.A.; Long, G.D.; Sullivan, K.M.; Gasparetto, C.; Chute, J.P.; Morris, A.; McDonald, C.; Waters-Pick, B.; et al. Umbilical cord blood expansion with nicotinamide provides long-term multilineage engraftment. J. Clin. Invest. 2014, 124, 3121–3128. [Google Scholar] [CrossRef] [Green Version]
- Cohen, S.; Roy, J.; Lachance, S.; Delisle, J.-S.; Marinier, A.; Busque, L.; Roy, D.-C.; Barabé, F.; Ahmad, I.; Bambace, N.; et al. Hematopoietic stem cell transplantation using single UM171-expanded cord blood: A single-arm, phase 1–2 safety and feasibility study. Lancet Haematol. 2019, 7, e134–e145. [Google Scholar] [CrossRef]
- Wagner, J.E., Jr.; Brunstein, C.G.; Boitano, A.E.; DeFor, T.E.; McKenna, D.; Sumstad, D.; Blazar, B.R.; Tolar, J.; Le, C.; Jones, J.; et al. Phase I/II Trial of StemRegenin-1 Expanded Umbilical Cord Blood Hematopoietic Stem Cells Supports Testing as a Stand-Alone Graft. Cell Stem Cell 2016, 18, 144–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horwitz, M.E.; Wease, S.; Blackwell, B.; Valcarcel, D.; Frassoni, F.; Boelens, J.J.; Nierkens, S.; Jagasia, M.; Wagner, J.E.; Kuball, J.; et al. Phase I/II Study of Stem-Cell Transplantation Using a Single Cord Blood Unit Expanded Ex Vivo with Nicotinamide. J. Clin. Oncol. 2019, 37, 367–374. [Google Scholar] [CrossRef]
- Cutler, C.; Multani, P.; Robbins, D.; Kim, H.T.; Le, T.; Hoggatt, J.; Pelus, L.M.; Desponts, C.; Chen, Y.B.; Rezner, B.; et al. Prostaglandin-modulated umbilical cord blood hematopoietic stem cell transplantation. Blood 2013, 122, 3074–3081. [Google Scholar] [CrossRef]
- Boisset, J.-C.; van Cappellen, W.; Andrieu-Soler, C.; Galjart, N.; Dzierzak, E.; Robin, C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 2010, 464, 116–120. [Google Scholar] [CrossRef]
- Bertrand, J.Y.; Chi, N.C.; Santoso, B.; Teng, S.; Stainier, D.Y.; Traver, D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 2010, 464, 108–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eilken, H.M.; Nishikawa, S.-I.; Schroeder, T. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature 2009, 457, 896–900. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, M.J.; Holmes, A.; Miles, C.; Dzierzak, E. Characterization of the first definitive hematopoietic stem cells in the AGM and liver of the mouse embryo. Immunity 1996, 5, 513–525. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, D.; Kulkeaw, K.; Mizuochi, C. TGF-beta-1 up-regulates extra-cellular matrix production in mouse hepatoblasts. Mech. Dev. 2013, 130, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, D.; Kulkeaw, K.; Mizuochi, C.; Horio, Y.; Okayama, S. Hepatoblasts comprise a niche for fetal liver erythropoiesis through cytokine production. Biochem. Biophys. Res. Commun. 2011, 410, 301–306. [Google Scholar] [CrossRef]
- Gerlach, J.; Jörg, C.; Thompson, R.L.; Gridelli, B.; Schmelzer, E. Effects of Delta-Like Noncanonical Notch Ligand 1 Expression of Human Fetal Liver Hepatoblasts on Hematopoietic Progenitors. Stem Cells Int. 2019, 2019, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Forman, S.J.; Bhatia, R. Expression of DLK1 in hematopoietic cells results in inhibition of differentiation and proliferation. Oncogene 2005, 24, 4472–4476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.C.; Kuo, H.M.; Wu, P.C.; Cheng, S.H.; Chang, T.T.; Chang, Y.C.; Kung, M.L.; Wu, D.C.; Chuang, J.H.; Tai, M.H. Soluble delta-like 1 homolog (DLK1) stimulates angiogenesis through Notch1/Akt/eNOS signaling in endothelial cells. Angiogenesis 2018, 21, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Smas, C.M.; Sul, H.S. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell 1993, 73, 725–734. [Google Scholar] [CrossRef]
- Notta, F.; Doulatov, S.; Laurenti, E.; Poeppl, A.; Jurisica, I.; Dick, J.E. Isolation of Single Human Hematopoietic Stem Cells Capable of Long-Term Multilineage Engraftment. Science 2011, 333, 218–221. [Google Scholar] [CrossRef] [PubMed]
- Flaveny, C.A.; Murray, I.A.; Chiaro, C.R.; Perdew, G.H. Ligand selectivity and gene regulation by the human aryl hydrocarbon receptor in transgenic mice. Mol. Pharm. 2009, 75, 1412–1420. [Google Scholar] [CrossRef] [Green Version]
- Subramaniam, A.; Žemaitis, K.; Talkhoncheh, M.S.; Yudovich, D.; Bäckström, A.; Debnath, S.; Chen, J.; Jain, M.V.; Galeev, R.; Gaetani, M.; et al. Lysine-specific demethylase 1A restricts ex vivo propagation of human HSCs and is a target of UM171. Blood 2020, 136, 2151–2161. [Google Scholar] [CrossRef]
- Chagraoui, J.; Girard, S.; Spinella, J.F.; Simon, L.; Bonneil, E.; Mayotte, N.; MacRae, T.; Coulombe-Huntington, J.; Bertomeu, T.; Moison, C.; et al. UM171 Preserves Epigenetic Marks that Are Reduced in Ex Vivo Culture of Human HSCs via Potentiation of the CLR3-KBTBD4 Complex. Cell Stem Cell 2021, 28, 48–62.e6. [Google Scholar] [CrossRef]
- Davis, T.A.; Loos, B.; Engelbrecht, A.M. AHNAK: The giant jack of all trades. Cell. Signal. 2014, 26, 2683–2693. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, I.; Ono, H.; Moritomo, T.; Kano, K.; Nakanishi, T.; Suda, T. Comparative gene expression analysis of zebrafish and mammals identifies common regulators in hematopoietic stem cells. Blood 2010, 115, e1–e9. [Google Scholar] [CrossRef]
- Emans, N.; Gorvel, J.P.; Walter, C.; Gerke, V.; Kellner, R.; Griffiths, G.; Gruenberg, J. Annexin II is a major component of fusogenic endosomal vesicles. J. Cell Biol. 1993, 120, 1357–1369. [Google Scholar] [CrossRef]
- Jung, Y.; Shiozawa, Y.; Wang, J.; Patel, L.R.; Havens, A.M.; Song, J.; Krebsbach, P.H.; Roodman, G.D.; Taichman, R.S. Annexin-2 is a regulator of stromal cell-derived factor–1/CXCL12 function in the hematopoietic stem cell endosteal niche. Exp. Hematol. 2011, 39, 151–166.e1. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.; Bhatti, D.L.; Lee, K.-W.; Medrihan, L.; Cheng, J.; Wei, J.; Zhong, P.; Yan, Z.; Kooiker, C.; Song, C.; et al. Ahnak scaffolds p11/Anxa2 complex and L-type voltage-gated calcium channel and modulates depressive behavior. Mol. Psychiatry 2020, 25, 1035–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castañón, M.J.; Walko, G.; Winter, L.; Wiche, G. Plectin–intermediate filament partnership in skin, skeletal muscle, and peripheral nerve. Histochem. Cell Biol. 2013, 140, 33–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Straub, B.K.; Boda, J.; Kuhn, C.; Schnoelzer, M.; Korf, U.; Kempf, T.; Spring, H.; Hatzfeld, M.; Franke, W.W. A novel cell-cell junction system: The cortex adhaerens mosaic of lens fiber cells. J. Cell Sci. 2003, 116, 4985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.; Yu, G.; Sarioglu, H.; Caballero-Martinez, A.; Schlott, F.; Ueffing, M.; Haase, H.; Peschel, C.; Krackhardt, A.M. Proteomic investigation of the interactome of FMNL1 in hematopoietic cells unveils a role in calcium-dependent membrane plasticity. J. Proteom. 2013, 78, 72–82. [Google Scholar] [CrossRef]

| Ingenuity Canonical Pathways | −log (p-Value) | Regulation | z Score | Ratio | Molecules |
|---|---|---|---|---|---|
| Role of NFAT in Regulation of the Immune Response | 3.11 | up | 1 | 0.072 | AKT3, CSNK1G1, CSNK1G2, GATA4, GNA13, GNAI3, GNAG2, GNG4, ITK, MAP2K1, ORAI1, PPP3R1, RAP1A |
| Role of BRCA1 in DNA Damage Response | 2.37 | up | 0.45 | 0.088 | ATM, ATR, BLM, FANCL, NBMN, PBRM1, UIMC1 |
| Role of NFAT in Cardiac Hypertrophy | 2.05 | up | 1.27 | 0.056 | AKT3, CACNA1A, GATA4, GNAI3, GNG2, GNG4, HDAC11, MAP2K1, PPP3R1, PRKAR2B, RAP1A, TGFBR2 |
| Superpathway of Methionine Degradation | 1.85 | up | 1 | 0.11 | CBS/CBSL, EEF1AKMT2, MAT2A, MCEE |
| Relaxin Signaling | 1.85 | up | 1.34 | 0.06 | AKT3, GNA13, GNAI3, GNG2, GNG4, MAP2K1, PDE6C, PRKAR2B, RAP1A |
| Protein Kinase A Signaling | 1.84 | up | 1 | 0.045 | ADD3, DUSP12, GNA13, GNAI3, GNG2, GNG4, GYS1, MAP2K1, PDE6C, PPP3R1, PRKAR2B, PTPN14, PTPRR, RAP1A, ROCK1, TCF3, TGFBR2, YWHAE |
| Ephrin Receptor Signaling | 1.76 | up | 1 | 0.056 | ADAM10, AKT3, GNA13, GNAI3, GNG2, GNG4, ITGA4, MAP2K1, RAP1A, ROCK1 |
| Telomerase Signaling | 1.71 | up | 0.45 | 0.065 | AKT3, HDAC11, IL2RB, MAP2K1, PPP2R3A, RAP1A, TERF2IP |
| PI3K/AKT Signaling | 1.7 | up | 1.41 | 0.061 | AKT3, EIF4E, GYS1, ITGA4, MAP2K1, PPP2R3A, RAP1A, YWHAE |
| Cardiac Hypertrophy Signaling (Enhanced) | 1.6 | up | 1.1 | 0.041 | AKT3, ATP2A2, CACNA1A, EIF4E, GATA4, GNA13, GNAI3, GNG2, HDAC11, IL10RA, IL2RB, ITGA4, MAP2K1, PDE6C, PPP3R1, PRKAR2B, RAP1A, ROCK1, TGFBR2, TNFSF10 |
| Insulin Receptor Signaling | 1.58 | up | 0.71 | 0.058 | AKT3, EIF4E, GYS1, MAP2K1, PRKAR2B, RAP1A, SOCS3, VAMP2 |
| Leukocyte Extravasation Signaling | 1.51 | down | −1.27 | 0.051 | BMX, F11R, FER, GNAI3, ITGA4, ITGAL, ITK, MMP24, RAP1A, ROCK1 |
| P2Y Purigenic Receptor Signaling Pathway | 1.36 | up | 1.63 | 0.055 | AKT3, GNAI3, GNG2, GNG4, MAP2K1, PRKAR2B, RAP1A |
| Neurotrophin/TRK Signaling | 1.35 | up | 1 | 0.066 | MAP2K1, NTRK1, RAP1A, SPRY1, SPRY2 |
| AMPK Signaling | 1.34 | up | 1.63 | 0.047 | AK4, AKT3, CHRNA3, EEF2K, GYS1, PBRM1, PFKFB3, PPP2R3A, PRKAR2B, RAB27A |
| IGF-1 Signaling | 1.3 | up | 1.34 | 0.058 | AKT3, MAP2K1, PRKAR2B, RAP1A, SOCS3, YWHAE |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nii, T.; Konno, K.; Matsumoto, M.; Bhukhai, K.; Borwornpinyo, S.; Sakai, K.; Hongeng, S.; Sugiyama, D. The Bioactive Peptide SL-13R Expands Human Umbilical Cord Blood Hematopoietic Stem and Progenitor Cells In Vitro. Molecules 2021, 26, 1995. https://doi.org/10.3390/molecules26071995
Nii T, Konno K, Matsumoto M, Bhukhai K, Borwornpinyo S, Sakai K, Hongeng S, Sugiyama D. The Bioactive Peptide SL-13R Expands Human Umbilical Cord Blood Hematopoietic Stem and Progenitor Cells In Vitro. Molecules. 2021; 26(7):1995. https://doi.org/10.3390/molecules26071995
Chicago/Turabian StyleNii, Takenobu, Katsuhiro Konno, Masaki Matsumoto, Kanit Bhukhai, Suparerk Borwornpinyo, Kazuhiro Sakai, Suradej Hongeng, and Daisuke Sugiyama. 2021. "The Bioactive Peptide SL-13R Expands Human Umbilical Cord Blood Hematopoietic Stem and Progenitor Cells In Vitro" Molecules 26, no. 7: 1995. https://doi.org/10.3390/molecules26071995






