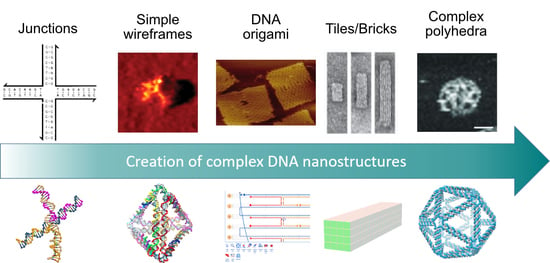
The Art of Designing DNA Nanostructures with CAD Software
Abstract
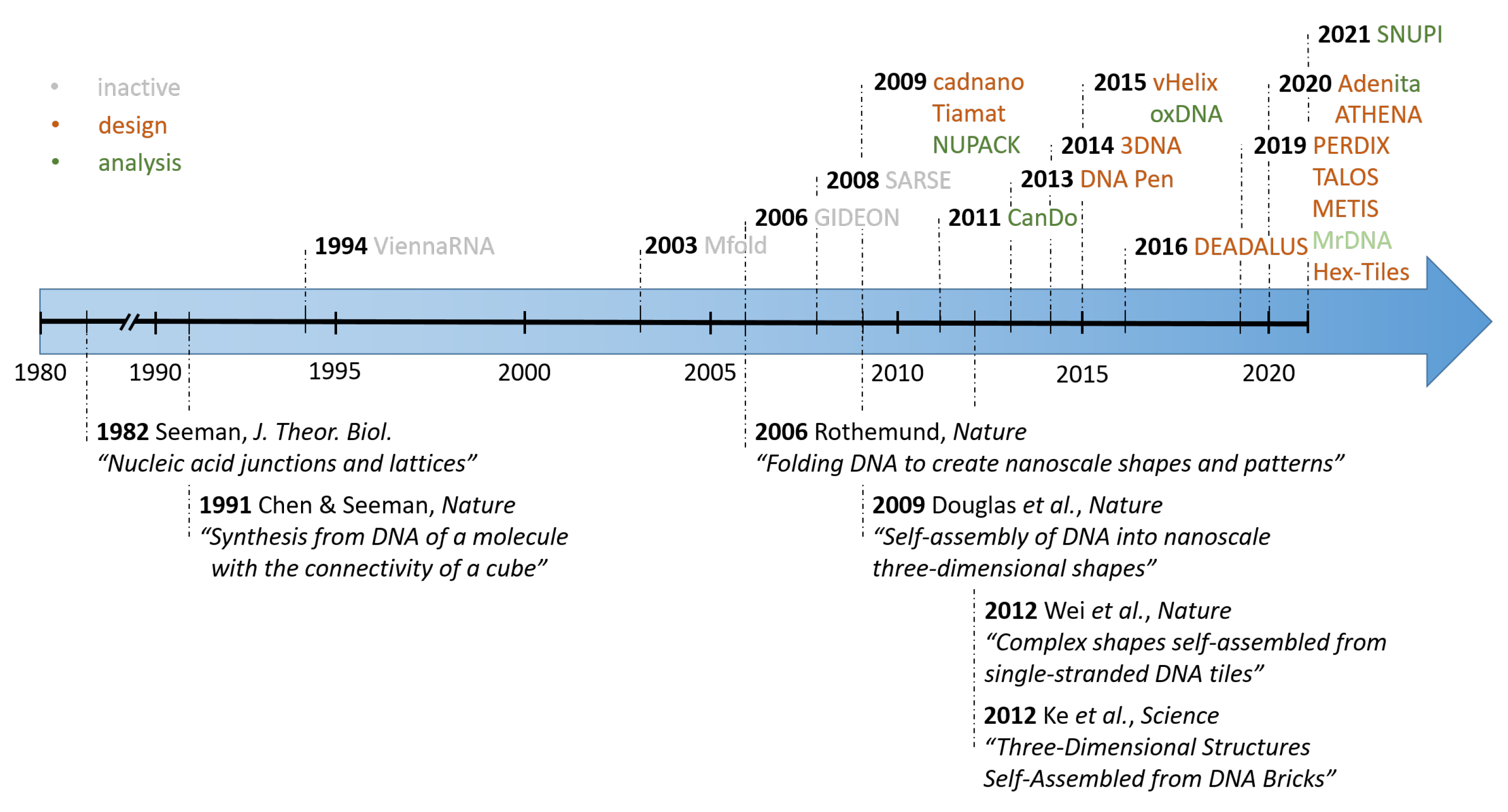
:1. Introduction
2. Scaffolded DNA Origami
2.1. Cadnano
2.2. Tiamat
2.3. vHelix
2.4. DAEDALUS, PERDIX, TALOS, METIS
3. DNA Tiles and DNA Bricks
- While offering the aforementioned design advantages, the lack of a central scaffold strand to template the assembly means that the self-assembly process is dependent upon a nucleation-and-growth mechanism [65]. Here, the local structure, topological connectivity, kinetic traps, and even stoichiometry between the hundreds or thousands of components are critical parameters and each can impact yields.
- Particularly for three-dimensional bricks, the process of translating an arbitrary design into a collection of hundreds or thousands of unique DNA oligonucleotide sequences is extraordinarily complex. The target structure is first rendered as a collection of voxels, each corresponding to an eight base pair segment of double stranded DNA, then connections between the voxels under the constraints of DNA geometry are applied, before each strand is populated with appropriate sequences, according to the original report from Ke et al. [50].
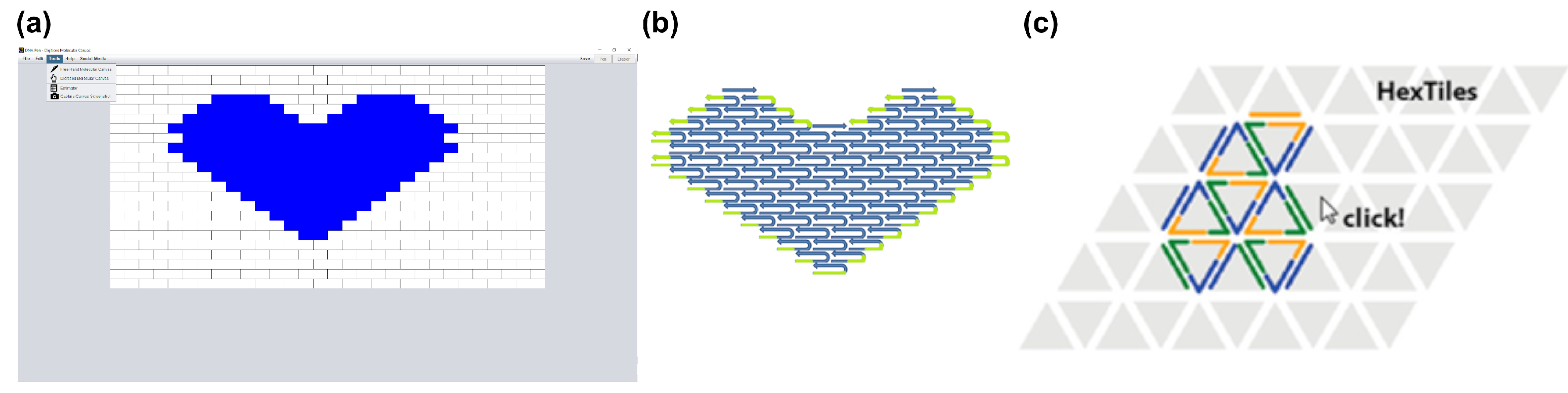
3.1. Two-Dimensional Tiles with DNA Pen
3.2. Three-Dimensional Bricks with 3DNA
4. Analysis
- Mfold was released in 2003 on its own web-server-based application and it is one of the oldest for computational molecular biology [86]. On this web server, several analysis tools for sequence analysis and the prediction of RNA and DNA folding can be found, including the Mfold software. In 2008, it was renamed to UNAfold. Mfold and UNAfold anticipate the folding of DNA and RNA strands through the prediction of the structure’s minimum free energy G [87].
- Nucleic Acid Package (NUPACK) is a design and analysis tool for the base pairing of one or multiple DNA sequences, released in 2010 [88]. The program is suitable for the design of nucleic acid sequences and their thermodynamic analysis. Therefore, it can be used for the evaluation of simple nucleic acid systems.
- Vienna RNA originally was released in 1994, providing a tool for the prediction of RNA secondary structures [13]. In 2011, the ViennaRNA software package was launched, including technical updates to the GUI and the underlying RNAlib. New tools for the assessment of RNA–RNA interactions as well as additional output information were added [89].
- CanDo (Computer-aided engineering for DNA origami) is a finite element modeling framework developed at MIT [90]. Originally, it was limited to model honeycomb and square lattice DNA assemblies that were designed using Cadnano, already proving its predictive power (Figure 10c). Later, it got reworked to model wireframe structures allowing for highly complex three-dimensional geometries and their flexibility that would be infeasible analytically [91]. It was later extended to enable lattice-free modelling [92] as well as long time-scale dynamics of DNA assemblies using Brownian Dynamics [93]. Later, CanDo launched its own online server, which even makes modelling to the atomic scale possible [94,95].
- OxDNA is a simulation code from the University of Oxford that implements a coarse-grained DNA model [96]. The code uses Monte Carlo and Molecular Dynamics simulations for determining the mechanical and thermodynamic properties of single- and double-stranded DNA and RNA (Figure 10a). Taking major and minor grooves into account and by adjusting the coaxial stacking and backbone-backbone interactions, it allows for more precise prediction of especially larger (kilobase-pair) structures. It has been reworked to OxDNA2, which allows for the adjustment of salt concentrations, and treats the interaction of consecutive adenine bases to consecutive thymine bases differently, a feature especially important in systems with flexible single-stranded regions [97]. This model can also be used to predict the involved forces when unraveling a DNA origami by force-induced melting, and it has been experimentally verified via Atomic Force Microscope (AFM), making it an interesting example for the flexibility of this software [98]. A recently-developed web browser-based visualization tool, oxView, provides a fast and user-friendly interface to the underlying code, and it includes additional modules for characterizing aspects, such as structural [99], and a tool, TacoxDNA, is also available for converting common CAD output files (e.g., from Cadnano or Tiamat) into representations that are suitable for simulation via OxDNA [100].
- Being released in 2019, MrDNA is a tool for the prediction of the structure and dynamics of DNA based systems [101]. The software features a fast multi-resolution model for the prediction of self-assembled DNA origami on an atomistic level in 30 min. or less. This allows for fast in situ simulations and saves a lot of time in a de novo design of complex DNA nanostructures.
- Finally, the newest addition to the pantheon of analysis tools, SNUPI (Structured NUcleic acids Programming Interface) renders near-atomically-precise analysis of DNA origami properties, such as shape, dynamic properties, and mechanical properties rigidity in a relatively quick processing time [102]. SNUPI functions as a standalone, downloadable program, and acts as a convenient companion tool for visualizing and analyzing structures that are designed within Cadnano. As input, standard JSON files along with an optional sequence file (in .csv format) for the scaffold are used. The analysis engine combines known, intrinsic properties of DNA molecules with sequence-specific geometric and mechanical properties that are determined by molecule dynamics simulations. Subsequently, this information is fed into a structural model to efficiently generate atomic-level information in a matter of minutes.
5. Discussion
6. Conclusions
7. Materials and Methods
7.1. DNA Origami
7.2. AFM Imaging
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AFM | Atomic Force Microscope |
| CAD | Computer-assisted Design |
| GUI | Graphical-user Interface |
| TEM | Transmission Electron Microscopy |
References
- Watson, J.D.; Crick, F.H.C. Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Seeman, N.C. Nucleic acid junctions and lattices. J. Theor. Biol. 1982, 99, 237–247. [Google Scholar] [CrossRef]
- Seeman, N.C. DNA in a material world. Nature 2003, 421, 427–431. [Google Scholar] [CrossRef]
- Smith, D.; Schüller, V.; Engst, C.; Rädler, J.; Liedl, T. Nucleic acid nanostructures for biomedical applications. Nanomedicine 2012, 8, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Linko, V. Challenges and Perspectives of DNA Nanostructures in Biomedicine. Angew. Chem. Int. Ed. 2020, 59, 15818–15833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, D.M.; Keller, A. DNA Nanostructures in the Fight Against Infectious Diseases. Adv. NanoBiomed Res. 2021, 1, 2000049. [Google Scholar] [CrossRef]
- Church, G.M.; Gao, Y.; Kosuri, S. Next-Generation Digital Information Storage in DNA. Science 2012, 337, 1628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldman, N.; Bertone, P.; Chen, S.; Dessimoz, C.; LeProust, E.M.; Sipos, B.; Birney, E. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature 2013, 494, 77–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Seeman, N.C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature 1991, 350, 631–633. [Google Scholar] [CrossRef] [PubMed]
- Winfree, E. Algorithmic Self-Assembly of DNA. In Proceedings of the 2006 International Conference on Microtechnologies in Medicine and Biology, Okinawa, Japan, 9–12 May 2006; p. 4. [Google Scholar] [CrossRef] [Green Version]
- Rothemund, P.W.K. Design of DNA origami. In Proceedings of the IEEE/ACM International Conference on Computer-Aided Design ICCAD-2005, Santa Clara, CA, USA, 6–10 November 2005; pp. 471–478. [Google Scholar] [CrossRef]
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [Green Version]
- Hofacker, I.L.; Fontana, W.; Stadler, P.F.; Bonhoeffer, L.S.; Tacker, M.; Schuster, P. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 1994, 125, 167–188. [Google Scholar] [CrossRef]
- Birac, J.J.; Sherman, W.B.; Kopatsch, J.; Constantinou, P.E.; Seeman, N.C. Architecture with GIDEON, a program for design in structural DNA nanotechnology. J. Mol. Graph. Model. 2006, 25, 470–480. [Google Scholar] [CrossRef] [Green Version]
- Andersen, E.S.; Dong, M.; Nielsen, M.M.; Jahn, K.; Lind-Thomsen, A.; Mamdouh, W.; Gothelf, K.V.; Besenbacher, F.; Kjems, J. DNA Origami Design of Dolphin-Shaped Structures with Flexible Tails. ACS Nano 2008, 2, 1213–1218. [Google Scholar] [CrossRef]
- Zhu, J.; Wei, B.; Yuan, Y.; Mi, Y. UNIQUIMER 3D, a software system for structural DNA nanotechnology design, analysis and evaluation. Nucleic Acids Res. 2009, 37, 2164–2175. [Google Scholar] [CrossRef] [Green Version]
- Tapio, K.; Bald, I. The potential of DNA origami to build multifunctional materials. Multifunct. Mater. 2020, 3, 032001. [Google Scholar] [CrossRef]
- Dey, S.; Fan, C.; Gothelf, K.V.; Li, J.; Lin, C.; Liu, L.; Liu, N.; Nijenhuis, M.A.D.; Saccà, B.; Simmel, F.C.; et al. DNA origami. Nat. Rev. Methods Primers 2021, 1, 1–24. [Google Scholar] [CrossRef]
- Douglas, S.M.; Dietz, H.; Liedl, T.; Högberg, B.; Graf, F.; Shih, W.M. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Voigt, N.V.; Gothelf, K.V.; Shih, W.M. Multilayer DNA Origami Packed on Hexagonal and Hybrid Lattices. J. Am. Chem. Soc. 2012, 134, 1770–1774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dietz, H.; Douglas, S.M.; Shih, W.M. Folding DNA into Twisted and Curved Nanoscale Shapes. Science 2009, 325, 725–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, D.; Pal, S.; Yang, Y.; Jiang, S.; Nangreave, J.; Liu, Y.; Yan, H. DNA Gridiron Nanostructures Based on Four-Arm Junctions. Science 2013, 339, 1412–1415. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Jiang, S.; Wu, S.; Li, Y.; Mao, C.; Liu, Y.; Yan, H. Complex wireframe DNA origami nanostructures with multi-arm junction vertices. Nat. Nanotechnol. 2015, 10, 779–784. [Google Scholar] [CrossRef]
- Benson, E.; Mohammed, A.; Gardell, J.; Masich, S.; Czeizler, E.; Orponen, P.; Högberg, B. DNA rendering of polyhedral meshes at the nanoscale. Nature 2015, 523, 441–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthies, M.; Agarwal, N.P.; Schmidt, T.L. Design and Synthesis of Triangulated DNA Origami Trusses. Nano Lett. 2016, 16, 2108–2113. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.M.; Schüller, V.; Forthmann, C.; Schreiber, R.; Tinnefeld, P.; Liedl, T. A structurally variable hinged tetrahedron framework from DNA origami. J. Nucleic Acids 2011, 2011, 360954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iinuma, R.; Ke, Y.; Jungmann, R.; Schlichthaerle, T.; Woehrstein, J.B.; Yin, P. Polyhedra Self-Assembled from DNA Tripods and Characterized with 3D DNA-PAINT. Science 2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veneziano, R.; Ratanalert, S.; Zhang, K.; Zhang, F.; Yan, H.; Chiu, W.; Bathe, M. Designer nanoscale DNA assemblies programmed from the top down. Science 2016, 352, 1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, E.S.; Dong, M.; Nielsen, M.M.; Jahn, K.; Subramani, R.; Mamdouh, W.; Golas, M.M.; Sander, B.; Stark, H.; Oliveira, C.L.P.; et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature 2009, 459, 73–76. [Google Scholar] [CrossRef] [Green Version]
- Han, D.; Pal, S.; Liu, Y.; Yan, H. Folding and cutting DNA into reconfigurable topological nanostructures. Nat. Nanotechnol. 2010, 5, 712–717. [Google Scholar] [CrossRef]
- Liedl, T.; Högberg, B.; Tytell, J.; Ingber, D.E.; Shih, W.M. Self-assembly of three-dimensional prestressed tensegrity structures from DNA. Nat. Nanotechnol. 2010, 5, 520–524. [Google Scholar] [CrossRef] [Green Version]
- Nickels, P.C.; Ke, Y.; Jungmann, R.; Smith, D.M.; Leichsenring, M.; Shih, W.M.; Liedl, T.; Högberg, B. DNA Origami Structures Directly Assembled from Intact Bacteriophages. Small 2014, 10, 1765–1769. [Google Scholar] [CrossRef]
- Marchi, A.N.; Saaem, I.; Vogen, B.N.; Brown, S.; LaBean, T.H. Toward Larger DNA Origami. Nano Lett. 2014, 14, 5740–5747. [Google Scholar] [CrossRef]
- Zhao, Z.; Yan, H.; Liu, Y. A route to scale up DNA origami using DNA tiles as folding staples. Angew. Chem. Int. Ed. Engl. 2010, 49, 1414–1417. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.; Rothemund, P.W.K. Self-assembly of two-dimensional DNA origami lattices using cation-controlled surface diffusion. Nat. Commun. 2014, 5, 4889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kocabey, S.; Kempter, S.; List, J.; Xing, Y.; Bae, W.; Schiffels, D.; Shih, W.M.; Simmel, F.C.; Liedl, T. Membrane-Assisted Growth of DNA Origami Nanostructure Arrays. ACS Nano 2015, 9, 3530–3539. [Google Scholar] [CrossRef] [PubMed]
- Wagenbauer, K.F.; Sigl, C.; Dietz, H. Gigadalton-scale shape-programmable DNA assemblies. Nature 2017, 552, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Hartl, C.; Frank, K.; Heuer-Jungemann, A.; Fischer, S.; Nickels, P.C.; Nickel, B.; Liedl, T. 3D DNA Origami Crystals. Adv. Mater. 2018, 30, 1800273. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.M.; Marblestone, A.H.; Teerapittayanon, S.; Vazquez, A.; Church, G.M.; Shih, W.M. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009, 37, 5001–5006. [Google Scholar] [CrossRef] [Green Version]
- Doty, D.; Lee, B.L.; Stérin, T. scadnano: A browser-based, scriptable tool for designing DNA nanostructures. arXiv 2020, arXiv:2005.11841. [Google Scholar]
- Williams, S.; Lund, K.; Lin, C.; Wonka, P.; Lindsay, S.; Yan, H. Tiamat: A Three-Dimensional Editing Tool for Complex DNA Structures. In DNA Computing; Lecture Notes in Computer Science; Goel, A., Simmel, F.C., Sosík, P., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 90–101. [Google Scholar] [CrossRef] [Green Version]
- Seeman, N.C. De Novo Design of Sequences for Nucleic Acid Structural Engineering. J. Biomol. Struct. Dyn. 1990, 8, 573–581. [Google Scholar] [CrossRef]
- Liedl, T. Pathfinder for DNA constructs. Nature 2015, 523, 412–413. [Google Scholar] [CrossRef]
- Euler, L. Solutio Problematis ad Geometriam Situs Pertinentis. 1741. Available online: https://scholarlycommons.pacific.edu/cgi/viewcontent.cgi?article=1052&context=euler-works (accessed on 1 February 2020).
- Jun, H.; Zhang, F.; Shepherd, T.; Ratanalert, S.; Qi, X.; Yan, H.; Bathe, M. Autonomously designed free-form 2D DNA origami. Sci. Adv. 2019, 5, eaav0655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jun, H.; Shepherd, T.R.; Zhang, K.; Bricker, W.P.; Li, S.; Chiu, W.; Bathe, M. Automated Sequence Design of 3D Polyhedral Wireframe DNA Origami with Honeycomb Edges. ACS Nano 2019, 13, 2083–2093. [Google Scholar] [CrossRef]
- Jun, H.; Wang, X.; Bricker, W.P.; Bathe, M. Automated sequence design of 2D wireframe DNA origami with honeycomb edges. Nat. Commun. 2019, 10, 5419. [Google Scholar] [CrossRef] [Green Version]
- Jun, H.; Wang, X.; Bricker, W.P.; Jackson, S.; Bathe, M. Rapid Prototyping of Wireframe Scaffolded DNA Origami using ATHENA. bioRxiv 2020, 940320. [Google Scholar] [CrossRef] [Green Version]
- Wei, B.; Dai, M.; Yin, P. Complex shapes self-assembled from single-stranded DNA tiles. Nature 2012, 485, 623–626. [Google Scholar] [CrossRef] [Green Version]
- Ke, Y.; Ong, L.L.; Shih, W.M.; Yin, P. Three-Dimensional Structures Self-Assembled from DNA Bricks. Science 2012, 338, 1177–1183. [Google Scholar] [CrossRef] [Green Version]
- Yin, P.; Hariadi, R.F.; Sahu, S.; Choi, H.M.T.; Park, S.H.; LaBean, T.H.; Reif, J.H. Programming DNA Tube Circumferences. Science 2008, 321, 824–826. [Google Scholar] [CrossRef] [Green Version]
- Ong, L.L.; Hanikel, N.; Yaghi, O.K.; Grun, C.; Strauss, M.T.; Bron, P.; Lai-Kee-Him, J.; Schueder, F.; Wang, B.; Wang, P.; et al. Programmable self-assembly of three-dimensional nanostructures from 10,000 unique components. Nature 2017, 552, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Ong, L.L.; Sun, W.; Song, J.; Dong, M.; Shih, W.M.; Yin, P. DNA brick crystals with prescribed depths. Nat. Chem. 2014, 6, 994–1002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, W.; Shen, J.; Zhao, Z.; Arellano, N.; Rettner, C.; Tang, J.; Cao, T.; Zhou, Z.; Ta, T.; Streit, J.K.; et al. Precise pitch-scaling of carbon nanotube arrays within three-dimensional DNA nanotrenches. Science 2020, 368, 874–877. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, A.; Frenkel, D. Numerical Evidence for Nucleated Self-Assembly of DNA Brick Structures. Phys. Rev. Lett. 2014, 112, 238103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobs, W.M.; Reinhardt, A.; Frenkel, D. Communication: Theoretical prediction of free-energy landscapes for complex self-assembly. J. Chem. Phys. 2015, 142, 021101. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, W.M.; Reinhardt, A.; Frenkel, D. Rational design of self-assembly pathways for complex multicomponent structures. Proc. Natl. Acad. Sci. USA 2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobs, W.M.; Frenkel, D. Self-assembly protocol design for periodic multicomponent structures. Soft Matter 2015, 11, 8930–8938. [Google Scholar] [CrossRef]
- Madge, J.; Miller, M.A. Design strategies for self-assembly of discrete targets. J. Chem. Phys. 2015, 143, 044905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reinhardt, A.; Frenkel, D. DNA brick self-assembly with an off-lattice potential. Soft Matter 2016, 12, 6253–6260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madge, J.; Miller, M.A. Optimising minimal building blocks for addressable self-assembly. Soft Matter 2017, 13, 7780–7792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wales, D.J. Atomic clusters with addressable complexity. J. Chem. Phys. 2017, 146, 054306. [Google Scholar] [CrossRef] [Green Version]
- Wayment-Steele, H.K.; Frenkel, D.; Reinhardt, A. Investigating the role of boundary bricks in DNA brick self-assembly. Soft Matter 2017, 13, 1670–1680. [Google Scholar] [CrossRef] [Green Version]
- Fonseca, P.; Romano, F.; Schreck, J.S.; Ouldridge, T.E.; Doye, J.P.K.; Louis, A.A. Multi-scale coarse-graining for the study of assembly pathways in DNA-brick self-assembly. J. Chem. Phys. 2018, 148, 134910. [Google Scholar] [CrossRef] [Green Version]
- Sajfutdinow, M.; Jacobs, W.M.; Reinhardt, A.; Schneider, C.; Smith, D.M. Direct observation and rational design of nucleation behavior in addressable self-assembly. Proc. Natl. Acad. Sci. USA 2018, 115, E5877–E5886. [Google Scholar] [CrossRef] [Green Version]
- Tian, C.; Li, X.; Liu, Z.; Jiang, W.; Wang, G.; Mao, C. Directed Self-Assembly of DNA Tiles into Complex Nanocages. Angew. Chem. Int. Ed. 2014, 53, 8041–8044. [Google Scholar] [CrossRef] [PubMed]
- Goyal, A.; Limbachiya, D.; Gupta, S.K.; Joshi, F.; Pritmani, S.; Sahai, A.; Gupta, M.K. DNA Pen: A Tool for Drawing on a Molecular Canvas. arXiv 2013, arXiv:1306.0369. [Google Scholar]
- Gerling, T.; Wagenbauer, K.F.; Neuner, A.M.; Dietz, H. Dynamic DNA devices and assemblies formed by shape-complementary, non–base pairing 3D components. Science 2015, 347, 1446–1452. [Google Scholar] [CrossRef] [PubMed]
- Matthies, M.; Agarwal, N.P.; Poppleton, E.; Joshi, F.M.; Šulc, P.; Schmidt, T.L. Triangulated Wireframe Structures Assembled Using Single-Stranded DNA Tiles. ACS Nano 2019, 13, 1839–1848. [Google Scholar] [CrossRef] [PubMed]
- Schiffels, D.; Liedl, T.; Fygenson, D.K. Nanoscale Structure and Microscale Stiffness of DNA Nanotubes. ACS Nano 2013, 7, 6700–6710. [Google Scholar] [CrossRef]
- Hariadi, R.F.; Yurke, B.; Winfree, E. Thermodynamics and kinetics of DNA nanotube polymerization from single-filament measurements. Chem. Sci. 2015, 6, 2252–2267. [Google Scholar] [CrossRef] [Green Version]
- Schuldt, C.; Schnauß, J.; Händler, T.; Glaser, M.; Lorenz, J.; Golde, T.; Käs, J.A.; Smith, D.M. Tuning Synthetic Semiflexible Networks by Bending Stiffness. Phys. Rev. Lett. 2016, 117, 197801. [Google Scholar] [CrossRef] [Green Version]
- Glaser, M.; Schnauß, J.; Tschirner, T.; Schmidt, B.U.S.; Moebius-Winkler, M.; Käs, J.A.; Smith, D.M. Self-assembly of hierarchically ordered structures in DNA nanotube systems. New J. Phys. 2016, 18, 055001. [Google Scholar] [CrossRef] [Green Version]
- Fu, T.J.; Seeman, N.C. DNA double-crossover molecules. Biochemistry 1993, 32, 3211–3220. [Google Scholar] [CrossRef] [PubMed]
- Winfree, E.; Liu, F.; Wenzler, L.A.; Seeman, N.C. Design and self-assembly of two-dimensional DNA crystals. Nature 1998, 394, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Sun, W.; Seeman, N.C. Designed Two-Dimensional DNA Holliday Junction Arrays Visualized by Atomic Force Microscopy. J. Am. Chem. Soc. 1999, 121, 5437–5443. [Google Scholar] [CrossRef]
- Yan, H.; LaBean, T.H.; Feng, L.; Reif, J.H. Directed nucleation assembly of DNA tile complexes for barcode-patterned lattices. Proc. Natl. Acad. Sci. USA 2003, 100, 8103–8108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothemund, P.W.K.; Ekani-Nkodo, A.; Papadakis, N.; Kumar, A.; Fygenson, D.K.; Winfree, E. Design and Characterization of Programmable DNA Nanotubes. J. Am. Chem. Soc. 2004, 126, 16344–16352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.K.; Joshi, F.; Limbachiya, D.; Gupta, M.K. 3DNA: A Tool for DNA Sculpting. arXiv 2014, arXiv:1405.4118. [Google Scholar]
- Kocabey, S.; Meinl, H.; MacPherson, I.S.; Cassinelli, V.; Manetto, A.; Rothenfusser, S.; Liedl, T.; Lichtenegger, F.S. Cellular Uptake of Tile-Assembled DNA Nanotubes. J. Nanomater. 2015, 5, 47–60. [Google Scholar] [CrossRef] [Green Version]
- Sellner, S.; Kocabey, S.; Nekolla, K.; Krombach, F.; Liedl, T.; Rehberg, M. DNA nanotubes as intracellular delivery vehicles in vivo. Biomaterials 2015, 53, 453–463. [Google Scholar] [CrossRef]
- Sellner, S.; Kocabey, S.; Zhang, T.; Nekolla, K.; Hutten, S.; Krombach, F.; Liedl, T.; Rehberg, M. Dexamethasone-conjugated DNA nanotubes as anti-inflammatory agents in vivo. Biomaterials 2017, 134, 78–90. [Google Scholar] [CrossRef]
- NanoBricks Software. Available online: http://molecular.systems/software (accessed on 1 February 2020).
- Ke, Y.; Bellot, G.; Voigt, N.V.; Fradkov, E.; Shih, W.M. Two design strategies for enhancement of multilayer–DNA-origami folding: Underwinding for specific intercalator rescue and staple-break positioning. Chem. Sci 2012, 3, 2587–2597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosinski, R.; Mukhortava, A.; Pfeifer, W.; Candelli, A.; Rauch, P.; Saccà, B. Sites of high local frustration in DNA origami. Nat. Commun. 2019, 10, 1061. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Markham, N.R.; Zuker, M. UNAFold: Software for nucleic acid folding and hybridization. Methods Mol. Biol. 2008, 453, 3–31. [Google Scholar] [CrossRef]
- Zadeh, J.N.; Steenberg, C.D.; Bois, J.S.; Wolfe, B.R.; Pierce, M.B.; Khan, A.R.; Dirks, R.M.; Pierce, N.A. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 2011, 32, 170–173. [Google Scholar] [CrossRef]
- Lorenz, R.; Bernhart, S.H.; Höner zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]
- Castro, C.E.; Kilchherr, F.; Kim, D.N.; Shiao, E.L.; Wauer, T.; Wortmann, P.; Bathe, M.; Dietz, H. A primer to scaffolded DNA origami. Nat. Methods 2011, 8, 221–229. [Google Scholar] [CrossRef]
- Kim, D.N.; Kilchherr, F.; Dietz, H.; Bathe, M. Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Res. 2012, 40, 2862–2868. [Google Scholar] [CrossRef]
- Pan, K.; Kim, D.N.; Zhang, F.; Adendorff, M.R.; Yan, H.; Bathe, M. Lattice-free prediction of three-dimensional structure of programmed DNA assemblies. Nat. Commun. 2014, 5, 5578. [Google Scholar] [CrossRef] [PubMed]
- Sedeh, R.S.; Pan, K.; Adendorff, M.R.; Hallatschek, O.; Bathe, K.J.; Bathe, M. Computing Nonequilibrium Conformational Dynamics of Structured Nucleic Acid Assemblies. J. Chem. Theory Comput. 2016, 12, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Pan, K.; Boulais, E.; Yang, L.; Bathe, M. Structure-based model for light-harvesting properties of nucleic acid nanostructures. Nucleic Acids Res. 2014, 42, 2159–2170. [Google Scholar] [CrossRef]
- Sun, W.; Boulais, E.; Hakobyan, Y.; Wang, W.L.; Guan, A.; Bathe, M.; Yin, P. Casting inorganic structures with DNA molds. Science 2014, 346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schreck, J.S.; Romano, F.; Zimmer, M.H.; Louis, A.A.; Doye, J.P.K. Characterizing DNA Star-Tile-Based Nanostructures Using a Coarse-Grained Model. ACS Nano 2016, 10, 4236–4247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snodin, B.E.K.; Randisi, F.; Mosayebi, M.; Šulc, P.; Schreck, J.S.; Romano, F.; Ouldridge, T.E.; Tsukanov, R.; Nir, E.; Louis, A.A.; et al. Introducing improved structural properties and salt dependence into a coarse-grained model of DNA. J. Chem. Phys. 2015, 142, 234901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engel, M.C.; Smith, D.M.; Jobst, M.A.; Sajfutdinow, M.; Liedl, T.; Romano, F.; Rovigatti, L.; Louis, A.A.; Doye, J.P.K. Force-Induced Unravelling of DNA Origami. ACS Nano 2018, 12, 6734–6747. [Google Scholar] [CrossRef]
- Poppleton, E.; Bohlin, J.; Matthies, M.; Sharma, S.; Zhang, F.; Šulc, P. Design, optimization and analysis of large DNA and RNA nanostructures through interactive visualization, editing and molecular simulation. Nucleic Acids Res. 2020, 48, e72. [Google Scholar] [CrossRef]
- Suma, A.; Poppleton, E.; Matthies, M.; Šulc, P.; Romano, F.; Louis, A.A.; Doye, J.P.K.; Micheletti, C.; Rovigatti, L. TacoxDNA: A user-friendly web server for simulations of complex DNA structures, from single strands to origami. J. Comput. Chem. 2019, 40, 2586–2595. [Google Scholar] [CrossRef] [PubMed]
- Maffeo, C.; Aksimentiev, A. MrDNA: A multi-resolution model for predicting the structure and dynamics of DNA systems. Nucleic Acids Res. 2020, 48, 5135–5146. [Google Scholar] [CrossRef]
- Lee, J.Y.; Lee, J.G.; Yun, G.; Lee, C.; Kim, Y.J.; Kim, K.S.; Kim, T.H.; Kim, D.N. Rapid Computational Analysis of DNA Origami Assemblies at Near-Atomic Resolution. ACS Nano 2021, 15, 1002–1015. [Google Scholar] [CrossRef]
- de Llano, E.; Miao, H.; Ahmadi, Y.; Wilson, A.J.; Beeby, M.; Viola, I.; Barisic, I. Adenita: Interactive 3D modelling and visualization of DNA nanostructures. Nucleic Acids Res. 2020, 48, 8269–8275. [Google Scholar] [CrossRef] [PubMed]
- Goodman, R.P.; Schaap, I.A.T.; Tardin, C.F.; Erben, C.M.; Berry, R.M.; Schmidt, C.F.; Turberfield, A.J. Rapid Chiral Assembly of Rigid DNA Building Blocks for Molecular Nanofabrication. Science 2005, 310, 1661–1665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Jiang, Q.; Zhao, X.; Zhao, R.; Wang, Y.; Wang, Y.; Liu, J.; Shang, Y.; Zhao, S.; Wu, T.; et al. Publisher Correction: A DNA nanodevice-based vaccine for cancer immunotherapy. Nat. Mater. 2020, 1–3. [Google Scholar] [CrossRef]
- Helmi, S.; Ziegler, C.; Kauert, D.J.; Seidel, R. Shape-Controlled Synthesis of Gold Nanostructures Using DNA Origami Molds. Nano Lett. 2014, 14, 6693–6698. [Google Scholar] [CrossRef]
- Gopinath, A.; Rothemund, P.W.K. Optimized Assembly and Covalent Coupling of Single-Molecule DNA Origami Nanoarrays. ACS Nano 2014, 8, 12030–12040. [Google Scholar] [CrossRef] [Green Version]
- Gállego, I.; Manning, B.; Prades, J.D.; Mir, M.; Samitier, J.; Eritja, R. DNA-Origami-Driven Lithography for Patterning on Gold Surfaces with Sub-10 nm Resolution. Adv. Mater. 2017, 29, 1603233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sajfutdinow, M.; Uhlig, K.; Prager, A.; Schneider, C.; Abel, B.; Smith, D.M. Nanoscale patterning of self-assembled monolayer (SAM)-functionalised substrates with single molecule contact printing. Nanoscale 2017, 9, 15098–15106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, F.D. Perceived Usefulness, Perceived Ease of Use, and User Acceptance of Information Technology. MIS Q. 1989, 13, 319–340. [Google Scholar] [CrossRef] [Green Version]
- Alwall, J.; Frederix, R.; Frixione, S.; Hirschi, V.; Maltoni, F.; Mattelaer, O.; Shao, H.S.; Stelzer, T.; Torrielli, P.; Zaro, M. The automated computation of tree-level and next-to-leading order differential cross sections, and their matching to parton shower simulations. J. High Energy Phys. 2014, 2014, 79, arXiv:1405.0301. [Google Scholar] [CrossRef] [Green Version]
- Wei, X.; Nangreave, J.; Jiang, S.; Yan, H.; Liu, Y. Mapping the Thermal Behavior of DNA Origami Nanostructures. J. Am. Chem. Soc. 2013, 135, 6165–6176. [Google Scholar] [CrossRef] [PubMed]
- Dunn, K.E.; Dannenberg, F.; Ouldridge, T.E.; Kwiatkowska, M.; Turberfield, A.J.; Bath, J. Guiding the folding pathway of DNA origami. Nature 2015, 525, 82–86. [Google Scholar] [CrossRef]
- Schneider, F.; Möritz, N.; Dietz, H. The sequence of events during folding of a DNA origami. Sci. Adv. 2019, 5, eaaw1412. [Google Scholar] [CrossRef] [Green Version]
- Sobczak, J.P.J.; Martin, T.G.; Gerling, T.; Dietz, H. Rapid Folding of DNA into Nanoscale Shapes at Constant Temperature. Science 2012, 338, 1458–1461. [Google Scholar] [CrossRef]








| CAD Software | Scope | Main Features | Website |
|---|---|---|---|
| Cadnano 1.0/2.0 (2009/2012) [39] | Lattice-based scaffolded DNA origami design | GUI allowing for design from scratch & manual manipulation of strands Lattice-based (honeycomb or square lattice) + 2.0 introduces undo button | cadnano.org/legacy cadnano.org (download) |
| Cadnano 2.5 (2018, beta) | github.com/cadnano/cadnano2.5 (download) | ||
| scadnano (2020) [40] | Lattice-based scaffolded DNA origami design | Similar to Cadnano script based online tool | scadnano.org (direct use via browser) |
| Tiamat (2009) [41] | Lattice and scaffold free DNA nanostructure design | + No geometrical constrains + corrects for: secondary structures, repetitions and GC-content − needs manual adjustment | yanlab.asu.edu/Resources.html (download) |
| vHelix (2015) [24] | Automated 3D wireframe DNA origami design | Automated 3D wireframe design + Scaffold automatically transverses trough every edge evenly − Restricted to designs equivalent to a sphere | vhelix.net (download, req. Maya) |
| DAEDALUS (2016) [28] | Fully automated 3D wireframe origami design | Automated 3D wireframe origami design + No geometrical restriction to a sphere + designs stable at low salt − no GUI | daedalus-dna-origami.org (download, req. Maya) |
| PERDIX (2019) [45] | Fully automated 2D wireframe origami design | Automated 2D wireframe origami design + Arbitrary large 2D constructs − no GUI | perdix-dna-origami.org (download, req. Maya) |
| TALOS (2019) [46] | Fully automated 3D wireframe origami design (higher stability) | Automated 3D wireframe origami design + Increased mechanical stability due to six-helix edges − Material intensive, requires high salt conc. | talos-dna-origami.org (download, req. Maya) |
| METIS (2019) [47] | Fully automated 2D wireframe origami design (higher stability) | Automated 2D wireframe origami design + Increased mechanical stability due to six-helix edges −Material intensive, requires high salt conc. − no GUI | metis-dna-origami.org (download, req. Maya) |
| ATHENA (2020) [48] | Fully automated 2D & 3D wireframe origami design | Combines all features of DEADALUS, PERDIX, TALOS, METIS in an interactive GUI | github.com/lcbb/Athena (download, req. Maya) |
| CAD Software | Scope | Main Features | Website |
|---|---|---|---|
| DNA Pen (2013) [67] | 2D tile-based DNA designs | Free hand drawn or digitalized 2D design + Automatic inclusion of poly-T chains to prevent base stacking − only planar structures | guptalab.org/dnapen (download) |
| 3DNA (2014) [79] | 3D tile-based DNA designs | Digitalized 3D design + Allows for arbitrarily large structures + Accounts for GC content & Hamming distance | guptalab.org/3dna/index.html (download) |
| Hex-tiles (2019) [69] | 2D Triangulated Wireframe Structures using DNA Tiles | Triangulated 2D Wireframe Structures without a scaffold + Allows for arbitrarily large structures + Rolled up sheets resemble 3D hollow tubes + physiological salt conditions | github.com/tls-dna/hex-tiles (download) |
| CAD Software | Scope | Main Features | Website |
|---|---|---|---|
| UNAfold (2008) [87] | DNA & RNA folding and hybridization prediction | Continuation of Mfold | www.unafold.org (download) |
| NUPACK (2010) [88] | DNA folding and hybridization prediction | Suitable for multiple strand analysis | nupack.org (download & direct use via browser) |
| ViennaRNA Package 2.0 (2011) [89] | RNA secondary structure prediction | RNA secondary structure prediction | www.tbi.univie.ac.at/RNA/#download (download) |
| CanDo (2011) [90] | 2D & 3D modeling of DNA nanostructures | Finite element modeling framework for DNA origami assemblies input: caDNAno or Tiamat | cando-dna-origami.org (submission via browser) |
| oxDNA/oxDNA2 (2015) [97] oxView (2020) [99] | 2D & 3D Coarse-grained modelling of DNA & RNA assemblies | Coarse-grained modelling of DNA/RNA for DNA origami assemblies Includes Monte Carlo and Molecular Dynamics simulations Easy to visualize via browser-based oxView | dna.physics.ox.ac.uk/index.php (download) sulcgroup.github.io/oxdna-viewer (direct use via browser) |
| TacoxDNA (2019) [100] | Web-based interface for converting common formats of DNA structures | input: XYZ coordinate file, cadnano, Tiamat, CanDo, oxDNA, PDB output: oxDNA, PDB | tacoxdna.sissa.it (direct use via browser) |
| MrDNA (2019) [101] | Fast analysis of DNA nano-structures with high resolution | Faster prediction of low- & high-resolution models at Near-Atomic Resolution Predicts 3D shape & equilibrium properties input: cadnano, vHelix, DAEDALUS, CanDo, oxDNA, PDB | gitlab.engr.illinois.edu/tbgl/tools/mrdna (download) |
| Adenita (2020) [103] | Universal approach for the design and/or analyisis of DNA nano-structures | Combines several previous approaches and also other molecular structures input: cadnano, vHelix, DAEDALUS | samson-connect.net/element/dda2a078-1ab6-96ba-0d14-ee1717632d7a.html (download, req. SAMSON) |
| SNUPI (2021) [102] | Rapid analysis of DNA Origami structures with high resolution | Rapid analysis due to a multiscale analysis framework Predicts 3D shape, equilibrium dynamic properties & mechanical rigidity input: cadnano | github.com/SSDL-SNU/SNUPI (download) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glaser, M.; Deb, S.; Seier, F.; Agrawal, A.; Liedl, T.; Douglas, S.; Gupta, M.K.; Smith, D.M. The Art of Designing DNA Nanostructures with CAD Software. Molecules 2021, 26, 2287. https://doi.org/10.3390/molecules26082287
Glaser M, Deb S, Seier F, Agrawal A, Liedl T, Douglas S, Gupta MK, Smith DM. The Art of Designing DNA Nanostructures with CAD Software. Molecules. 2021; 26(8):2287. https://doi.org/10.3390/molecules26082287
Chicago/Turabian StyleGlaser, Martin, Sourav Deb, Florian Seier, Amay Agrawal, Tim Liedl, Shawn Douglas, Manish K. Gupta, and David M. Smith. 2021. "The Art of Designing DNA Nanostructures with CAD Software" Molecules 26, no. 8: 2287. https://doi.org/10.3390/molecules26082287
APA StyleGlaser, M., Deb, S., Seier, F., Agrawal, A., Liedl, T., Douglas, S., Gupta, M. K., & Smith, D. M. (2021). The Art of Designing DNA Nanostructures with CAD Software. Molecules, 26(8), 2287. https://doi.org/10.3390/molecules26082287