Predicting Absorption-Distribution Properties of Neuroprotective Phosphine-Borane Compounds Using In Silico Modeling and Machine Learning
Abstract
:1. Introduction
2. Results
2.1. Absorption
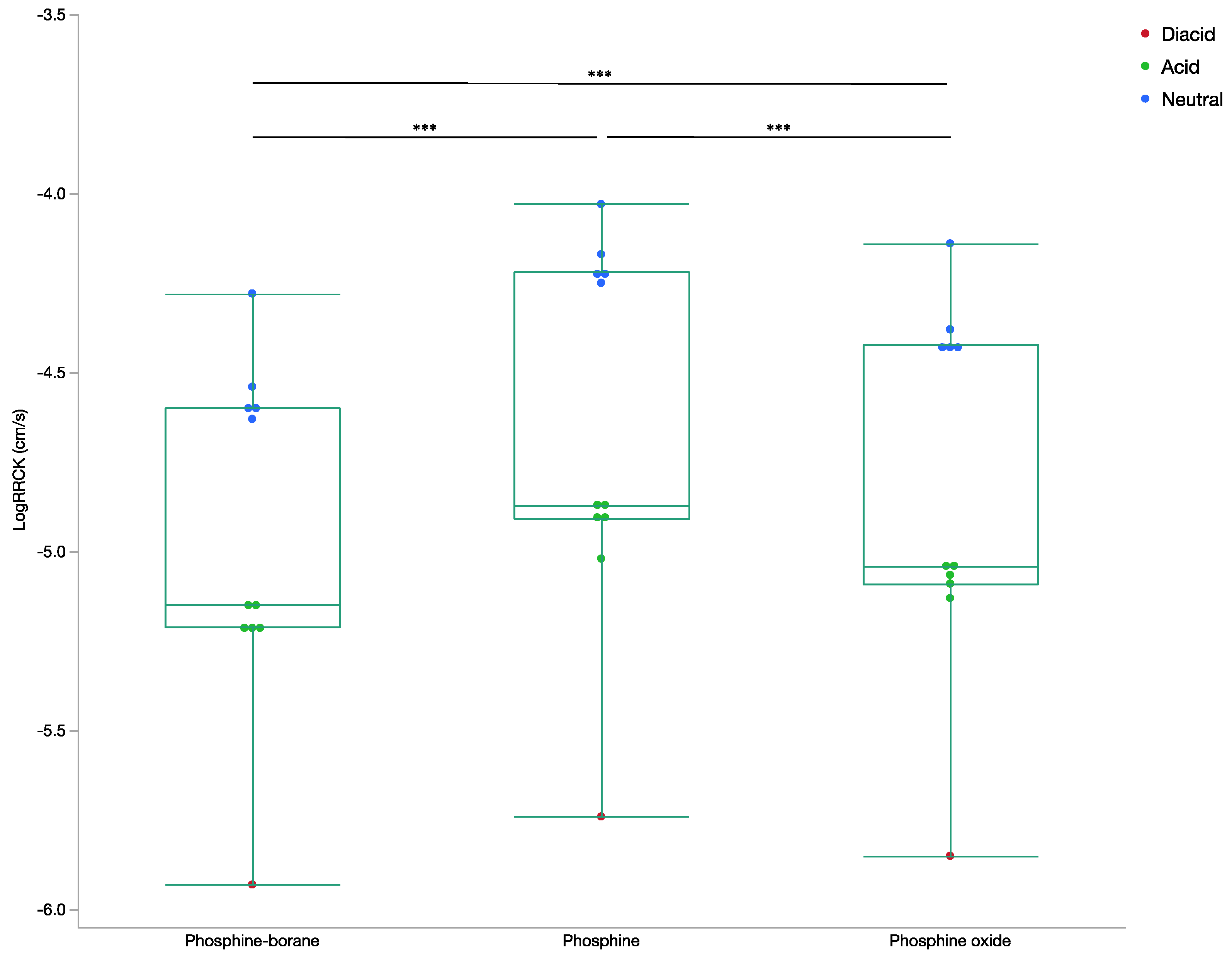
2.1.1. Cellular Permeability
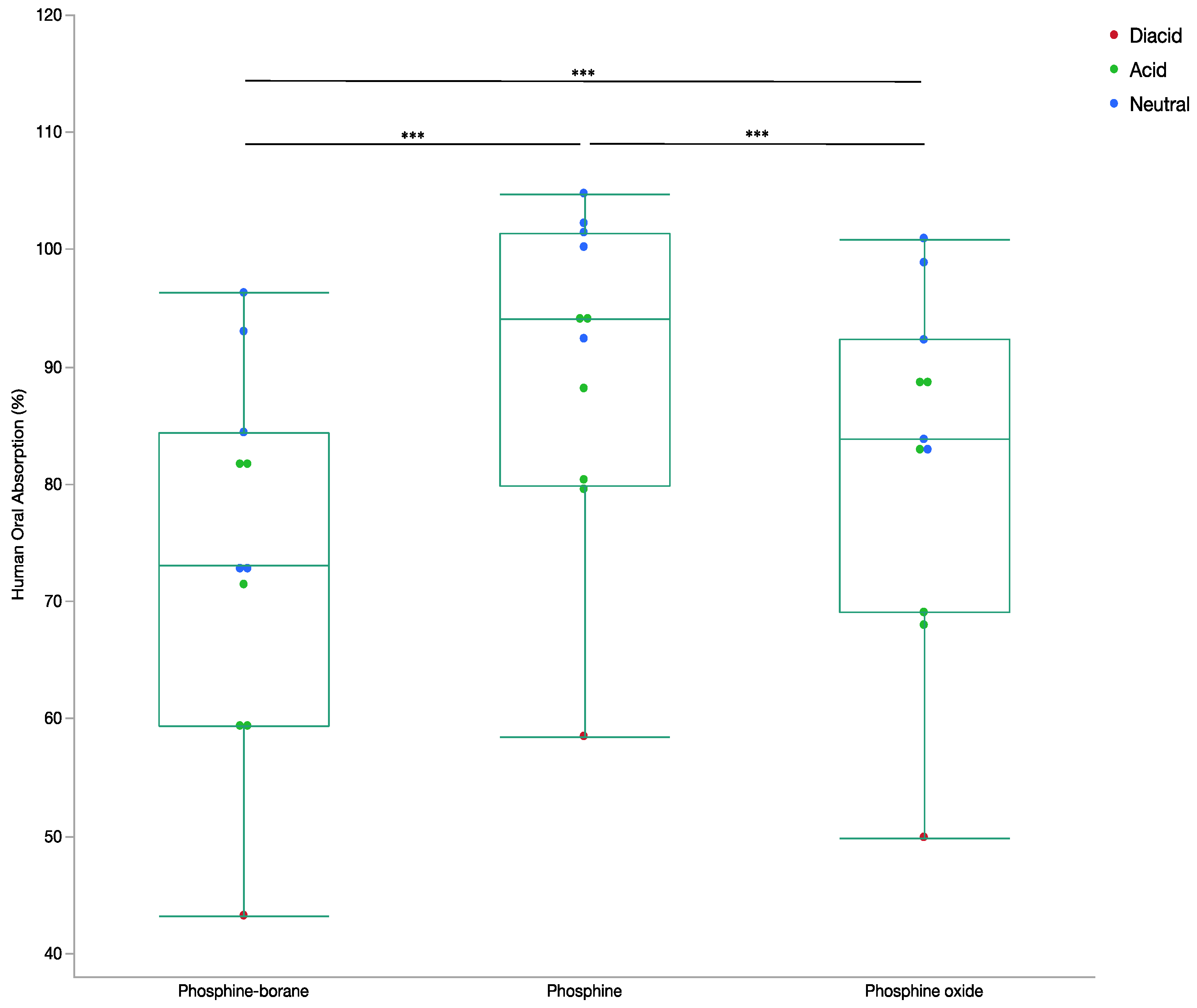
2.1.2. Oral Absorption
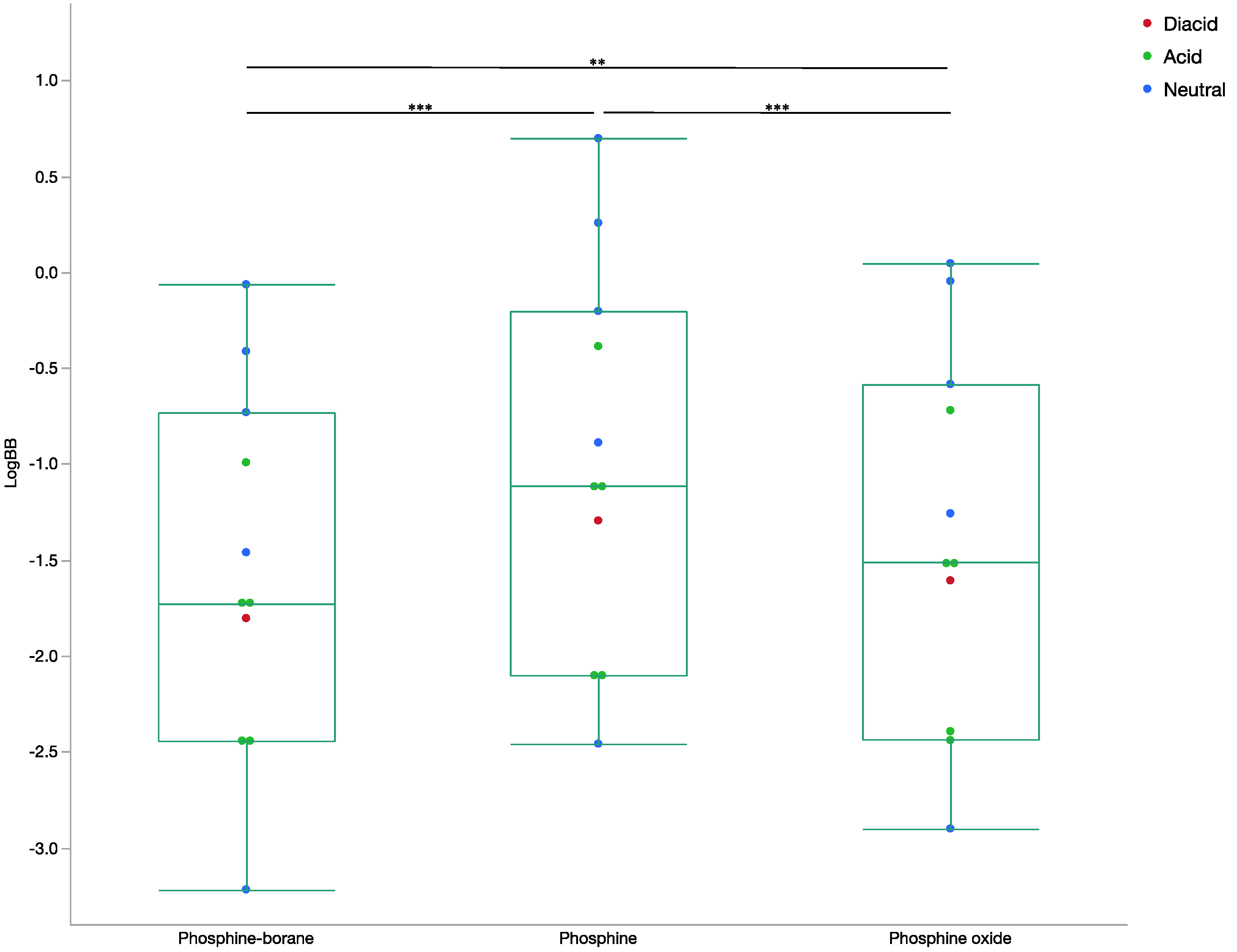
2.1.3. Blood-Brain Barrier Absorption
2.2. Distribution
2.2.1. Octanol/Water Coefficient
2.2.2. LogKHSA
3. Discussion
4. Materials and Methods
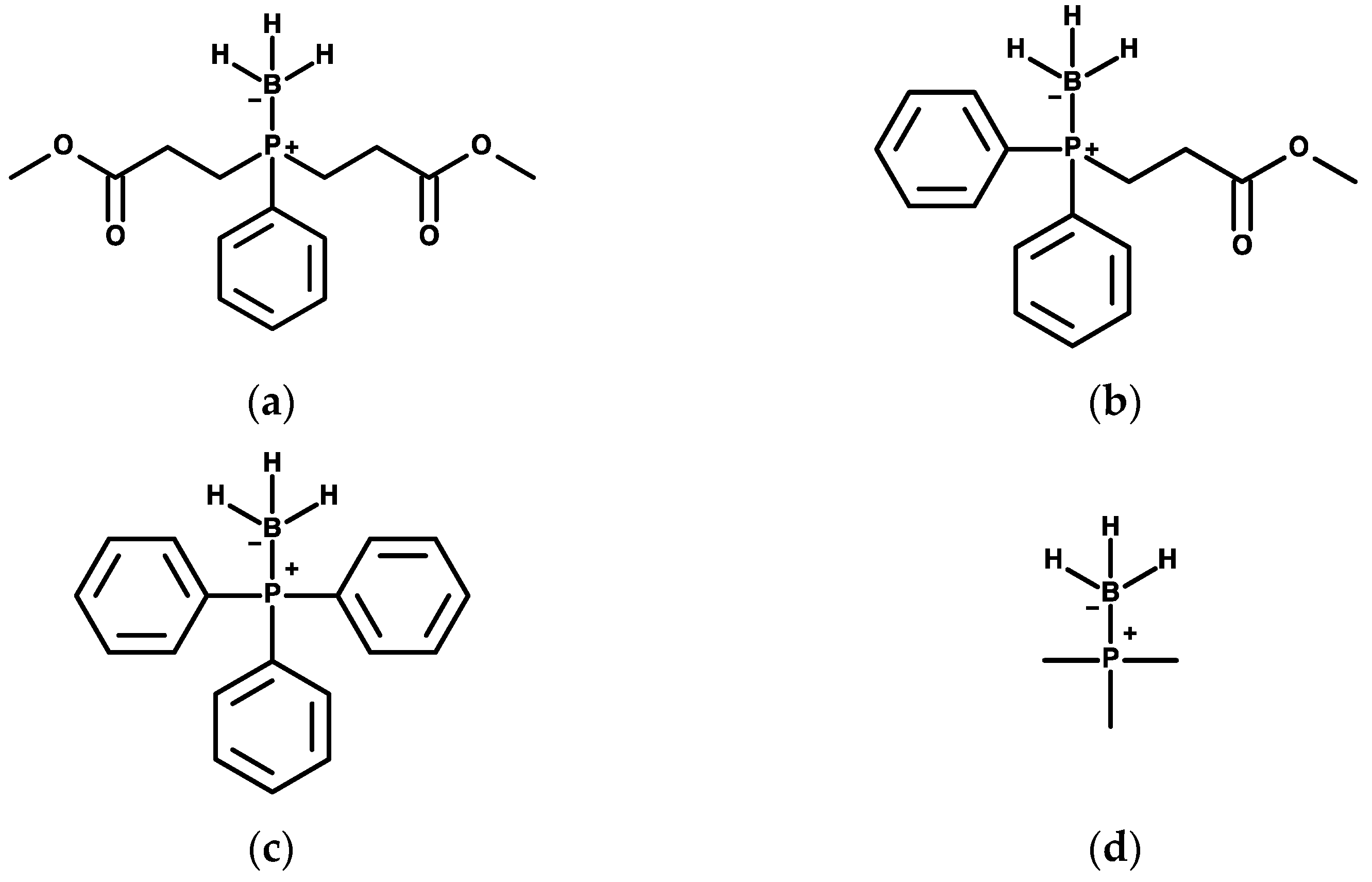
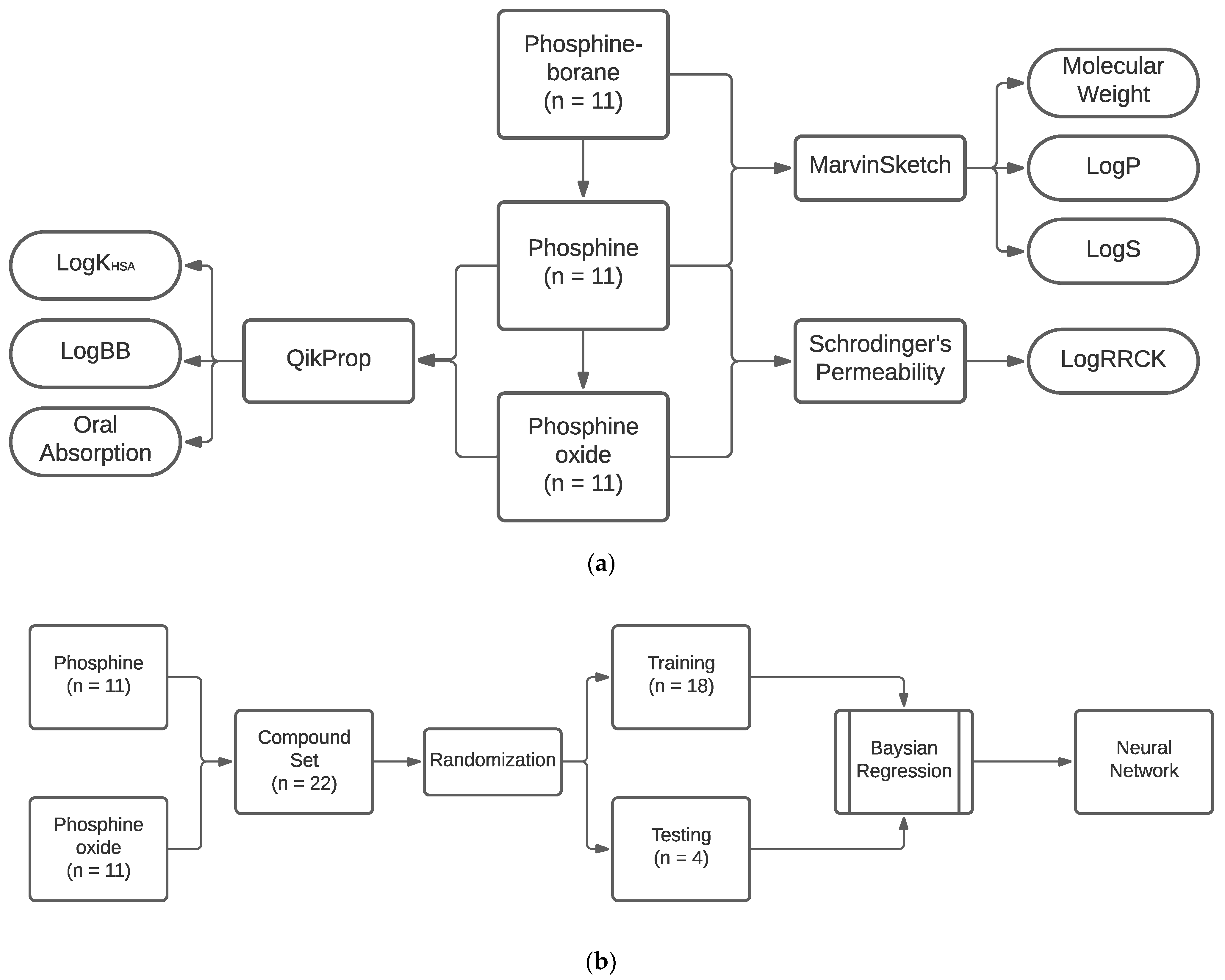
4.1. Compounds Tested
4.2. Absorption
4.3. Distribution
4.4. Neural Network Training
4.5. Statistics
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Almasieh, M.; Lieven, C.J.; Levin, L.A.; Di Polo, A. A cell-permeable phosphine-borane complex delays retinal ganglion cell death after axonal injury through activation of the pro-survival extracellular signal-regulated kinases 1/2 pathway. J. Neurochem. 2011, 118, 1075–1086. [Google Scholar] [CrossRef]
- Behbehani, R. Clinical Approach to Optic Neuropathies; Clinical Ophthalmology: Auckland, New Zealand, 2007; Volume 1, p. 233. [Google Scholar]
- Syc-Mazurek, S.B.; Fernandes, K.A.; Wilson, M.P.; Shrager, P.; Libby, R.T. Together JUN and DDIT3 (CHOP) control retinal ganglion cell death after axonal injury. Mol. Neurodegener. 2017, 12, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lieven, C.J.; Hoegger, M.J.; Schlieve, C.R.; Levin, L.A. Retinal ganglion cell axotomy induces an increase in intracellular superoxide anion. Invest. Ophthalmol. Vis. Sci. 2006, 47, 1477–1485. [Google Scholar] [CrossRef] [Green Version]
- Kanamori, A.; Catrinescu, M.-M.; Kanamori, N.; Mears, K.A.; Beaubien, R.; Levin, L.A. Superoxide is an associated signal for apoptosis in axonal injury. Brain 2010, 133, 2612–2625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geiger, L.; Kortuem, K.; Alexejun, C.; Levin, L. Reduced redox state allows prolonged survival of axotomized neonatal retinal ganglion cells. Neuroscience 2002, 109, 635–642. [Google Scholar] [CrossRef]
- Swanson, K.I.; Schlieve, C.R.; Lieven, C.J.; Levin, L.A. Neuroprotective effect of sulfhydryl reduction in a rat optic nerve crush model. Investig. Ophthalmol. Vis. Sci. 2005, 46, 3737–3741. [Google Scholar] [CrossRef] [PubMed]
- Schlieve, C.R.; Tam, A.; Nilsson, B.L.; Lieven, C.J.; Raines, R.T.; Levin, L.A. Synthesis and characterization of a novel class of reducing agents that are highly neuroprotective for retinal ganglion cells. Exp. Eye Res. 2006, 83, 1252–1259. [Google Scholar] [CrossRef] [PubMed]
- Catrinescu, M.-M.; Chan, W.; Mahammed, A.; Gross, Z.; Levin, L.A. Superoxide signaling and cell death in retinal ganglion cell axotomy: Effects of metallocorroles. Exp. Eye Res. 2012, 97, 31–35. [Google Scholar] [CrossRef] [Green Version]
- Dmitrenko, O.; Thorpe, C.; Bach, R.D. Mechanism of SN2 disulfide bond cleavage by phosphorus nucleophiles. Implications for biochemical disulfide reducing agents. J. Org. Chem. 2007, 72, 8298–8307. [Google Scholar] [CrossRef] [Green Version]
- Niemuth, N.J.; Thompson, A.F.; Crowe, M.E.; Lieven, C.J.; Levin, L.A. Intracellular disulfide reduction by phosphine-borane complexes: Mechanism of action for neuroprotection. Neurochem. Int. 2016, 99, 24–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, S.S.; Mijalkovic, J.; Borrelli, K.; Jacobson, M.P. Testing physical models of passive membrane permeation. J. Chem. Inf. Modeling 2012, 52, 1621–1636. [Google Scholar] [CrossRef] [Green Version]
- Leung, S.S.; Sindhikara, D.; Jacobson, M.P. Simple predictive models of passive membrane permeability incorporating size-dependent membrane-water partition. J. Chem. Inf. Modeling 2016, 56, 924–929. [Google Scholar] [CrossRef] [PubMed]
- US Dept Health Human Services (Ed.) Toxicology Profile for Boron; Agency for Toxic Substances and Diseases Registry: Atlanta, GA, USA, 2010.
- Dourson, M.; Maier, A.; Meek, B.; Renwick, A.; Ohanian, E.; Poirier, K. Boron tolerable intake. Biol. Trace Elem. Res. 1998, 66, 453–463. [Google Scholar] [CrossRef]
- Viswanadhan, V.N.; Ghose, A.K.; Revankar, G.R.; Robins, R.K. Atomic physicochemical parameters for three dimensional structure directed quantitative structure-activity relationships. 4. Additional parameters for hydrophobic and dispersive interactions and their application for an automated superposition of certain naturally occurring nucleoside antibiotics. J. Chem. Inf. Comput. Sci. 1989, 29, 163–172. [Google Scholar]
- Huehnchen, P.; Springer, A.; Kern, J.; Kopp, U.; Kohler, S.; Alexander, T.; Hiepe, F.; Meisel, A.; Boehmerle, W.; Endres, M. Bortezomib at therapeutic doses poorly passes the blood–brain barrier and does not impair cognition. Brain Commun. 2020, 2, fcaa021. [Google Scholar] [CrossRef]
- Ku, W.W.; Chapin, R.E.; Moseman, R.F.; Brink, R.E.; Pierce, K.D.; Adams, K.Y. Tissue disposition of boron in male Fischer rats. Toxicol. Appl. Pharmacol. 1991, 111, 145–151. [Google Scholar] [CrossRef]
- FDA (Ed.) VELCADE® (Bortezomib) for Injection, for Subcutaneous or Intravenous Use; U.S. Food and Drug Administration: Silver Spring, MD, USA, 2019. [Google Scholar]
- FDA (Ed.) EUCRISA™ (Crisaborole) Ointment, for Topical Use; U.S. Food and Drug Administration: Silver Spring, MD, USA, 2016. [Google Scholar]
- Griffith, D.C.; Loutit, J.S.; Morgan, E.E.; Durso, S.; Dudley, M.N. Phase 1 Study of the Safety, Tolerability, and Pharmacokinetics of the β-Lactamase Inhibitor Vaborbactam (RPX7009) in Healthy Adult Subjects. Antimicrob. Agents Chemother. 2016, 60, 6326–6332. [Google Scholar] [CrossRef] [Green Version]
- Lloyd-Jones, G.C.; Taylor, N.P. Mechanism of phosphine borane deprotection with amines: The effects of phosphine, solvent and amine on rate and efficiency. Chem. Eur. J. 2015, 21, 5423–5428. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Di, L.; Kerns, E.H.; Fan, K.; McConnell, O.J.; Carter, G.T. High throughput artificial membrane permeability assay for blood–brain barrier. Eur. J. Med. Chem. 2003, 38, 223–232. [Google Scholar] [CrossRef]
- Campbell, S.D.; Regina, K.J.; Kharasch, E.D. Significance of lipid composition in a blood-brain barrier–mimetic PAMPA assay. J. Biomol. Screen. 2014, 19, 437–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busher, J.T. Serum Albumin and Globulin. In The History, Physical, and Laboratory Examinations, 3rd ed.; Kenneth Walker, D.H.W.H., Ed.; Butterworths: Boston, MA, USA, 1990. [Google Scholar]
- Rezai, T.; Yu, B.; Millhauser, G.L.; Jacobson, M.P.; Lokey, R.S. Testing the conformational hypothesis of passive membrane permeability using synthetic cyclic peptide diastereomers. J. Am. Chem. Soc. 2006, 128, 2510–2511. [Google Scholar] [CrossRef] [PubMed]
- Klopman, G.; Li, J.-Y.; Wang, S.; Dimayuga, M. Computer automated log P calculations based on an extended group contribution approach. J. Chem. Inf. Comput. Sci. 1994, 34, 752–781. [Google Scholar] [CrossRef]
- Bloch, D. Computer software review. Review of PHYSPROP database (version 1.0). J. Chem. Inf. Comput. Sci. 1995, 35, 328–329. [Google Scholar] [CrossRef]
- Margaret, S.; Morton, D. The Digestive System, 2nd ed.; Churchill Livingstone: London, UK, 2010. [Google Scholar]
- Garcia, A.; Fry, N.A.; Karimi, K.; Liu, C.-C.; Apell, H.-J.; Rasmussen, H.H.; Clarke, R.J. Extracellular allosteric Na+ binding to the Na+, K+-ATPase in cardiac myocytes. Biophys. J. 2013, 105, 2695–2705. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Measure | Phase | Oral Absorption | LogBB | LogKHSA |
|---|---|---|---|---|
| Correlation | Training | 0.979 | 0.995 | 0.916 |
| Test | 0.895 | 0.968 | 0.958 | |
| Total | 0.964 | 0.993 | 0.920 | |
| Mean Absolute | Training | 2.33 | 0.14 | 0.37 |
| Error | Test | 5.73 | 0.08 | 0.32 |
| Total | 2.95 | 0.13 | 0.36 |
| Oral Absorption | LogBB | LogKHSA | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Measure | Training (SD) | Testing (SD) | Total | Training (SD) | Testing (SD) | Total | Training (SD) | Testing (SD) | Total |
| LogRRCK | −4.75 (0.53) | −4.70 (0.44) | 0.87 | −4.77 (0.52) | −4.62 (0.50) | 0.61 | −4.78 (0.51) | −4.55 (0.50) | 0.42 |
| Molecular Weight | 324.4 (140.7) | 273.3 (15.2) | 0.49 | 334.7 (129.2) | 226 (90.4) | 0.13 | 327.0 (126.5) | 261.3 (142.1) | 0.37 |
| LogP | 3.52 (3.18) | 3.02 (1.70) | 0.77 | 3.83 (2.97) | 1.59 (2.29) | 0.17 | 3.45 (3.15) | 3.34 (2.05) | 0.95 |
| LogS | −1.51 (3.88) | −1.20 (2.48) | 0.88 | −1.85 (3.83) | 0.32 (1.94) | 0.29 | 1.61 (3.80) | −0.73 (2.99) | 0.67 |
| Oral Absorption | 86.0 (15.3) | 90.5 (16.6) | 0.61 | ||||||
| LogBB | −1.31 (1.02) | −0.47 (0.66) | 0.13 | ||||||
| LogKHSA | −0.02 (1.07) | 0.23 (0.82) | 0.67 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Remtulla, R.; Das, S.K.; Levin, L.A. Predicting Absorption-Distribution Properties of Neuroprotective Phosphine-Borane Compounds Using In Silico Modeling and Machine Learning. Molecules 2021, 26, 2505. https://doi.org/10.3390/molecules26092505
Remtulla R, Das SK, Levin LA. Predicting Absorption-Distribution Properties of Neuroprotective Phosphine-Borane Compounds Using In Silico Modeling and Machine Learning. Molecules. 2021; 26(9):2505. https://doi.org/10.3390/molecules26092505
Chicago/Turabian StyleRemtulla, Raheem, Sanjoy Kumar Das, and Leonard A. Levin. 2021. "Predicting Absorption-Distribution Properties of Neuroprotective Phosphine-Borane Compounds Using In Silico Modeling and Machine Learning" Molecules 26, no. 9: 2505. https://doi.org/10.3390/molecules26092505





