Secondary Metabolites of Purpureocillium lilacinum
Abstract
1. Introduction
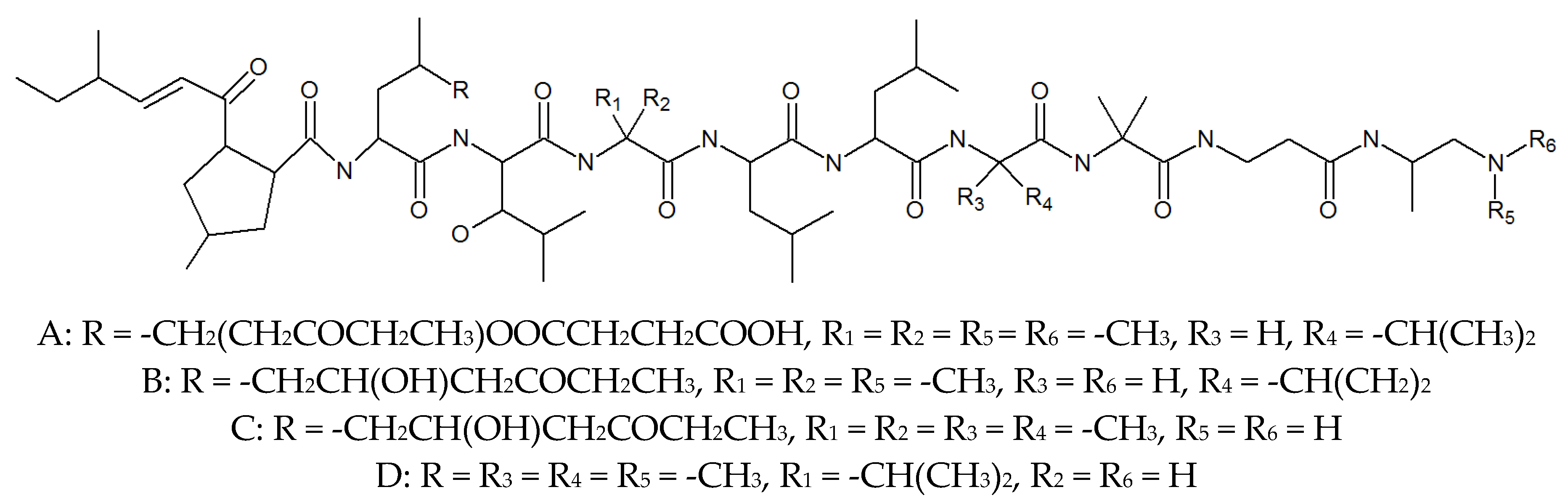
2. Leucinostatins
3. PK Metabolites
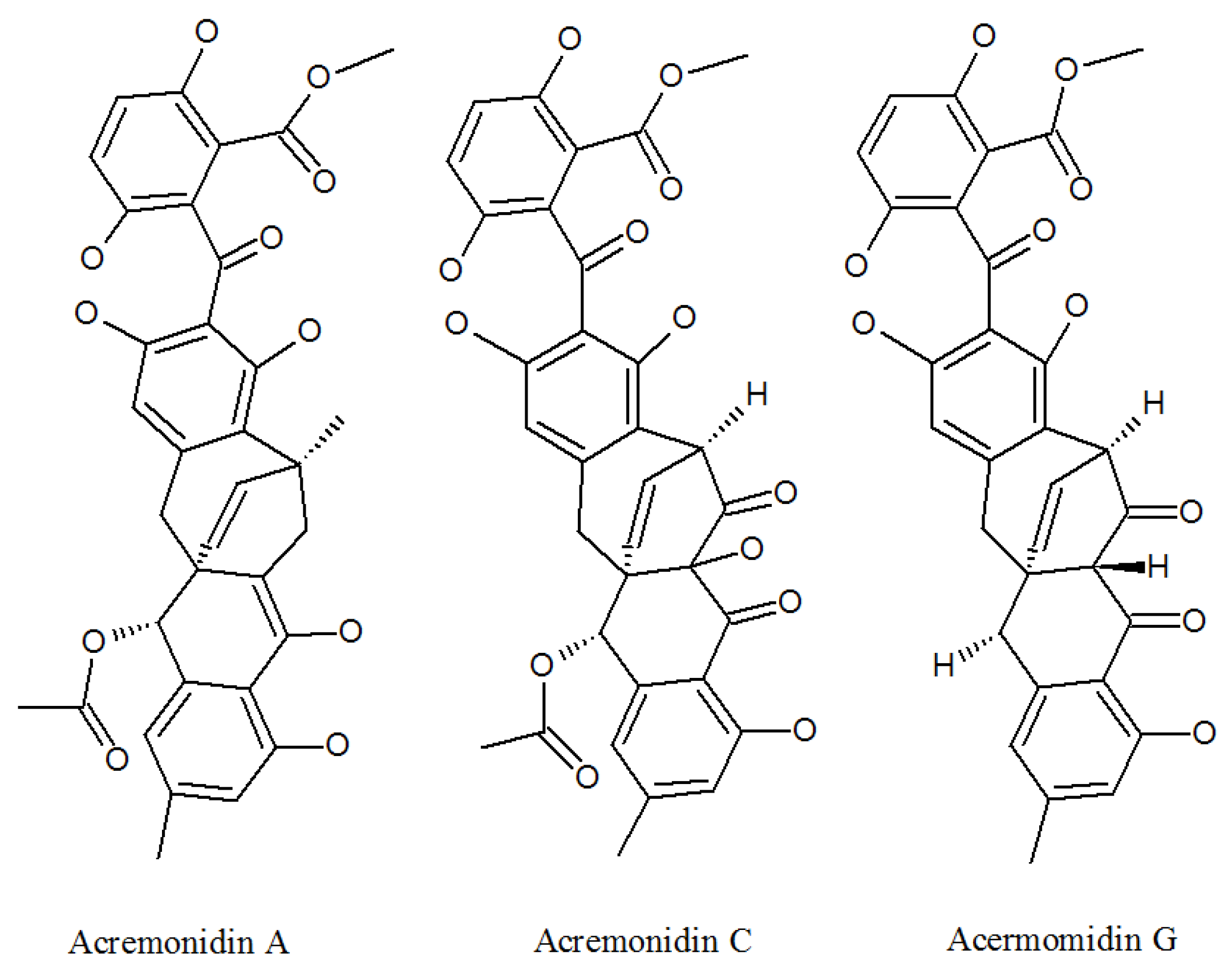
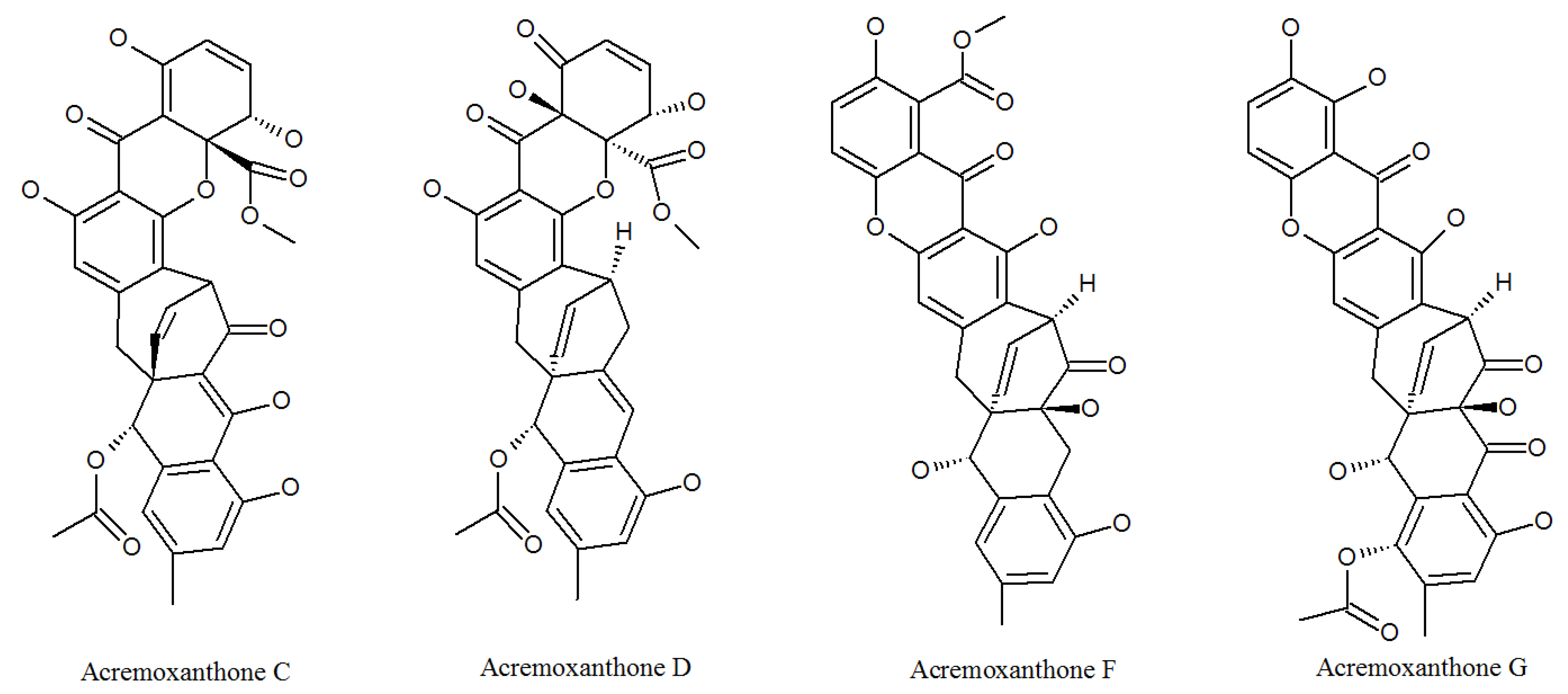
3.1. Acremonidins and Acremoxanthones
3.2. Paecilomide
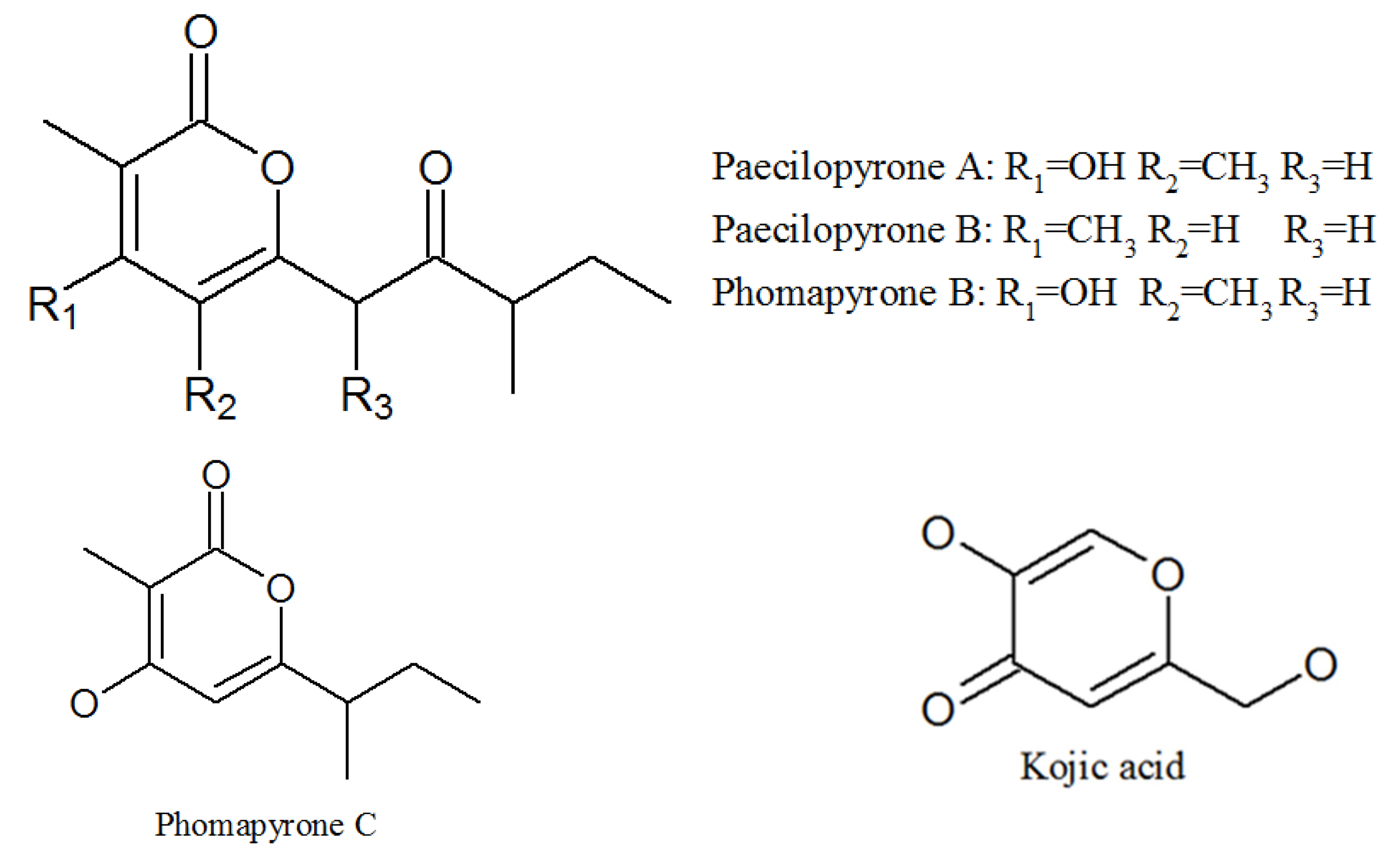
3.3. Pyrones
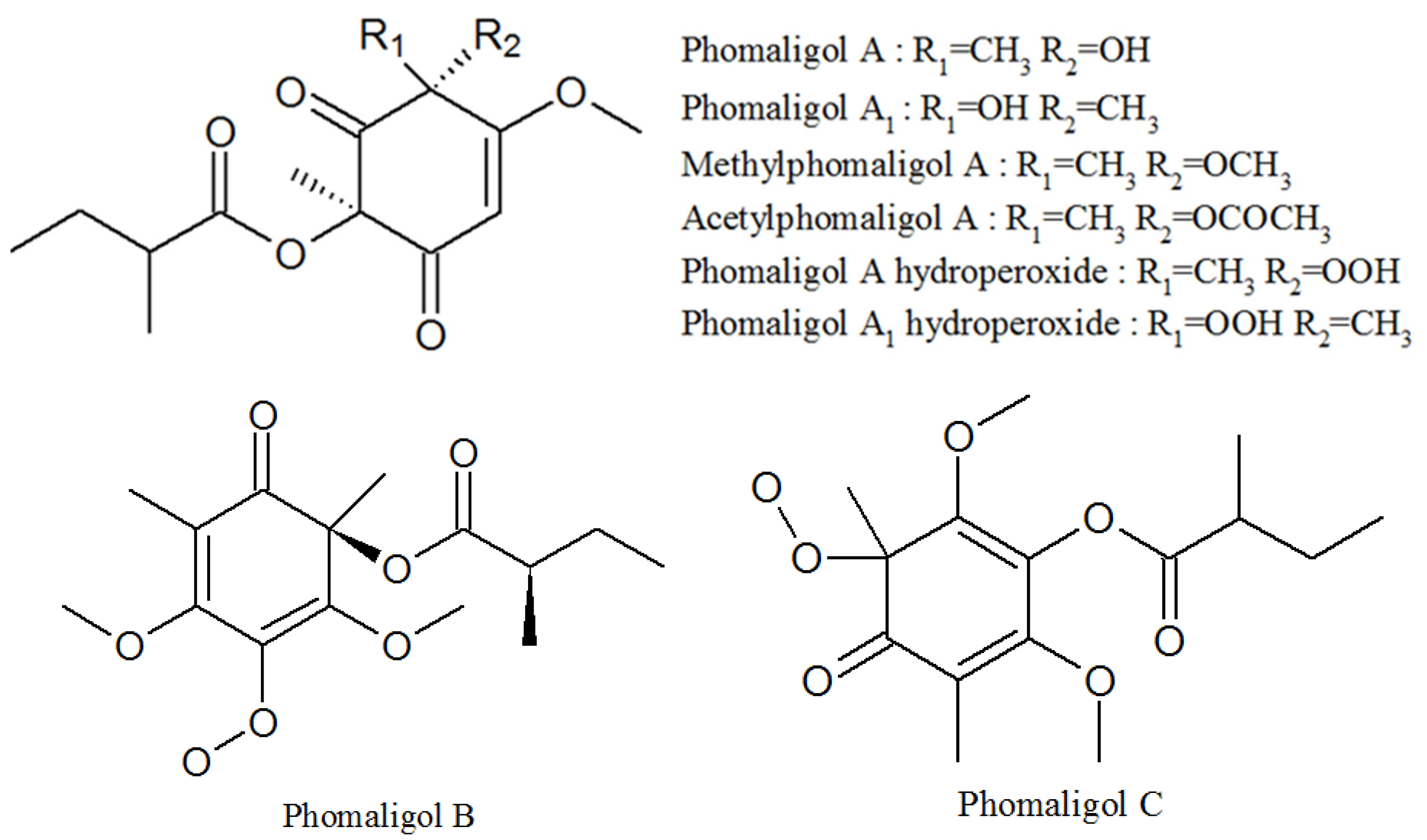
3.4. Phomaligols
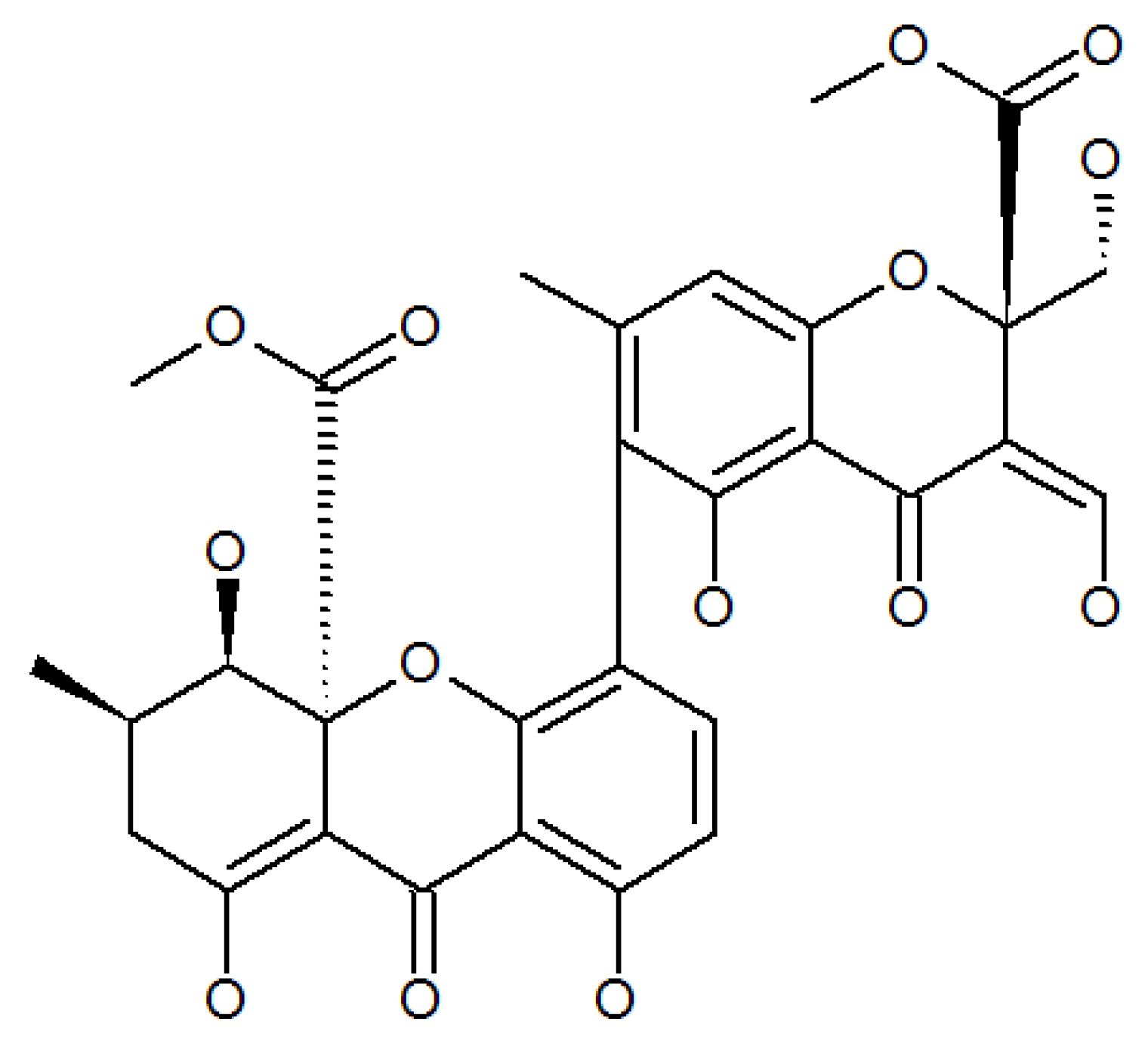
3.5. Pigment
4. Other Compounds
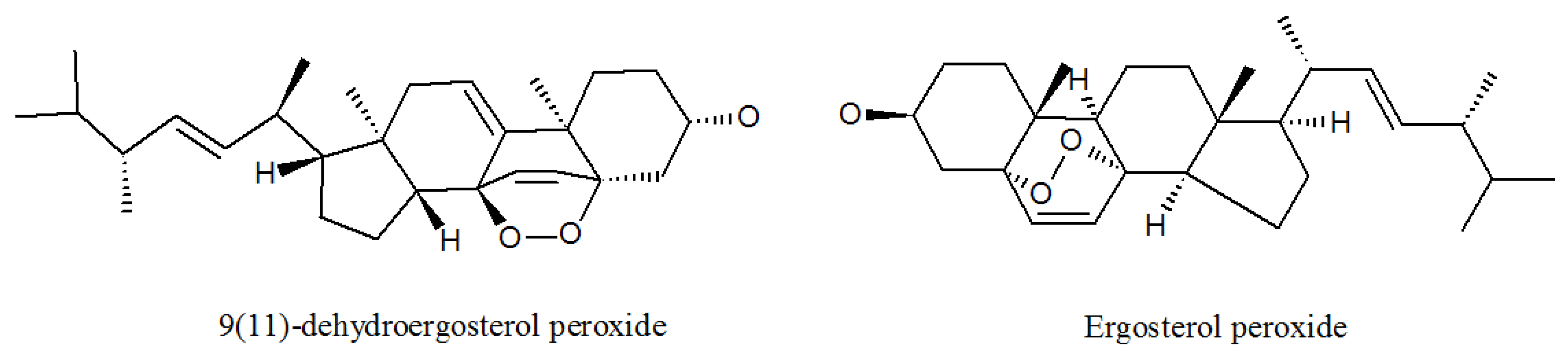
4.1. Ergosterols
4.2. Cerebrosides
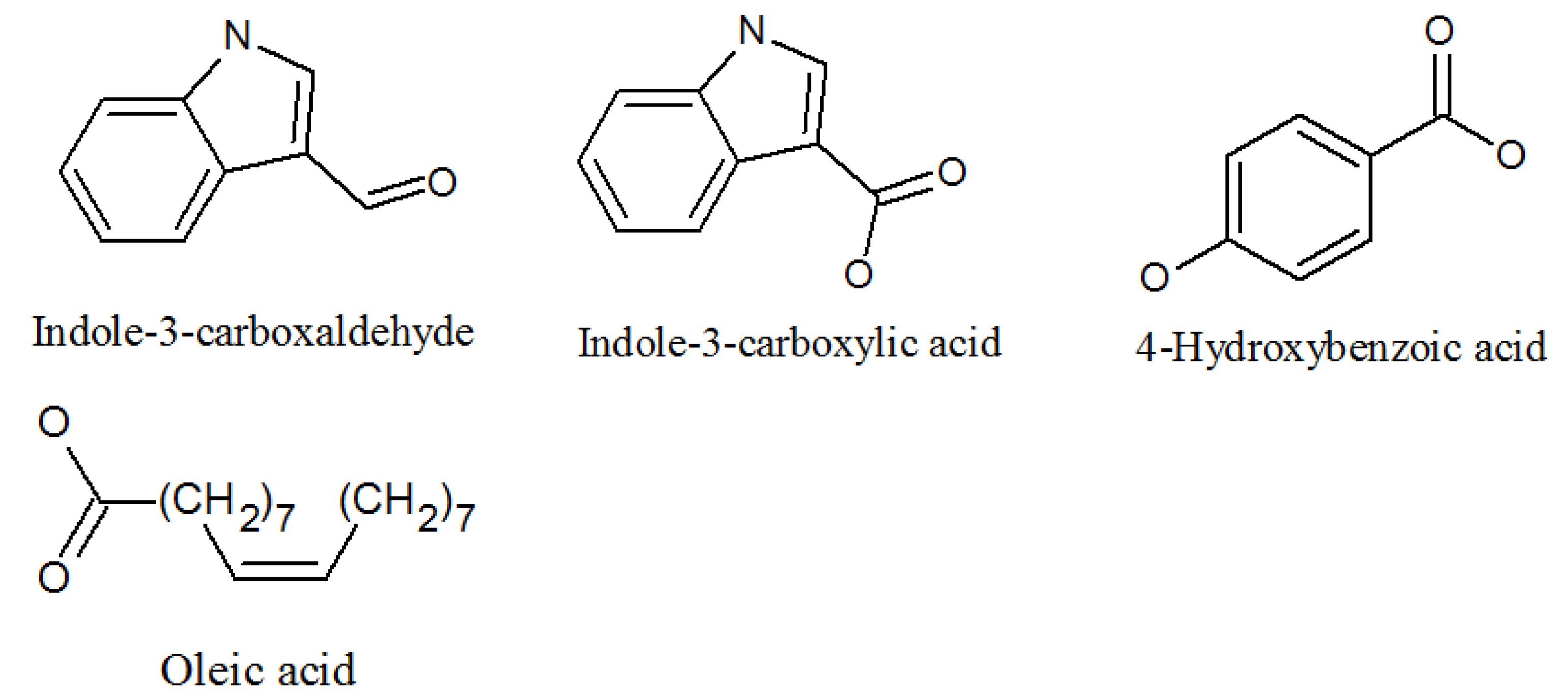
4.3. Paecilaminols and Others
| Metabolites | CAS. No | Material Source | Biological Activity |
|---|---|---|---|
| Leucinostatin A | 76600-38-9 | P. lilacinus ZBY-1 from deep sea water | Inhibited prostate cancer cells [29], nematocidal activity [26], activity against Gram-positive bacteria [27]. |
| Leucinostatin B | 159544-15-7 | Culture medium of P. lilacinm | Treatment of systemic candidiasis, nematocidal activity [26], activity against Gram-positive bacteria [27]. |
| Leucinostatin C | 110483-88-0 | Culture medium of P. lilacinm | Drug-related side-effects and adverse reactions activity against Gram-positive bacteria [27], nematocidal activity [26]. |
| Leucinostatin D | 100334-47-2 | Cultivated, mycelia complex of P. marquandii | Activity against Gram-positive bacteria [27], nematocidal activity [26]. |
| Leucinostatin F | Culture medium of P. lilacinm | Unknown | |
| Leucinostatin H | 109539-58-4 | Culture medium of P. lilacinm | Nematocidal activity [26]. |
| Leucinostatin K | 109539-57-3 | Culture medium of P. lilacinm | Nematocidal activity [26]. |
| Leucinostatin Y | Mycelia, cultivated complex of P. linacinus 40-H-28 | Preferential cytotoxicity to cancer cells under glucose-deprived conditions and inhibition of mitochondrial function [32]. | |
| Acremoxanthone C | 1360445-63-1P | Cultivated, mycelia complex of P. lilacinm | Cytotoxicity and 20 s proteasome inhibitory activity; high affinity with human calmodulin biosensors [37]; anti-oomycete activities [38]; exhibited anti-Bacillus cereus, antibacterial, antifungal, antiplasmodial, and cytotoxic activity; Gram-positive bacteria [36]. |
| Acremoxanthone D | 1360445-62-0P | Cultivated, mycelia complex of P. lilacinm | Moderate 20 s proteasome inhibitory activity [37]. |
| Acremoxanthone F | 1882150-25-5P | Cultivated, mycelia complex of P. lilacinm | Antimalarial activity against plasmodium falciparum K1 strain and multidrug-resistant strain [39]. |
| Acremoxanthone G | 1882150-26-6P | Cultivated, mycelia complex of P. lilacinm | Antimalarial activity against plasmodium falciparum K1 strain and multidrug-resistant strain [39]. |
| Acremonidin A | 701914-77-4P | Cultivated, mycelia complex of P. lilacinm | Moderate activity Against Gram-positive bacteria [36]. |
| Acremonidin C | 701914-79-6P | Cultivated, mycelia complex of P. lilacinm | Antibacterial activity [36]. |
| Acremonidin G | 1882150-23-3P | P. lilacinus ZBY-1 from deep sea water | Anti-enterococcus faecium activity [39]. |
| Paecilomide | 1538575-22-2P | Cultivated, mycelia complex of P. lilacinm | Acetylcholinesterase inhibitor [41]. |
| 9(11)-dehydroergosterolperoxide | 91579717 | P. lilacinus ZBY-1 from deep sea water | Cytotoxic effect [51]. |
| Ergosterol peroxide | 2061-64-5 | P. lilacinus ZBY-1 from deep sea water | Exhibits antimycobacterial, trypanocidal, and antineoplastic activities [51]. |
| (22E,24R)-5α, 6α-epoxy-3β-hydroxyergosta-22-ene-7-one | P. lilacinus ZBY-1 from deep sea water | Inhibitory effect of human cancer K562, MCF-7, HL-60, and BGC-823 cells [50]. | |
| Cerebroside A | 115681-40-8 | P. lilacinus ZBY-1 from deep sea water | Induction of cell growth, differentiation, and apoptosis in animals [56]. |
| Cerebroside B | 88642-46-0 | P. lilacinus ZBY-1 from deep sea water | Causes disease such as fusariosis, colitis, and apnea |
| Cerebroside C | 98677-33-9 | P. lilacinus ZBY-1 from deep sea water | Activity of cell wall-active; antibiotics; induction of cell growth, differentiation, and apoptosis in animals [55]. |
| Cerebroside D | 113773-89-0 | P. lilacinus ZBY-1 from deep sea water | Activity of cell wall-active antibiotics [55]. |
| Paecilopyrone A | 1173292-70-0 | Cultivated, mycelia complex of P. lilacinm | Unknown |
| Paecilopyrone B | 1173292-71-1 | Same as above | Unknown |
| Phomapyrone B | 157744-25-7 | Same as above | Unknown |
| Micropyrone | 54682570 | Same as above | Unknown |
| Phomapyrone C | 157744-26-8 | Same as above | Unknown |
| Kojic acid | 501-30-4 | Same as above | Antibacterial activities; tyrosinase inhibitory activity [44]. |
| Phomaligol A | 152204-32-5 | Same as above | Unknown |
| Phomaligol A1 | 152053-11-7 | Same as above | Unknown |
| Methylphomaligol A | 152159-01-8 | Same as above | Unknown |
| Acetylphomaligol A | 1173292-72-2 | Same as above | Unknown |
| Phomaligol A hydroperoxide | 181798-75-4 | Same as above | Unknown |
| Phomaligol A1 hydroperoxide | 182072-72-6 | Same as above | Unknown |
| Phomaligol B | 1173292-73-3 | Same as above | Unknown |
| Phomaligol C | 1173292-74-4 | Same as above | Unknown |
| Paecilaminol | 540770-33-0 | Same as above | Inhibits human cancer cell K562, MCF-7, HL-60, and BGC-823 cells [50]. |
| Paecilaminol Hydrochloride | 1650570-79-8 | Same as above | Inhibits human cancer cell K562, MCF-7, HL-60, and BGC-823 cells |
| Me myristate | 124-10-7 | Same as above | Medical carrier [50]. |
| Me linoleate | 112-63-0 | Same as above | Exhibited cytotoxic antibacterial activities against Bacillus subtilis and Staphylococcus aureus [59]. |
| Indole-3-carboxaldehyde | 487-89-8 | Same as above | Antimicrobial properties [62]. |
| Indolyl-3-carboxylic acid | 771-50-6 | Same as above | Potential in vitro antimalarial, anticancer activity [63]. |
| 4-hydroxybenzoic acid | 99-96-7 | Same as above | Inhibits LPS-induced protein [64]. |
| Purpureone | 2231079-10-8P | Mycelium of P. lilacinm | Antileishmanial activity; antibacterial activity [49]. |
5. Biosynthesis of Secondary Metabolites in Purpureocillium lilacinum
6. Problems and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luangsa-Ard, J.; Houbraken, J.; van Doorn, T.; Hong, S.B.; Borman, A.M.; Hywel-Jones, N.L.; Samson, R.A. Purpureocillium, a new genus for the medically important Paecilomyces lilacinus. FEMS Microbiol. Lett. 2011, 321, 141–149. [Google Scholar] [CrossRef]
- Sampson, R.A. Paecilomyces and Some Allied Hyphomycetes. Cent. Voor Schimmelcultures 1975, 64, 174. [Google Scholar] [CrossRef]
- Srilakshmi, A.; Sai Gopal, D.V.R.; Narasimha, G. Impact of bioprocess parameters on cellulase production by Purpureocillium lilacinum isolated from forest soil. Int. J. Pharma Bio Sci. 2017, 8, 157–165. [Google Scholar] [CrossRef]
- Zhu, Y.; Ai, D.; Zhang, W. Difference of soil microbiota in perennial ryegrass turf before and after turning green using high-throughput sequencing technology. Res. J. BioTechnol. 2017, 12, 50–60. [Google Scholar]
- Redou, V.; Navarri, M.; Meslet-Cladiere, L.; Barbier, G.; Burgaud, G. Species richness and adaptation of marine fungi from deep-subseafloor sediments. Appl. Environ. Microbiol. 2015, 81, 3571–3583. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhang, C.; Fan, H.; Guo, Z.; Yang, H.; Chen, M.; Han, J.; Cao, Y.; Xu, J.; Zhang, K.; et al. An efficient gene disruption system for the nematophagous fungus Purpureocillium lavendulum. Fungal. Biol. 2019, 123, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.D.; Carneiro, R.M.D.G.; Faria, M.; Souza, D.A.; Monnerat, R.G.; Lopes, R.B. Evaluation of Pochonia chlamydosporia and Purpureocillium lilacinum for suppression of Meloidogyne enterolobii on tomato and banana. J. Nematol. 2017, 49, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Gine, A.; Sorribas, F.J. Effect of plant resistance and BioAct WG (Purpureocillium lilacinum strain 251) on Meloidogyne incognita in a tomato-cucumber rotation in a greenhouse. Pest Manag. Sci. 2017, 73, 880–887. [Google Scholar] [CrossRef]
- Cavello, I.A.; Hours, R.A.; Rojas, N.L.; Cavalitto, S.F. Purification and characterization of a keratinolytic serine protease from Purpureocillium lilacinum LPS # 876. Process. Biochem. 2013, 48, 972–978. [Google Scholar] [CrossRef]
- Desaeger, J.A.; Watson, T.T. Evaluation of new chemical and biological nematicides for managing Meloidogyne javanica in tomato production and associated double-crops in Florida. Pest Manag. Sci. 2019, 75, 3363–3370. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Li, Y.; Li, Y.; Cao, H.; Mao, Z.; Ling, J.; Yang, Y.; Xie, B. Functional genetic analysis of the leucinostatin biosynthesis transcription regulator lcsL in Purpureocillium lilacinum using CRISPR-Cas9 technology. Appl. Microbiol. Biotechnol. 2019, 103, 6187–6194. [Google Scholar] [CrossRef]
- Wang, M.; Zhou, H.; Fu, Y.; Wang, C. The Antifungal Activities of the Fungus 36–1 to Several Plant Pathogens. Chin. J. Biol. Control. 1996, 12, 20–23. [Google Scholar]
- Li, F.; Chen, J.; Shi, H.; Liu, B. Anatgoinstic effect of biocontrol fungus, Paecilomyces lilacinus strain NH-PL-3 and its mechainsm against Fusairum oxyspourm. J. Plant Prot. 2005, 32, 373–378. [Google Scholar]
- Hotaka, D.; Amnuaykanjanasin, A.; Maketon, C.; Siritutsoontorn, S.; Maketon, M. Efficacy of Purpureocillium lilacinum CKPL-053 in controlling Thrips palmi (Thysanoptera: Thripidae) in orchid farms in Thailand. Appl. Entomol. Zool. 2015, 50, 317–329. [Google Scholar] [CrossRef]
- Yoder, J.A.; Fisher, K.A.; Dobrotka, C.J. A report on Purpureocillium lilacinum found naturally infecting the predatory mite, Balaustium murorum (Parasitengona: Erythraeidae). Int. J. Acarol. 2018, 44, 139–145. [Google Scholar] [CrossRef]
- Deng, J.X.; Paul, N.C.; Sang, H.K.; Lee, J.H.; Hwang, Y.S.; Yu, S.H. First Report on Isolation of Penicillium adametzioides and Purpureocillium lilacinum from Decayed Fruit of Cheongsoo Grapes in Korea. Mycobiology 2012, 40, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.-N.; Wang, H.; Hsueh, P.-R.; Meis, J.F.; Chen, H.; Xu, Y.-C. Endophthalmitis caused by Purpureocillium lilacinum. J. Microbiol. Immunol. Infect. 2019, 52, 170–171. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Liu, Z.; Lin, R.; Li, E.; Mao, Z.; Ling, J.; Yang, Y.; Yin, W.-B.; Xie, B. Biosynthesis of antibiotic leucinostatins in bio-control fungus Purpureocillium lilacinum and their inhibition on Phytophthora revealed by genome mining. PLoS Pathog. 2016, 12, e1005685. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B.; Bethe, B.; Hofs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. Chembiochem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, J.; Chen, C.; Teng, J.; Wang, C.; Luo, D. Structure and biosynthesis of fumosorinone, a new protein tyrosine phosphatase 1B inhibitor firstly isolated from the entomogenous fungus Isaria fumosorosea. Fungal Genet. Biol. 2015, 81, 191–200. [Google Scholar] [CrossRef]
- Yurchenko, A.N.; Girich, E.V.; Yurchenko, E.A. Metabolites of Marine Sediment-Derived Fungi: Actual Trends of Biological Activity Studies. Mar. Drugs 2021, 19, 88. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Q.; Xu, K.; Liu, X.-M.; Zhang, P. A Systematic Review on Secondary Metabolites of Paecilomyces Species: Chemical Diversity and Biological Activity. Planta Medica 2020, 86, 805–821. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Lin, Y.C.; She, Z.G.; Du, D.S.; Chan, W.L.; Zheng, Z.H. Paeciloxanthone, a new cytotoxic xanthone from the marine mangrove fungus Paecilomyces sp. (Tree1–7). J. Asian Nat. Prod. Res. 2008, 10, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Weng, Q.; Zhang, X.; Chen, W.; Hu, Q. Secondary Metabolites and the Risks of Isaria fumosorosea and Isaria farinosa. Molecules 2019, 24, 664. [Google Scholar] [CrossRef]
- Zhang, X.; Hu, Q.; Weng, Q. Secondary metabolites (SMs) of Isaria cicadae and Isaria tenuipes. RSC Adv. 2019, 9, 172–184. [Google Scholar] [CrossRef]
- Mikami, Y.; Yazawa, K.; Fukushima, K.; Arai, T.; Samson, R.A.J.M. Paecilotoxin production in clinical or terrestrial isolates of Paecilomyces lilacinus strains. Mycopathologia 1989, 108, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Rossi, C.; Tuttobello, L.; Ricci, M.; Casinovi, C.G.; Radios, L. Leucinostatin D, a novel peptide antibiotic from Paecilomyces marquandii. J. Antibiot. 1987, 40, 130–133. [Google Scholar] [CrossRef][Green Version]
- Momose, I.; Onodera, T.; Doi, H.; Adachi, H.; Iijima, M.; Yamazaki, Y.; Sawa, R.; Kubota, Y.; Igarashi, M.; Kawada, M. Leucinostatin Y: A Peptaibiotic Produced by the Entomoparasitic Fungus Purpureocillium lilacinum 40-H-28. J. Nat. Prod. 2019, 82, 1120–1127. [Google Scholar] [CrossRef]
- Kawada, M.; Inoue, H.; Momose, I.; Masuda, T.; Ikeda, D. 188 POSTER Leucinostatins suppress prostate cancer cell growth through the tumour-stromal cell interactions. Eur. J. Cancer Suppl. 2008, 6, 59–60. [Google Scholar] [CrossRef]
- Kawada, M.; Inoue, H.; Ohba, S.I.; Masuda, T.; Momose, I.; Ikeda, D. Leucinostatin A inhibits prostate cancer growth through reduction of insulin-like growth factor-I expression in prostate stromal cells. Int. J. Cancer 2010, 126, 810–818. [Google Scholar] [CrossRef]
- Ricci, M.; Blasi, P.; Giovagnoli, S.; Perioli, L.; Vescovi, C.; Rossi, C. Leucinostatin-A loaded nanospheres: Characterization and in vivo toxicity and efficacy evaluation. Int. J. Pharm. 2004, 275, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Arai, T.; Mikami, Y.; Fukushima, K.; Utsumi, T.; Yazawa, K. A new antibiotic, leucinostatin, derived from Penicillium lilacinum. J. Antibiot. 1973, 26, 157–161. [Google Scholar] [CrossRef]
- Fukushima, K.; Arai, T.; Mori, Y.; Tsuboi, M.; Suzuki, M. Studies on peptide antibiotics, leucinostatins. I. Separation, physico-chemical properties and biological activities of leucinostatins A and B. J. Antibiot. 1983, 36, 1606–1612. [Google Scholar] [CrossRef] [PubMed]
- Ishiyama, A.; Otoguro, K.; Iwatsuki, M.; Namatame, M.; Nishihara, A.; Nonaka, K.; Kinoshita, Y.; Takahashi, Y.; Masuma, R.; Shiomi, K.; et al. In vitro and in vivo antitrypanosomal activities of three peptide antibiotics: Leucinostatin A and B, alamethicin I and tsushimycin. J. Antibiot. 2009, 62, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Madariaga-Mazón, A.; González-Andrade, M.; González, M.d.C.; Glenn, A.E.; Cerda-García-Rojas, C.M.; Mata, R. Absolute Configuration of Acremoxanthone C, a Potent Calmodulin Inhibitor from Purpureocillium lilacinum. J. Nat. Prod. 2013, 76, 1454–1460. [Google Scholar] [CrossRef] [PubMed]
- Ayers, S.; Graf, T.N.; Adcock, A.F.; Kroll, D.J.; Shen, Q.; Swanson, S.M.; Matthew, S.; de Blanco, E.; Wani, M.C.; Darveaux, B.A.; et al. Cytotoxic xanthone-anthraquinone heterodimers from an unidentified fungus of the order Hypocreales (MSX 17022). J. Antibiot. 2012, 65, 3–8. [Google Scholar] [CrossRef]
- Melendez-Gonzalez, C.; Muria-Gonzalez, M.J.; Anaya, A.L.; Hernandez-Bautista, B.E.; Hernandez-Ortega, S.; Gonzalez, M.C.; Glenn, A.E.; Hanlin, R.T.; Macias-Rubalcava, M.L. Acremoxanthone E, a Novel Member of Heterodimeric Polyketides with a Bicyclo[3.2.2]nonene Ring, Produced by Acremonium camptosporum W. Gams (Clavicipitaceae) Endophytic Fungus. Chem. Biodivers. 2015, 12, 133–147. [Google Scholar] [CrossRef]
- Isaka, M.; Palasarn, S.; Auncharoen, P.; Komwijit, S.; Gareth Jones, E.B. Acremoxanthones A and B, novel antibiotic polyketides from the fungus Acremonium sp. BCC 31806. Tetrahedron Lett. 2009, 50, 284–287. [Google Scholar] [CrossRef]
- He, H.; Bigelis, R.; Solum, E.H.; Greenstein, M.; Carter, G.T. Acremonidins, new polyketide-derived antibiotics produced by Acremonium sp., LL-Cyan 416. J. Antibiot. 2003, 56, 923–930. [Google Scholar] [CrossRef]
- Teles, A.P.C.; Takahashi, J.A. Paecilomide, a new acetylcholinesterase inhibitor from Paecilomyces lilacinus. Microbiol. Res. 2013, 168, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Tumiatti, V.; Bolognesi, M.L.; Minarini, A.; Rosini, M.; Milelli, A.; Matera, R.; Melchiorre, C. Progress in acetylcholinesterase inhibitors for Alzheimer’s disease: An update. Expert Opin. Ther. Pat. 2008, 18, 387–401. [Google Scholar] [CrossRef]
- Kulshreshtha, A.; Piplani, P. Current pharmacotherapy and putative disease-modifying therapy for Alzheimer’s disease. Neurol. Sci. 2016, 37, 1403–1435. [Google Scholar] [CrossRef]
- Seibert, S.F.; Eguereva, E.; Krick, A.; Kehraus, S.; Voloshina, E.; Raabe, G.; Fleischhauer, J.; Leistner, E.; Wiese, M.; Prinz, H.; et al. Polyketides from the marine-derived fungus Ascochyta salicorniae and their potential to inhibit protein phosphatases. Org. Biomol. Chem. 2006, 4, 2233–2240. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Xie, W.; Zhao, Y.; Lv, X.; Yang, H.; Zeng, Q.; Zheng, Z.; Yang, X. Synthesis, antimicrobial, moisture absorption and retention activities of kojic acid-grafted konjac glucomannan oligosaccharides. Polymers 2019, 11, 1979. [Google Scholar] [CrossRef]
- Li, X.; Jeong, J.H.; Lee, K.T.; Rho, J.R.; Choi, H.D.; Kang, J.S.; Son, B.W. γ-pyrone derivatives, kojic acid methyl ethers from a marine-derived fungus Altenaria sp. Arch. Pharmacal Res. 2003, 26, 532–534. [Google Scholar] [CrossRef]
- Elbandy, M.; Shinde, P.B.; Hong, J.; Bae, K.S.; Kim, M.A.; Lee, S.M.; Jung, J.H. α-pyrones and yellow pigments from the sponge-derived fungus Paecilomyces lilacinus. Bull. Korean Chem. Soc. 2009, 30, 188–192. [Google Scholar] [CrossRef]
- Pedras, M.S.C.; Taylor, J.L.; Morales, V.M. Phomaligin A and other yellow pigments in Phoma lingam and P. wasabiae. Phytochemistry 1995, 38, 1215–1222. [Google Scholar] [CrossRef]
- Soga, O.; Iwamoto, H.; Hata, K.; Maeba, R.; Takuwa, A.; Fujiwara, T.; Hsu, Y.H.; Nakayama, M. New oxidation product of was abidienone-A. Agric. Biol. Chem. 1988, 52, 865–866. [Google Scholar] [CrossRef]
- Lenta, B.N.; Ngatchou, J.; Frese, M.; Ladoh-Yemeda, F.; Voundi, S.; Nardella, F.; Michalek, C.; Wibberg, D.; Ngouela, S.; Tsamo, E.; et al. Purpureone, an antileishmanial ergochrome from the endophytic fungus Purpureocillium lilacinum. Z. Für Nat. B J. Chem. Sci. 2016, 71, 1159–1167. [Google Scholar] [CrossRef]
- Cui, X.; Li, C.; Wu, C.; Hua, W.; Cui, C.; Zhu, T.; Gu, Q.-Q. Metabolites of Paecilomyces lilacinus zby-1 from deep-sea water and their antitumor activity. J. Int. Pharm. Res 2013, 40, 177–186. [Google Scholar]
- Wu, H.; Yang, F.; Li, L.; Rao, Y.K.; Ju, T.; Wong, W.; Hsieh, C.; Pivkin, M.V.; Hua, K.; Wu, S. Ergosterol peroxide from marine fungus Phoma sp. induces ROS-dependent apoptosis and autophagy in human lung adenocarcinoma cells. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, S.; Roh, H.-S.; Song, S.-S.; Ryoo, R.; Pang, C.; Baek, K.-H.; Kim, K.H. Cytotoxic constituents from the sclerotia of Poria cocos against human lung adenocarcinoma cells by inducing mitochondrial apoptosis. Cells 2018, 7, 116. [Google Scholar] [CrossRef] [PubMed]
- Umemura, K.; Tanino, S.; Nagatsuka, T.; Koga, J.; Iwata, M.; Nagashima, K.; Amemiya, Y. Cerebroside elicitor confers resistance to Fusarium disease in various plant species. Phytopathology 2004, 94, 813–818. [Google Scholar] [CrossRef] [PubMed]
- Hua, W.; Miao, Y.; Chen, J.; You, Y.; Sun, C.; Yuan, L.; Xu, L. Identification and growth characterization of a cerebroside producing Termitomyces clypeatus CTM-1. Shengwu Jiagong Guocheng 2015, 13, 67–73. [Google Scholar] [CrossRef]
- Wicklow, D.T.; Joshi, B.K.; Gamble, W.R.; Gloer, J.B.; Dowd, P.F. Antifungal metabolites (monorden, monocillin IV, and cerebrosides) from Humicola fuscoatra traaen NRRL 22980, a mycoparasite of Aspergillus flavus sclerotia. Appl. Environ. Microbiol. 1998, 64, 4482–4484. [Google Scholar] [CrossRef] [PubMed]
- Koga, J. Induction of Rice Disease Resistance by Fungal Sphingolipids and Bile Acids. Am. Chem. Soc. 2009, 237, 100. [Google Scholar]
- Ui, H.; Shiomi, K.; Suzuki, H.; Hatano, H.; Morimoto, H.; Yamaguchi, Y.; Masuma, R.; Sakamoto, K.; Kita, K.; Miyoshi, H.; et al. Paecilaminol, a new NADH-fumarate reductase inhibitor, produced by Paecilomyces sp. FKI-0550. J. Antibiot. 2006, 59, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Guo, J.; Liu, W. The synthesis and property research of nanoparticles based on hydrophobic modified inulin. Shandong Huagong 2015, 44, 10–15. [Google Scholar]
- Khentoul, H.; Bensouici, C.; Reyes, F.; Albanese, D.; Sarri, D.; Ratiba, M.; Fadila, B.; Seghiri, R.; Boumaza, O. Chemical constituents and HRESI-MS analysis of an Algerian endemic plant—Verbascum atlanticum Batt—extracts and their antioxidant activity. Nat. Prod. Res. 2019, 34, 3008–3012. [Google Scholar] [CrossRef]
- Abd-Ellatif, A.E.S.; Abdel-Razek, A.S.; Hamed, A.; Soltan, M.M.; Soliman, H.S.M.; Shaaban, M. Bioactive compounds from marine Streptomyces sp.: Structure identification and biological activities. Vietnam. J. Chem. 2019, 57, 628–635. [Google Scholar] [CrossRef]
- Goncalves-de-Albuquerque, C.F.; Silva, A.R.; Burth, P.; Castro-Faria, M.V.; Castro-Faria-Neto, H.C. Acute Respiratory Distress Syndrome: Role of Oleic Acid-Triggered Lung Injury and Inflammation. Mediat. Inflamm. 2015, 2015, 1–9. [Google Scholar] [CrossRef]
- Balaz, M.; Kudlickova, Z.; Vilkova, M.; Imrich, J.; Balazova, L.; Daneu, N. Mechanochemical synthesis and isomerization of N-substituted indole-3-carboxaldehyde oximes. Molecules 2019, 24, 3347. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yu, J.; Yao, X. Application of Indole-3-Carboxaldehyde in Preparation of Cosmetic or Drug for Treating Atopic Dermatitis. CN110368385A, 25 October 2019. China National Intellectual Property Administration, Beijing, China. [Google Scholar]
- Kim, H.; Kim, S.Y.; Sim, G.Y.; Ahn, J.-H. Synthesis of 4-Hydroxybenzoic Acid Derivatives in Escherichia coli. J. Agric. Food Chem. 2020, 68, 9743–9749. [Google Scholar] [CrossRef]
- Hurtado-Barroso, S.; Quifer-Rada, P.; Marhuenda-Munoz, M.; de Alvarenga, J.F.R.; Tresserra-Rimbau, A.; Lamuela-Raventos, R.M. Increase of 4-hydroxybenzoic, a bioactive phenolic compound, after an organic intervention diet. Antioxidants 2019, 8, 340. [Google Scholar] [CrossRef] [PubMed]
- Maia, N.J.L.; Correa, J.A.F.; Rigotti, R.T.; da Silva Junior, A.A.; Luciano, F.B. Combination of natural antimicrobials for contamination control in ethanol production. World J. Microbiol. Biotechnol. 2019, 35, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Prasad, P.; Varshney, D.; Adholeya, A. Whole genome annotation and comparative genomic analyses of bio-control fungus Purpureocillium lilacinum. BMC Genom. 2015, 16, 1004. [Google Scholar] [CrossRef]
- Khaldi, N.; Seifuddin, F.T.; Turner, G.; Haft, D.; Nierman, W.C.; Wolfe, K.H.; Fedorova, N.D. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 2010, 47, 736–741. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Muller, R.; Wohlleben, W.; et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef]
- Takeda, K.; Kemmoku, K.; Satoh, Y.; Ogasawara, Y.; Shinya, K.; Dairi, T.J.A.C.B. N-Phenylacetylation and Nonribosomal Peptide Synthetases with Substrate Promiscuity for Biosynthesis of Heptapeptide Variants, JBIR-78 and JBIR-95. ACS Chem. Biol. 2017, 12, 1813. [Google Scholar] [CrossRef]
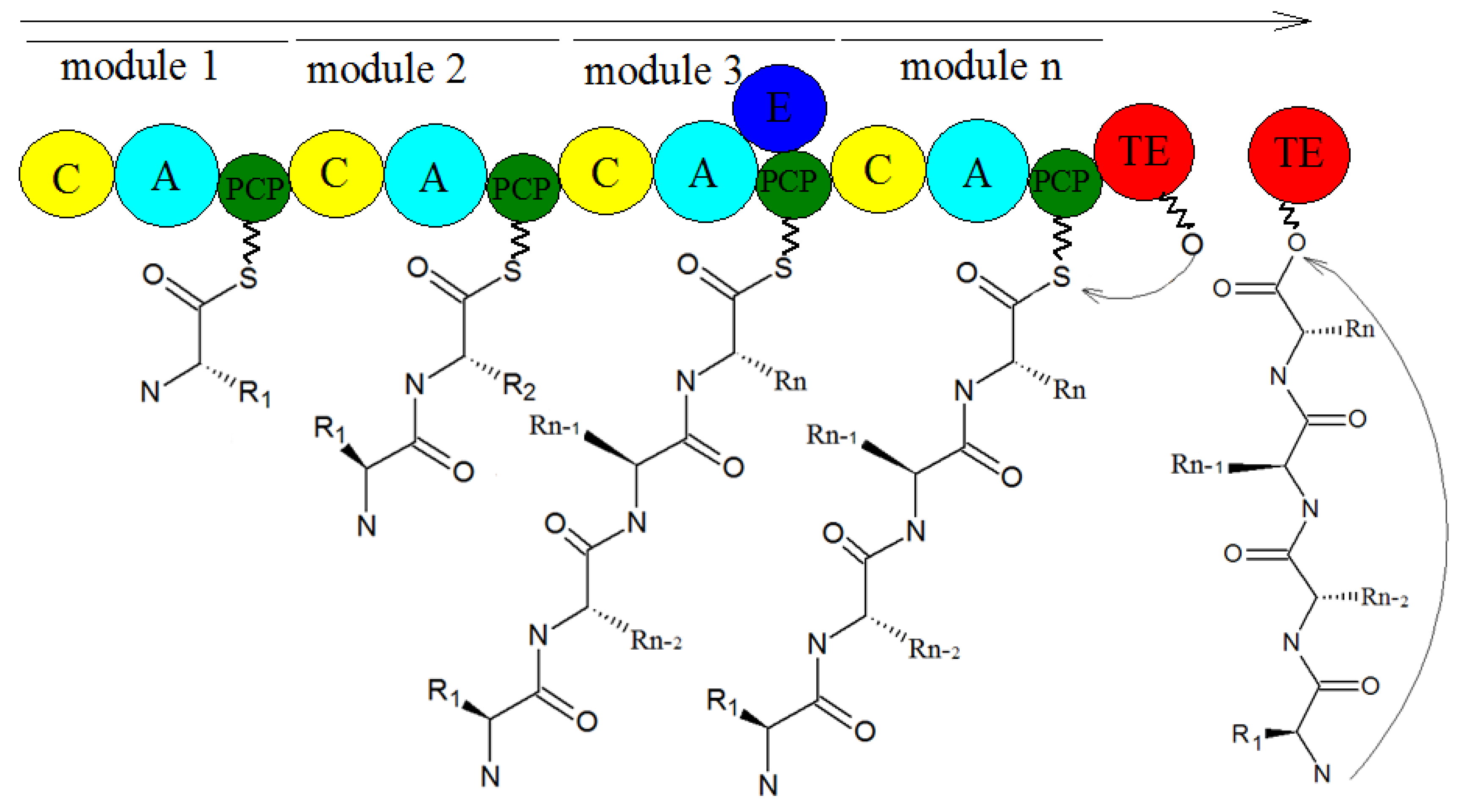
- Han, M.; Chen, J.; Qiao, Y.; Zhu, P. Advances in the nonribosomal peptide synthetases. Yaoxue Xuebao 2018, 53, 1080–1089. [Google Scholar] [CrossRef]
- Sung, C.T.; Chang, S.; Entwistle, R.; Ahn, G.; Lin, T.; Petrova, V.; Yeh, H.; Praseuth, M.B.; Chiang, Y.; Oakley, B.R.; et al. Overexpression of a three-gene conidial pigment biosynthetic pathway in Aspergillus nidulans reveals the first NRPS known to acetylate tryptophan. Fungal Genet. Biol. 2017, 101, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Payne, J.A.E.; Schoppet, M.; Hansen, M.H.; Cryle, M.J. Diversity of nature’s assembly lines—recent discoveries in non-ribosomal peptide synthesis. Mol. Biosyst. 2017, 13, 9–22. [Google Scholar] [CrossRef]
- Sun, Y.-H.; Deng, Z.-X. Polyketides and combinatorial biosynthetic approaches. Zhongguo Kangshengsu Zazhi 2006, 31, 6. [Google Scholar]
- Bloudoff, K.; Fage, C.D.; Marahiel, M.A.; Schmeing, T.M. Structural and mutational analysis of the nonribosomal peptide synthetase heterocyclization domain provides insight into catalysis. Proc. Natl. Acad. Sci. USA 2017, 114, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Strieker, M.; Tanovic, A.; Marahiel, M.A. Nonribosomal peptide synthetases: Structures and dynamics. Curr. Opin. Struct. Biol. 2010, 20, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Hertweck, C. The Biosynthetic Logic of Polyketide Diversity. Angew. Chem. Int. Ed. 2009, 48, 4688–4716. [Google Scholar] [CrossRef] [PubMed]
- Schaeberle, T.F. Biosynthesis of alpha-pyrones. Beilstein J. Org. Chem. 2016, 12, 571–588. [Google Scholar] [CrossRef]
- Sun, J.; Bu, J.; Cui, G.; Ma, Y.; Zhao, H.; Mao, Y.; Zeng, W.; Guo, J.; Huang, L. Accumulation and biosynthetic of curcuminoids and terpenoids in turmeric rhizome in different development periods. Zhongguo Zhong Yao Za Zhi 2019, 44, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Hocquette, A.; Grondin, M.; Bertout, S.; Mallié, M. Les champignons des genres Acremonium, Beauveria, Chrysosporium, Fusarium, Onychocola, Paecilomyces, Penicillium, Scedosporium et Scopulariopsis responsables de hyalohyphomycoses. J. De Mycol. Médicale 2005, 15, 136–149. [Google Scholar] [CrossRef]
- Okhravi, N.; Lightman, S. Clinical manifestations, treatment and outcome of Paecilomyces lilacinus infections. Clin. Microbiol. Infect. 2007, 13, 554. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.; Hu, Q. Secondary Metabolites of Purpureocillium lilacinum. Molecules 2022, 27, 18. https://doi.org/10.3390/molecules27010018
Chen W, Hu Q. Secondary Metabolites of Purpureocillium lilacinum. Molecules. 2022; 27(1):18. https://doi.org/10.3390/molecules27010018
Chicago/Turabian StyleChen, Wei, and Qiongbo Hu. 2022. "Secondary Metabolites of Purpureocillium lilacinum" Molecules 27, no. 1: 18. https://doi.org/10.3390/molecules27010018
APA StyleChen, W., & Hu, Q. (2022). Secondary Metabolites of Purpureocillium lilacinum. Molecules, 27(1), 18. https://doi.org/10.3390/molecules27010018






