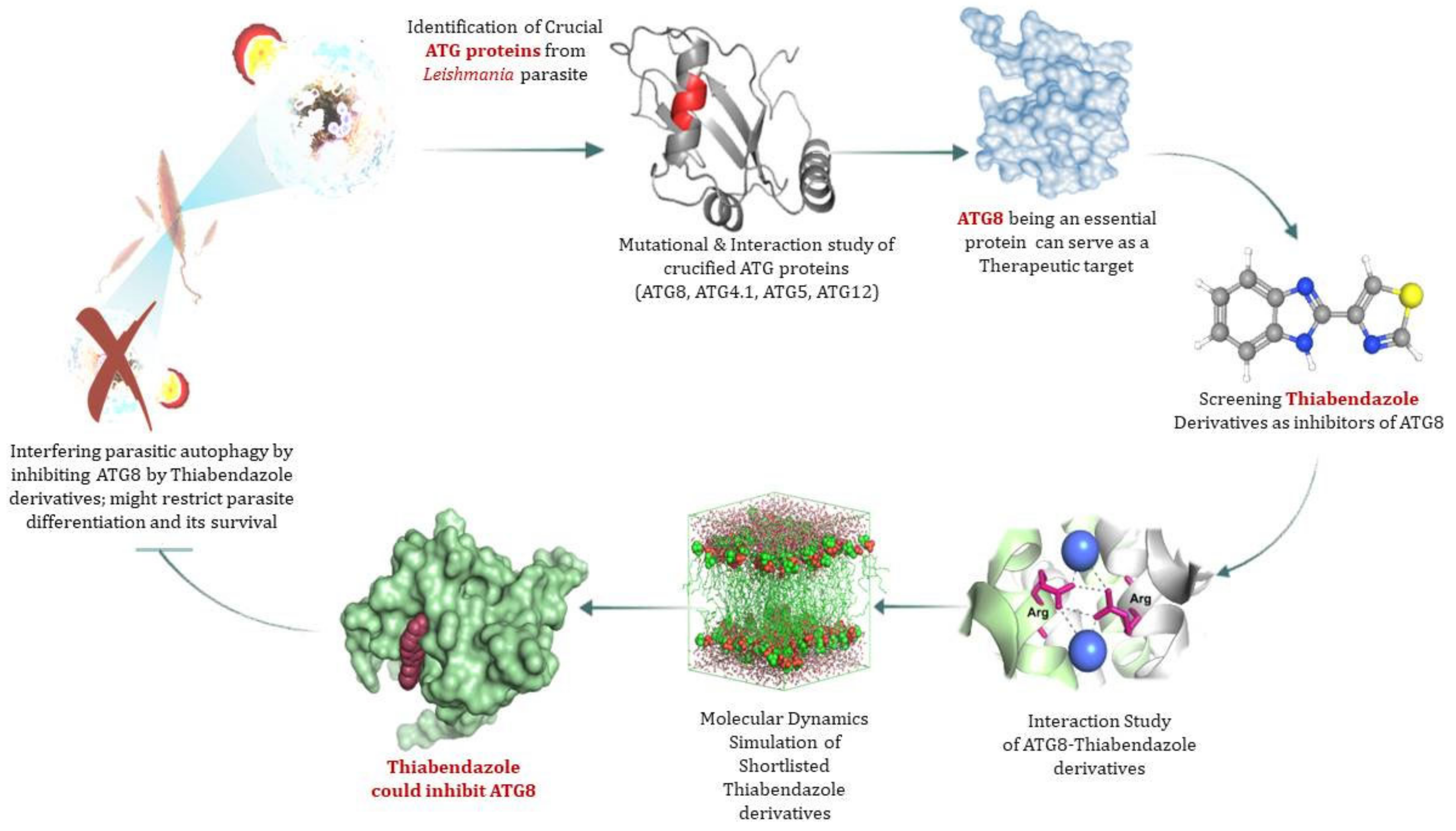
Infection Dynamics of ATG8 in Leishmania: Balancing Autophagy for Therapeutics
Abstract
:1. Introduction
2. Methodology
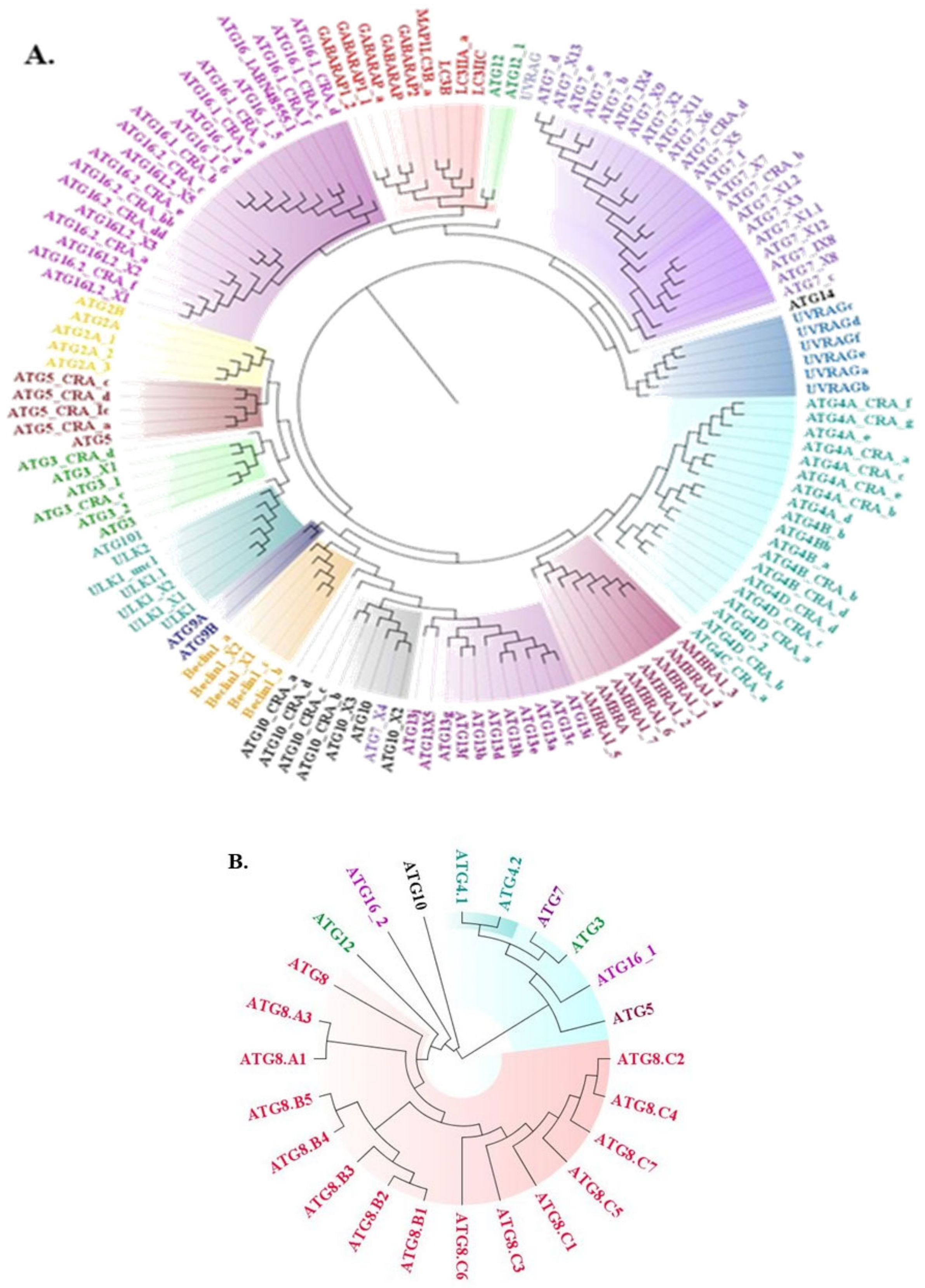
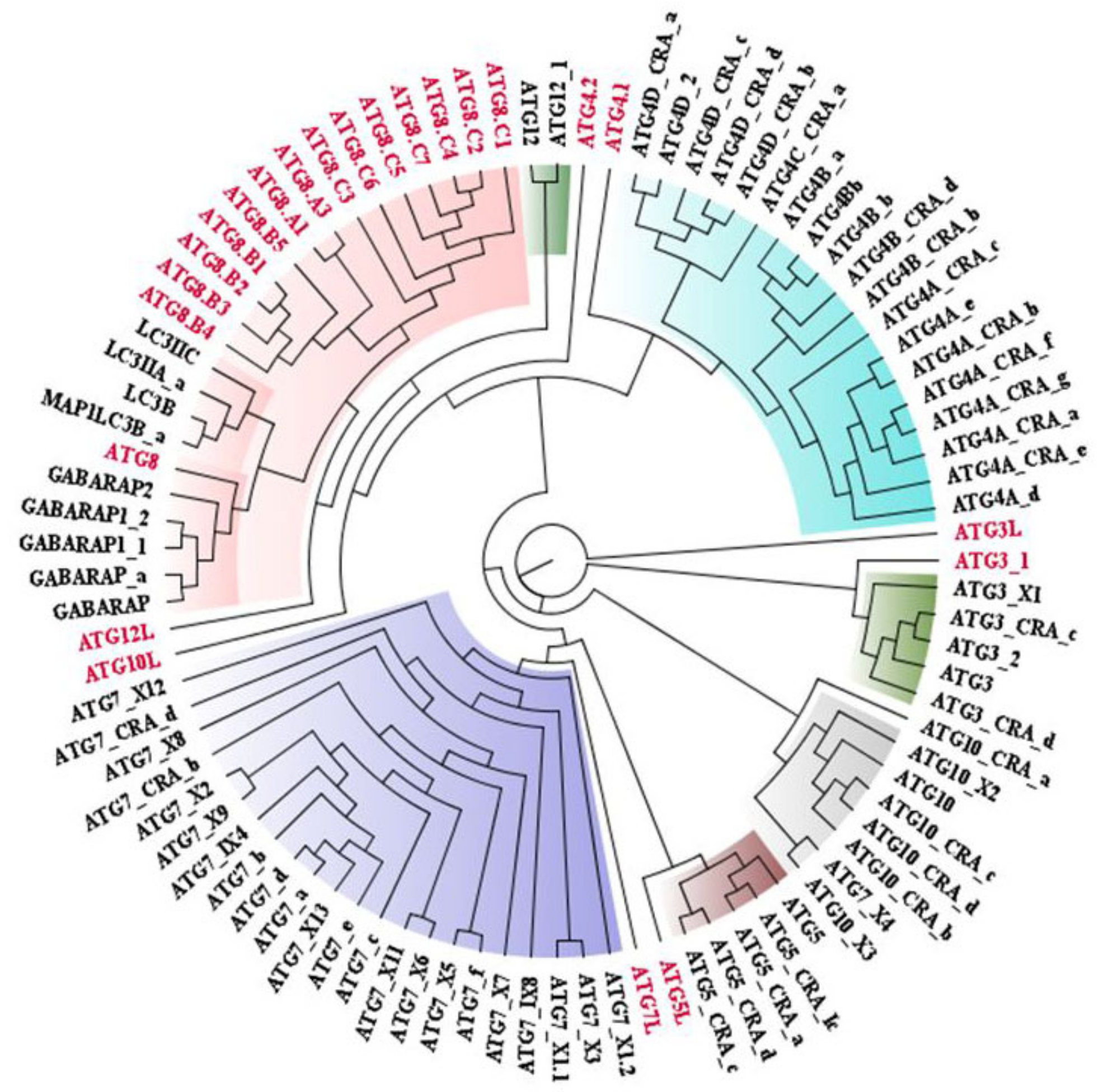
2.1. Phylogenetic Tree Construction for Sequential Analysis
2.1.1. Sequence Acquisition
2.1.2. Multiple Sequence Alignment (MSA)
2.1.3. Phylogenetic Tree Construction
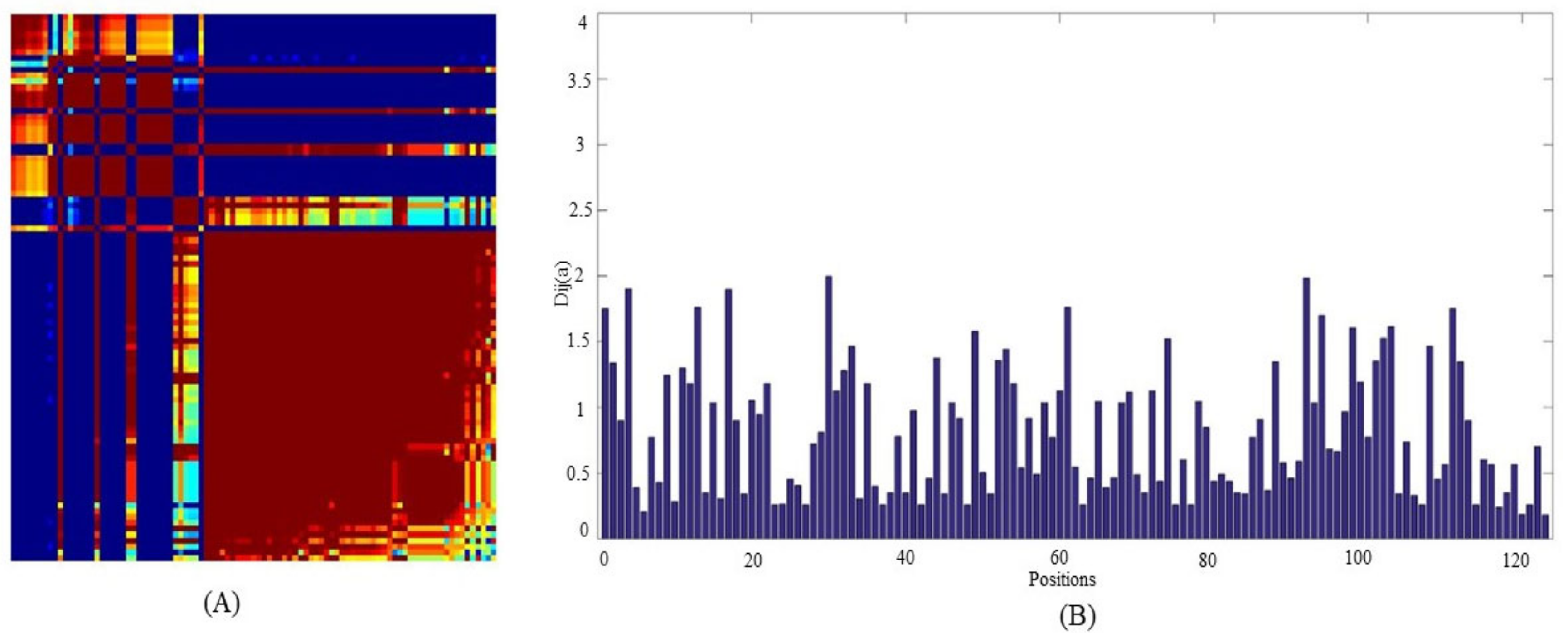
2.2. Statistical Coupling Analysis (SCA) for Positional Conservedness Analysis
2.3. Structural and Functional Residue Validation
2.4. Effect of Conserved Amino Acid Substitution on Protein
2.4.1. SDM (Site Directed Mutator)
2.4.2. PROVEAN (Protein Variation Effect Analyzer)
2.4.3. SODA
2.4.4. Arpeggio
2.5. Structure Prediction, Validation, and Interaction studies
2.6. Virtual Screening
2.7. Molecular Docking
2.8. Molecular Dynamics Simulation
3. Results
3.1. Conservedness Analysis of ATG Proteins
3.1.1. Sequential Conservedness Analysis
3.1.2. Positional Conservedness Analysis
3.2. Structural and Functional Residue Validation
3.3. Effect of Conserved Amino Acid Substitution on Protein
3.4. Computational Structure Prediction, Validation and Molecular Dynamics Simulation
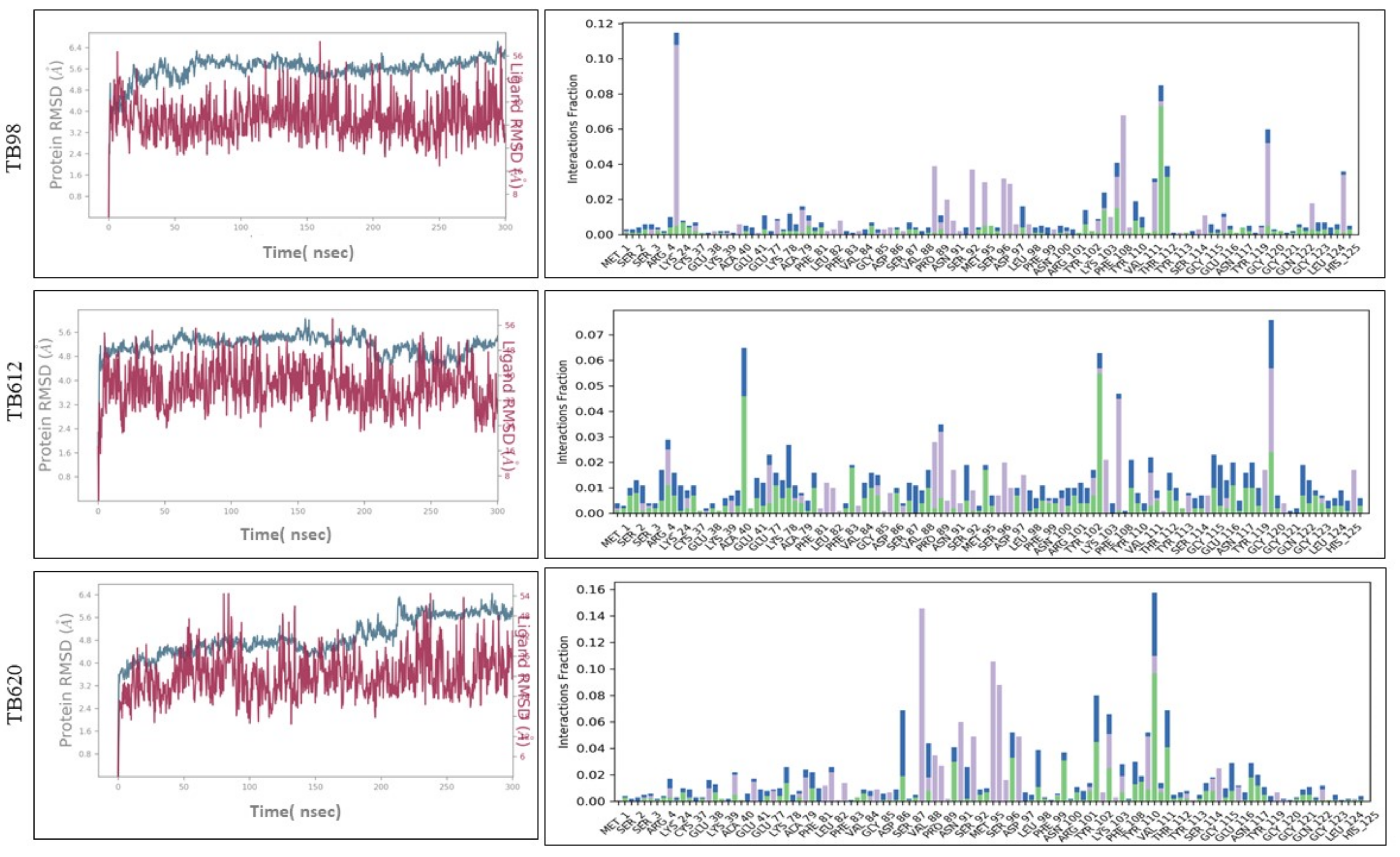
3.5. Molecular Dynamics Simulation of Proteins
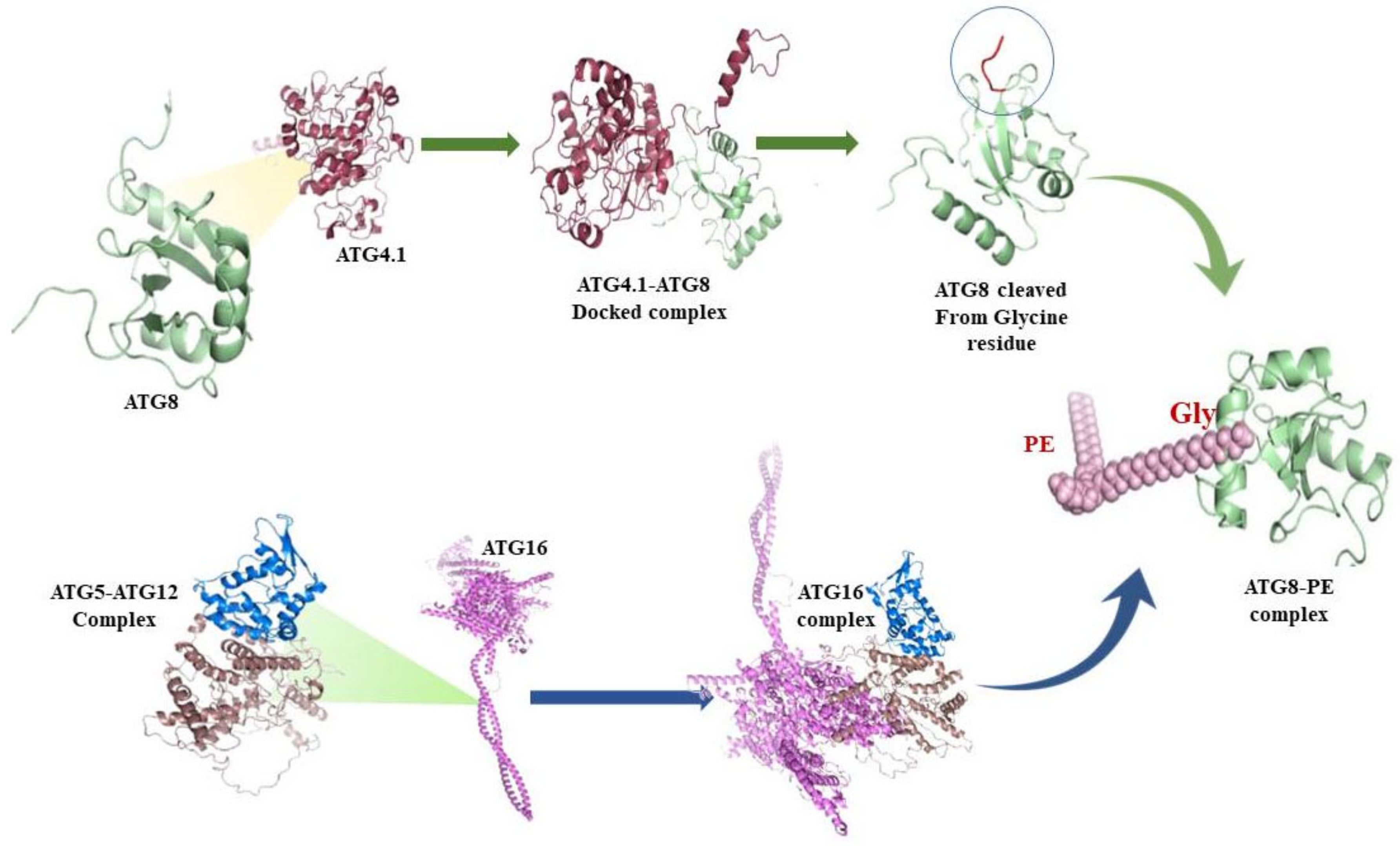
3.6. Protein-Protein interactions
3.7. ATG8 Can Be a Novel Target for Therapeutics against Leishmaniasis
3.8. Virtual Screening of Thiabendazole Derivatives against ATG8
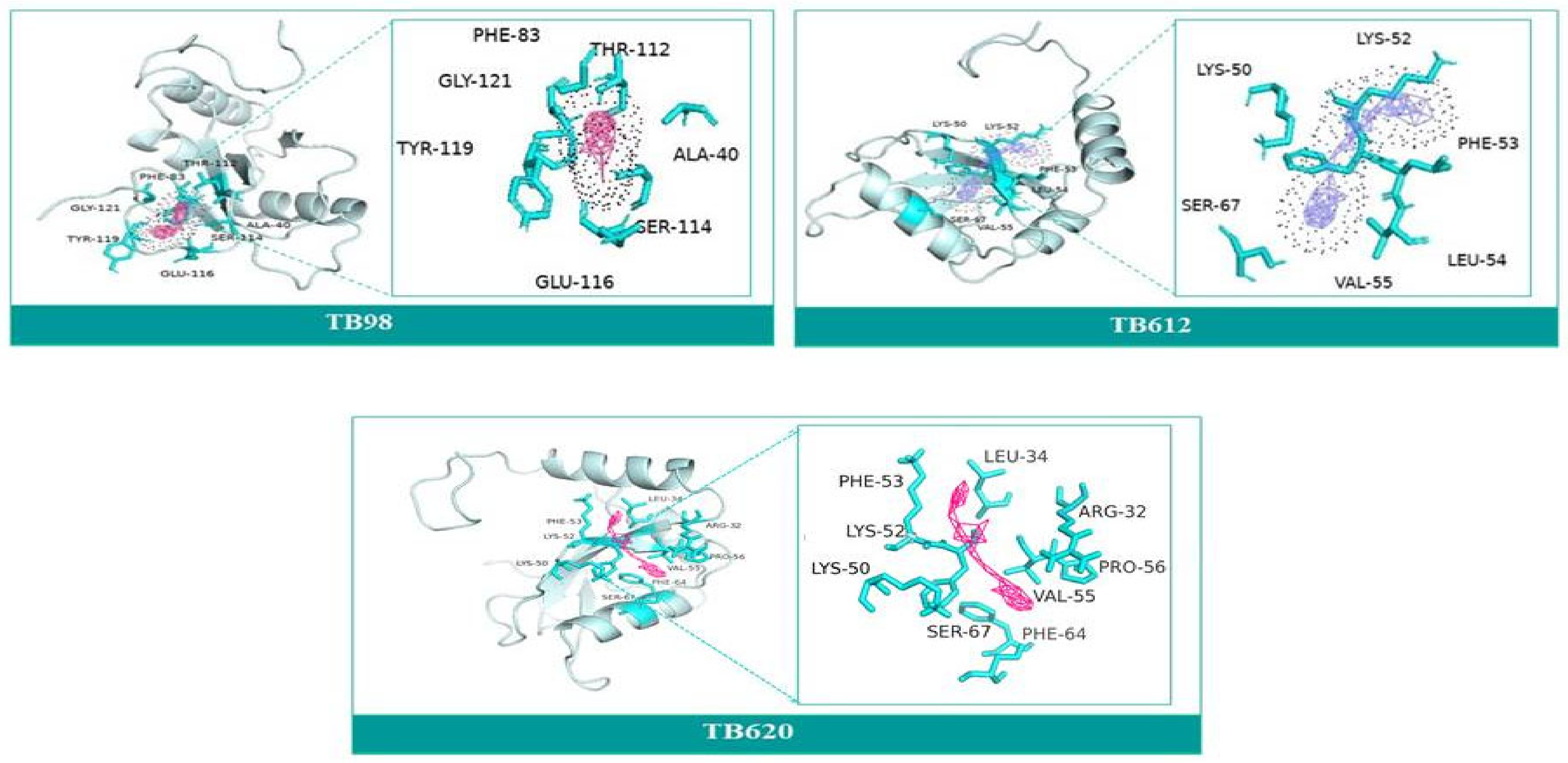
3.9. Molecular Docking and Molecular Dynamics Simulation (MDS)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| ATG | Autophagy Related Proteins |
| ULK1 | unc-51-like kinase 1 |
| PI3K | phosphatidylinositol (PI) 3-kinase complex |
| VPS | Vacuolar Protein Sorting |
| WIPI | WD repeat domain phosphoinositide interacting protein 2 |
| DFCP1 | Double FYVE containing protein 1 |
| LC3 | Microtubule-associated proteins 1A/1B light chain 3 |
| SCA | Statistical Coupling Analysis |
| SDM | Site Directed Mutator |
| PROVEAN | Protein Variation Effect Analyzer |
| SCA | Statistical Coupling Analysis |
| PE | Phosphatidylethanolamine |
References
- Read, A.; Hurwitz, I.; Durvasula, R. Leishmaniasis: An update on a neglected tropical disease. In Dynamic Models of Infectious Diseases; Springer: New York, NY, USA, 2013; pp. 95–138. [Google Scholar]
- Alvar, J.; Yactayo, S.; Bern, C. Leishmaniasis and poverty. Trends Parasitol. 2006, 22, 552–557. [Google Scholar] [CrossRef] [PubMed]
- Croft, S.L.; Sundar, S.; Fairlamb, A.H. Drug resistance in leishmaniasis. Clin. Microbiol. Rev. 2006, 19, 111–126. [Google Scholar] [CrossRef] [Green Version]
- Egerton, J.R. The ovicidal and larvicidal effect of thiabendazole on various helminth species. Tex. Rep. Biol. Med. 1969, 27 (Suppl. S2), 561–580. [Google Scholar]
- Bouazizi-Ben Messaoud, H.; Guichard, M.; Lawton, P.; Delton, I.; Azzouz-Maache, S. Changes in lipid and fatty acid composition during intramacrophagic transformation of Leishmania donovani complex promastigotes into amastigotes. Lipids 2017, 52, 433–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef]
- Veras, P.S.T.; De Menezes, J.P.B.; Dias, B.R.S. Deciphering the role played by autophagy in Leishmania infection. Front. Immunol. 2019, 10, 2523. [Google Scholar] [CrossRef]
- Zachari, M.; Ganley, I.G. The mammalian ULK1 complex and autophagy initiation. Essays Biochem. 2017, 61, 585–596. [Google Scholar]
- Kotani, T.; Kirisako, H.; Koizumi, M.; Ohsumi, Y.; Nakatogawa, H. The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc. Natl. Acad. Sci. USA 2018, 115, 10363–10368. [Google Scholar] [CrossRef] [Green Version]
- Geng, J.; Baba, M.; Nair, U.; Klionsky, D.J. Quantitative analysis of autophagy-related protein stoichiometry by fluorescence microscopy. J. Cell Biol. 2008, 182, 129–140. [Google Scholar] [CrossRef]
- Romanov, J.; Walczak, M.; Ibiricu, I.; Schüchner, S.; Ogris, E.; Kraft, C.; Martens, S. Mechanism and functions of membrane binding by the Atg5–Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012, 31, 4304–4317. [Google Scholar] [CrossRef]
- Thomas, S.A.; Nandan, D.; Kass, J.; Reiner, N.E. Countervailing, time-dependent effects on host autophagy promote intracellular survival of Leishmania. J. Biol. Chem. 2018, 293, 2617–2630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Besteiro, S.; Williams, R.A.; Morrison, L.S.; Coombs, G.H.; Mottram, J.C. Endosome sorting and autophagy are essential for differentiation and virulence of Leishmania major. J. Biol. Chem. 2006, 281, 11384–11396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maruyama, T.; Noda, N.N. Autophagy-regulating protease Atg4: Structure, function, regulation and inhibition. J. Antibiot. 2018, 71, 72–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giri, S.; Shaha, C. Leishmania donovani parasite requires Atg8 protein for infectivity and survival under stress. Cell Death Dis. 2019, 10, 1–19. [Google Scholar] [CrossRef]
- Williams, R.A.; Tetley, L.; Mottram, J.C.; Coombs, G.H. Cysteine peptidases CPA and CPB are vital for autophagy and differentiation in Leishmania mexicana. Mol. Microbiol. 2006, 61, 655–674. [Google Scholar] [CrossRef]
- Williams, R.A.; Woods, K.L.; Juliano, L.; Mottram, J.C.; Coombs, G.H. Characterization of unusual families of ATG8-like proteins and ATG12 in the protozoan parasite Leishmania major. Autophagy 2009, 5, 159–172. [Google Scholar] [CrossRef] [Green Version]
- Tan, H.H.; Goh, C.L. Parasitic Skin Infections in the Elderly. Drugs Aging 2001, 18, 165–176. [Google Scholar] [CrossRef]
- Davidse, L.C.; Flach, W. Interaction of thiabendazole with fungal tubulin. Biochim. Biophys. Acta-Gen. Subj. 1978, 543, 82–90. [Google Scholar] [CrossRef]
- Jasra, N.; Sanyal, S.N.; Khera, S. Effect of thiabendazole and fenbendazole on glucose uptake and carbohydrate metabolism in Trichuris globulosa. Vet. Parasitol. 1990, 35, 201–209. [Google Scholar] [CrossRef]
- Vijayakumar, K.; Cho, G.W. Autophagy: An evolutionarily conserved process in the maintenance of stem cells and aging. Cell Biochem. Funct. 2019, 37, 452–458. [Google Scholar] [CrossRef]
- Büssow, K.; Hoffmann, S.; Sievert, V. ORFer–retrieval of protein sequences and open reading frames from GenBank and storage into relational databases or text files. BMC Bioinform. 2002, 3, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sievers, F.; Higgins, D.G. Clustal omega. Curr. Protoc. Bioinform. 2014, 48, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R. Interpreting the universal phylogenetic tree. Proc. Natl. Acad. Sci. USA 2000, 97, 8392–8396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Süel, G.M.; Lockless, S.W.; Wall, M.A.; Ranganathan, R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Biol. 2003, 10, 59–69. [Google Scholar] [CrossRef]
- Nichols, S.E.; Hernández, C.X.; Wang, Y.; McCammon, J.A. Structure-based network analysis of an evolved G protein-coupled receptor homodimer interface. Protein Sci. 2013, 22, 745–754. [Google Scholar] [CrossRef] [Green Version]
- Robert, M.; Sayed, S.; Aguayo, C.; Menon, R.; Channak, K.; Vander Valk, C.; Neely, C.; Tsou, T.; Mandeville, J.; Reed, J.H. OSSIE: Open Source SCA for Researchers. In Proceedings of the SDR Forum Technical Conference, Phoenix, AZ, USA, 15–18 November 2004; Volume 47. [Google Scholar]
- Glaser, F.; Pupko, T.; Paz, I.; Bell, R.E.; Bechor-Shental, D.; Martz, E.; Ben-Tal, N. ConSurf: Identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 2003, 19, 163–164. [Google Scholar] [CrossRef] [Green Version]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef] [Green Version]
- Worth, C.L.; Preissner, R.; Blundell, T.L. SDM—a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res. 2011, 39 (Suppl. S2), W215–W222. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef] [Green Version]
- Paladin, L.; Piovesan, D.; Tosatto, S.C. SODA: Prediction of protein solubility from disorder and aggregation propensity. Nucleic Acids Res. 2017, 45, W236–W240. [Google Scholar] [CrossRef] [PubMed]
- Jubb, H.C.; Higueruelo, A.P.; Ochoa-Montaño, B.; Pitt, W.R.; Ascher, D.B.; Blundell, T.L. Arpeggio: A web server for calculating and visualising interatomic interactions in protein structures. J. Mol. Biol. 2017, 429, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Eswar, N.; Eramian, D.; Webb, B.; Shen, M.Y.; Sali, A. Protein structure modeling with MODELLER. In Structural Proteomics; Humana Press: Totowa, NJ, USA, 2008; pp. 145–159. [Google Scholar]
- Kim, D.E.; Chivian, D.; Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004, 32 (Suppl. S2), W526–W531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, T.; Chen, L.; Shi, M.; Niu, J.; Zhang, X.; Yang, X.; Gao, J. Developing novel methods to image and visualize 3D genomes. Cell Biol. Toxicol. 2018, 34, 367–380. [Google Scholar] [CrossRef] [Green Version]
- De Vries, S.J.; Van Dijk, M.; Bonvin, A.M. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 2010, 5, 883–897. [Google Scholar] [CrossRef] [Green Version]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Vajda, S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef]
- Laskowski, R.A. PDBsum: Summaries and analyses of PDB structures. Nucleic Acids Res. 2001, 29, 221–222. [Google Scholar] [CrossRef]
- Song, W.J.; Sontz, P.A.; Ambroggio, X.I.; Tezcan, F.A. Metals in protein–protein interfaces. Annu. Rev. Biophys. 2014, 43, 409–431. [Google Scholar] [CrossRef]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Bryant, S.H. PubChem substance and compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef]
- Walters, W.P. Going further than Lipinski’s rule in drug design. Expert Opin. Drug Discov. 2012, 7, 99–107. [Google Scholar] [CrossRef]
- Ravi, L.; Kannabiran, K. A Handbook on Protein-Ligand Docking Tool: AutoDock 4. Innovare J. Med. Sci. 2016, 4, 28–33. [Google Scholar]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput.-Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, H.; Huang, W.; He, G.; Peng, C.; Wu, F.; Ouyang, L. Molecular dynamics simulation of tryptophan hydroxylase-1: Binding modes and free energy analysis to phenylalanine derivative inhibitors. Int. J. Mol. Sci. 2013, 14, 9947–9962. [Google Scholar] [CrossRef] [PubMed]
- Bowers, K.J.; Chow, D.E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL, USA, 11–17 November 2006; p. 43. [Google Scholar]
- Rubinsztein, D.C.; Codogno, P.; Levine, B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 2012, 11, 709–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cull, B.; Prado Godinho, J.L.; Fernandes Rodrigues, J.C.; Frank, B.; Schurigt, U.; Williams, R.A.; Coombs, G.H.; Mottram, J.C. Glycosome turnover in Leishmania major is mediated by autophagy. Autophagy 2014, 10, 2143–2157. [Google Scholar] [CrossRef] [Green Version]



| Amino Acid Substitution | ΔΔG | Stability | PROVEAN Score | Impact on Protein | SODA Score | Solubility | |
|---|---|---|---|---|---|---|---|
| ATG8 | R71T | −1.94 | Decrease | −5.825 | Deleterious | −14.878 | Less Soluble |
| P56E | −1.07 | Decrease | −7.694 | Deleterious | −2.13 | Less Soluble | |
| R18P | −3.81 | Decrease | −6.641 | Deleterious | −1.685 | Less Soluble | |
| ATG5 | P110C | 0.29 | Increase | −6.250 | Deleterious | −50.412 | Less Soluble |
| P89K | −1.04 | Decrease | −7.000 | Deleterious | −2.053 | Less Soluble | |
| D47P | −0.74 | Decrease | −7.000 | Deleterious | −2.279 | Less Soluble | |
| ATG12 | P41K | −1.04 | Decrease | −3.098 | Deleterious | −60.883 | Less Soluble |
| P152N | 0.91 | Increase | −6.968 | Deleterious | −13.018 | Less Soluble | |
| E178T | −2.00 | Decrease | −5.600 | Deleterious | −15.209 | Less Soluble | |
| N179R | −1.99 | Decrease | −5.200 | Deleterious | −1.375 | Less Soluble | |
| ATG4.1 | D133T | 0.03 | Increase | −6.980 | Deleterious | −12.363 | Less Soluble |
| P213G | −1.13 | Decrease | −4.467 | Deleterious | −213.32 | Less Soluble | |
| G244I | −2.29 | Decrease | −10.00 | Deleterious | −17.447 | Less Soluble | |
| P265K | −0.88 | Decrease | −7.987 | Deleterious | −11.794 | Less Soluble |
| ATG4.1 | ATG8 | Interactions | |
|---|---|---|---|
| Interacting Residues | Arg276 | Val88 | Hydrogen Bond |
| Arg276 | Pro89 | Hydrogen Bond | |
| Arg73 | Glu116 | Hydrogen Bond and Salt Bridge | |
| Arg73 | Asn117 | Hydrogen Bond | |
| Cys75 | Gly120 | Hydrogen Bond | |
| ATG5 | ATG12 | Interactions | |
| Interacting Residues | Glu125 | Arg186 | Hydrogen Bond and Salt Bridge |
| Pro269 | Lys29 | Hydrogen Bond | |
| Lys131 | Glu191 | Hydrogen Bond and Salt Bridge | |
| Arg146 | Glu191 | Hydrogen Bond and Salt Bridge | |
| Arg146 | Glu45 | Hydrogen Bond and Salt Bridge | |
| Glu128 | Asp184 | Hydrogen Bond | |
| Lys127 | Glu22 | Hydrogen Bond and Salt Bridge | |
| Tyr115 | Gly182 | Hydrogen Bond | |
| Gln175 | His80 | Hydrogen Bond | |
| Gln175 | Arg79 | Hydrogen Bond |
| Derivatives | IUPAC Name | Structure of Derivative | Binding Energy | Molecular Weight | LogP | H-Donor | H-Acceptor |
|---|---|---|---|---|---|---|---|
| TB98 | N-(1H-Benzimidazol-2-yl)-4,5,6,7-tetrahydro-[1,3]thiazolo[5,4-c]pyridin-2-amine |  | −8.1 | 271.34 | 2.2 | 3 | 5 |
| TB612 | 1-[3-(1H-Benzimidazol-2-yl)propyl]-2-methyl-3-[[4-(trifluoromethyl)-1,3-thiazol-2-yl]methyl]guanidine | 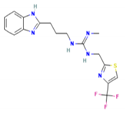 | −8.4 | 396.4 | 2.9 | 3 | 7 |
| TB620 | 2-[3-(1H-Benzimidazol-2-yl)propyl]-1-ethyl-3-[2-[4-(trifluoromethyl)-1,3-thiazol-2-yl]ethyl]guanidine | 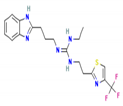 | −8.6 | 424.5 | 3.7 | 3 | 7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guhe, V.; Anjum, F.; Shafie, A.; Hassan, M.I.; Pasupuleti, V.R.; Singh, S. Infection Dynamics of ATG8 in Leishmania: Balancing Autophagy for Therapeutics. Molecules 2022, 27, 3142. https://doi.org/10.3390/molecules27103142
Guhe V, Anjum F, Shafie A, Hassan MI, Pasupuleti VR, Singh S. Infection Dynamics of ATG8 in Leishmania: Balancing Autophagy for Therapeutics. Molecules. 2022; 27(10):3142. https://doi.org/10.3390/molecules27103142
Chicago/Turabian StyleGuhe, Vrushali, Farah Anjum, Alaa Shafie, Md Imtaiyaz Hassan, Visweswara Rao Pasupuleti, and Shailza Singh. 2022. "Infection Dynamics of ATG8 in Leishmania: Balancing Autophagy for Therapeutics" Molecules 27, no. 10: 3142. https://doi.org/10.3390/molecules27103142







