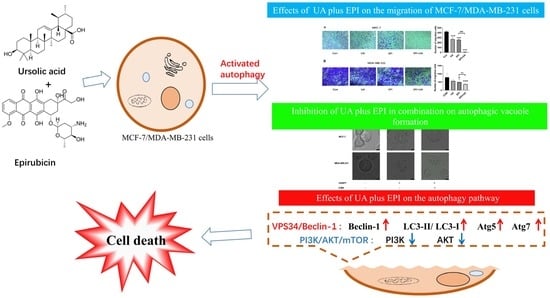
Ursolic Acid Enhances the Sensitivity of MCF-7 and MDA-MB-231 Cells to Epirubicin by Modulating the Autophagy Pathway
Abstract
:1. Introduction
2. Results
2.1. Effects of UA and EPI on the Viability of MCF-7/MDA-MB-231 Cells
2.2. Effect of UA on the Sensitivity of MCF-7/MDA-MB-231 Cells to EPI
2.3. Effect of UA and EPI on the Migration of MCF-7/MDA-MB-231 Cells
2.4. Autophagy Is Involved in the Treatment of UA plus EPI in Combination in MCF-7/MDA-MB-231 Cells
2.5. UA Promotes EPI-Sensitivity of MCF-7/MDA-MB-231 Cells by Modulating the Autophagy Pathway
3. Discussion
4. Materials and Methods
4.1. Reagents and Instruments
4.2. Cell Culture
4.3. Cell Viability Assessment
4.4. Caspase 3 Activity Assay
4.5. Transwell Assay
4.6. The Effects of the Combination of EPI with UA on Autophagy in MCF-7 and MDA-MB-231 Cells
4.7. Quantification of Monodansylcadaverine Cell Labeling
4.8. Western Blot Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, P.; Du, Y.; Wang, J. Indentification of breast cancer subtypes sensitive to HCQ-induced autophagy inhibition. Pathol. Res. Pract. 2019, 215, 152609. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.M.; Yusuf, S.W.; Ewer, M.S. Anthracycline-induced cardiotoxicity and the cardiac-sparing effect of liposomal formulation. Int. J. Nanomed. 2007, 2, 567–583. [Google Scholar]
- Marinello, J.; Delcuratolo, M.; Capranico, G. Anthracyclines as Topoisomerase II Poisons: From Early Studies to New Perspectives. Int. J. Mol. Sci. 2018, 19, 3480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, W.-L.; Chen, J.; Wang, Y.-P.; Zheng, H. Autophagy protects breast cancer cells from epirubicin-induced apoptosis and facilitates epirubicin-resistance development. Autophagy 2011, 7, 1035–1044. [Google Scholar] [CrossRef] [Green Version]
- Klionsky, D.J.; Emr, S.D. Autophagy as a Regulated Pathway of Cellular Degradation. Science 2000, 290, 1717–1721. [Google Scholar] [CrossRef]
- Gallagher, L.E.; Williamson, L.E.; Chan, E.Y.W. Advances in Autophagy Regulatory Mechanisms. Cells 2016, 5, 24. [Google Scholar] [CrossRef]
- Liang, X.H.; Jackson, S.; Seaman, M.; Brown, K.; Kempkes, B.; Hibshoosh, H.; Levine, B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999, 402, 672–676. [Google Scholar] [CrossRef]
- Moretti, L.; Yang, E.S.; Kim, K.W.; Lu, B. Autophagy signaling in cancer and its potential as novel target to improve anticancer therapy. Drug. Resist. Update 2007, 10, 135–143. [Google Scholar] [CrossRef]
- Pooladanda, V.; Bandi, S.; Mondi, S.R.; Gottumukkala, K.M.; Godugu, C. Nimbolide epigenetically regulates autophagy and apoptosis in breast cancer. Toxicol. In Vitro 2018, 51, 114–128. [Google Scholar] [CrossRef]
- Rejhová, A.; Opattová, A.; Čumová, A.; Slíva, D.; Vodička, P. Natural compounds and combination therapy in colorectal cancer treatment. Eur. J. Med. Chem. 2018, 144, 582–594. [Google Scholar] [CrossRef]
- Liu, H.C.; Chiang, C.C.; Lin, C.H.; Chen, C.S.; Wei, C.W.; Lin, S.Y.; Yiang, G.T.; Yu, Y.L. Anti-cancer therapeutic benefit of red guava extracts as a potential therapy in combination with doxorubicin or targeted therapy for triple-negative breast cancer cells. Int. J. Med. Sci. 2020, 17, 1015–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, E.W.C.; Soon, C.Y.; Tan, J.B.L.; Wong, S.K.; Hui, Y.W. Ursolic acid: An overview on its cytotoxic activities against breast and colorectal cancer cells. J. Integr. Med. 2019, 17, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Navin, R.; Kim, S.M. Therapeutic interventions using ursolic acid for cancer treatment. Med. Chem. 2016, 6, 339–344. [Google Scholar] [CrossRef] [Green Version]
- Shanmugam, M.K.; Dai, X.; Kumar, A.P.; Tan, B.K.H.; Sethi, G.; Bishayee, A. Ursolic acid in cancer prevention and treatment: Molecular targets, pharmacokinetics and clinical studies. Biochem. Pharmacol. 2013, 85, 1579–1587. [Google Scholar] [CrossRef] [Green Version]
- Lewinska, A.; Adamczyk-Grochala, J.; Kwasniewicz, E.; Deregowska, A.; Wnuk, M. Ursolic acid-mediated changes in glycolytic pathway promote cytotoxic autophagy and apoptosis in phenotypically different breast cancer cells. Apoptosis 2017, 22, 800–815. [Google Scholar] [CrossRef] [Green Version]
- Kondo, Y.; Kanzawa, T.; Sawaya, R.; Kondo, S. The role of autophagy in cancer development and response to therapy. Nat. Rev. Cancer 2005, 5, 726–734. [Google Scholar] [CrossRef]
- Pan, Z.; Xie, Y.; Bai, J.; Lin, Q.; Cui, X.; Zhang, N. Bufalin suppresses colorectal cancer cell growth through promoting autophagy in vivo and in vitro. RSC Adv. 2018, 8, 38910–38918. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Na, J.; Lee, M.S.; Cha, E.Y.; Sul, J.Y.; Park, J.B.; Lee, J.S. Combination of pristimerin and paclitaxel additively induces autophagy in human breast cancer cells via ERK1/2 regulation. Mol. Med. Rep. 2018, 18, 4281–4288. [Google Scholar] [CrossRef]
- Raje, S.; Pandav, K.; Barthwal, R. Dual mode of binding of anti cancer drug epirubicin to G-quadruplex [d-(TTAGGGT)]4 containing human telomeric DNA sequence induces thermal stabilization. Bioorg. Med. Chem. 2019, 27, 115131. [Google Scholar] [CrossRef]
- Khasraw, M.; Bell, R.; Dang, C. Epirubicin: Is it like doxorubicin in breast cancer? A clinical review. Breast 2012, 21, 142–149. [Google Scholar] [CrossRef]
- Uzoigwe, J.; Sauter, E.R. Cancer prevention and treatment using combination therapy with plant- and animal-derived compounds. Expert Rev. Clin. Pharmacol. 2012, 5, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Yu, Y.; Xu, C.; Xiong, H.; Yang, S.; Yao, J. LMWH and its derivatives represent new rational for cancer therapy: Construction strategies and combination therapy. Drug Discov. Today 2019, 24, 2096–2104. [Google Scholar] [CrossRef] [PubMed]
- Aires, V.; Limagne, E.; Cotte, A.K.; Latruffe, N.; Ghiringhelli, F.; Delmas, D. Resveratrol metabolites inhibit human metastatic colon cancer cells progression and synergize with chemotherapeutic drugs to induce cell death. Mol. Nutr. Food Res. 2013, 57, 1170–1181. [Google Scholar] [CrossRef] [PubMed]
- Colla, A.R.S.; Rosa, J.M.; Cunha, M.P.; Rodrigues, A.L.S. Anxiolytic-like effects of ursolic acid in mice. Eur. J. Pharmacol. 2015, 758, 171–176. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Zhang, X.; Cui, L.; Wang, L.; Liu, H.; Ji, H.; Du, Y. Ursolic acid promotes the neuroprotection by activating Nrf2 pathway after cerebral ischemia in mice. Brain Res. 2013, 1497, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Xavier, C.P.R.; Lima, C.F.; Pedro, D.F.N.; Wilson, J.M.; Kristiansen, K.; Pereira-Wilson, C. Ursolic acid induces cell death and modulates autophagy through JNK pathway in apoptosis-resistant colorectal cancer cells. J. Nutr. Biochem. 2013, 24, 706–712. [Google Scholar] [CrossRef] [Green Version]
- Zhi, X.; Zhong, Q. Autophagy in cancer. F1000prime Rep. 2015, 7, 18. [Google Scholar] [CrossRef] [Green Version]
- Bhat, P.; Kriel, J.; Priya, B.S.; Basappa; Shivananju, N.S.; Loss, B. Modulating autophagy in cancer therapy: Advancements and challenges for cancer cell death sensitization. Biochem. Pharmacol. 2018, 147, 170–182. [Google Scholar] [CrossRef]
- Ma, Y. Chapter 13—Role of Autophagy in Cancer Therapy. In Autophagy: Cancer, Other Pathologies, Inflammation, Immunity, Infection, and Aging; Hayat, M.A., Ed.; Academic Press: San Diego, CA, USA, 2016; pp. 231–251. [Google Scholar]
- Yang, K.L.; Khoo, B.Y.; Ong, M.T.; Yoong, I.C.K.; Sreeramanan, S. In vitro anti-breast cancer studies of LED red light therapy through autophagy. Breast Cancer 2021, 28, 60–66. [Google Scholar] [CrossRef]
- Funderburk, S.F.; Wang, Q.J.; Yue, Z. The Beclin 1–VPS34 complex—At the crossroads of autophagy and beyond. Trends Cell Biol. 2010, 20, 355–362. [Google Scholar] [CrossRef] [Green Version]
- Leng, S.; Hao, Y.; Du, D.; Xie, S.; Hong, L.; Gu, H.; Zhu, X.; Zhang, J.; Fan, D.; Kung, H.-F. Ursolic acid promotes cancer cell death by inducing Atg5-dependent autophagy. Int. J. Cancer 2013, 133, 2781–2790. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Wang, W.; Lu, L.; Wang, L.; Chen, X.; Guo, R.; Li, S.; Jiang, J. Inactivation of Beclin-1-dependent autophagy promotes ursolic acid-induced apoptosis in hypertrophic scar fibroblasts. Exp. Derm. 2018, 27, 58–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, J.F.; Shi, Z.M.; Zou, J.; Li, X.L. Inhibition of PI3K/AKT/mTOR signaling pathway promotes autophagy of articular chondrocytes and attenuates inflammatory response in rats with osteoarthritis. Biomed. Pharmacother. 2017, 89, 1252–1261. [Google Scholar] [CrossRef]
- Chang, L.; Graham, P.H.; Hao, J.; Ni, J.; Bucci, J.; Cozzi, P.J.; Kearsley, J.H.; Li, Y. PI3K/Akt/mTOR pathway inhibitors enhance radiosensitivity in radioresistant prostate cancer cells through inducing apoptosis, reducing autophagy, suppressing NHEJ and HR repair pathways. Cell Death Dis. 2014, 5, e1437. [Google Scholar] [CrossRef]
- Deng, F.; Ma, Y.X.; Liang, L.; Zhang, P.; Feng, J. The pro-apoptosis effect of sinomenine in renal carcinoma via inducing autophagy through inactivating PI3K/AKT/mTOR pathway. Biomed. Pharmacother. 2018, 97, 1269–1274. [Google Scholar] [CrossRef]
- Meng, Y.; Lin, Z.-M.; Ge, N.; Zhang, D.-L.; Huang, J.; Kong, F. Ursolic Acid Induces Apoptosis of Prostate Cancer Cells via the PI3K/Akt/mTOR Pathway. Am. J. Chin. Med. 2015, 43, 1471–1486. [Google Scholar] [CrossRef]
- Singh, S.S.; Vats, S.; Chia, A.Y.Q.; Tan, T.Z.; Deng, S.; Ong, M.S.; Arfuso, F.; Yap, C.T.; Goh, B.C.; Sethi, G.; et al. Dual role of autophagy in hallmarks of cancer. Oncogene 2018, 37, 1142–1158. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Shen, Z. An optimized in vitro assay for screening compounds that stimulate liver cell glucose utilization with low cytotoxicity. J. Pharmacol. Toxicol. Met. 2007, 56, 58–62. [Google Scholar] [CrossRef]







| Agents | Concentration (µg/mL)/Cell Viability (%) | ||
|---|---|---|---|
| UA (0 μM) | UA (5 μM) | UA (10 μM) | |
| EPI (0 μM) | 100 ± 5.84 | 98.91 ± 6.79 | 93.78 ± 4.53 |
| EPI (0.5 μM) | 92.83 ± 2.37 | 79.02 ± 4.54 *# | 73.76 ± 4.74 **#∆ |
| EPI (1 μM) | 83.40 ± 0.83 | 60.10 ± 1.13 ***###∆∆ | 44.19 ± 3.14 ***###∆∆∆ |
| EPI (5 μM) | 68.67 ± 5.69 *** | 43.63 ± 2.82 ***###∆∆ | 34.85 ± 5.90 ***###∆∆∆ |
| Agents | Concentration (µg/mL)/Cell Viability (%) | ||
|---|---|---|---|
| UA (0 μM) | UA (10 μM) | UA (20 μM) | |
| EPI (0 μM) | 100 ± 1.40 | 97.50 ± 4.23 | 90.12 ± 3.70 |
| EPI (1 μM) | 91.24 ± 4.26 | 63.12 ± 4.91 ***###∆∆∆ | 53.68 ± 4.70 ***###∆∆∆ |
| EPI (5 μM) | 63.80 ± 1.48 *** | 44.49 ± 3.99 ***###∆∆ | 34.49 ± 1.24 ***###∆∆∆ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Zhang, P.; Jiang, H.; Sun, B.; Luo, H.; Jia, A. Ursolic Acid Enhances the Sensitivity of MCF-7 and MDA-MB-231 Cells to Epirubicin by Modulating the Autophagy Pathway. Molecules 2022, 27, 3399. https://doi.org/10.3390/molecules27113399
Wang Z, Zhang P, Jiang H, Sun B, Luo H, Jia A. Ursolic Acid Enhances the Sensitivity of MCF-7 and MDA-MB-231 Cells to Epirubicin by Modulating the Autophagy Pathway. Molecules. 2022; 27(11):3399. https://doi.org/10.3390/molecules27113399
Chicago/Turabian StyleWang, Zhennan, Pingping Zhang, Huan Jiang, Bing Sun, Huaizhi Luo, and Aiqun Jia. 2022. "Ursolic Acid Enhances the Sensitivity of MCF-7 and MDA-MB-231 Cells to Epirubicin by Modulating the Autophagy Pathway" Molecules 27, no. 11: 3399. https://doi.org/10.3390/molecules27113399
APA StyleWang, Z., Zhang, P., Jiang, H., Sun, B., Luo, H., & Jia, A. (2022). Ursolic Acid Enhances the Sensitivity of MCF-7 and MDA-MB-231 Cells to Epirubicin by Modulating the Autophagy Pathway. Molecules, 27(11), 3399. https://doi.org/10.3390/molecules27113399