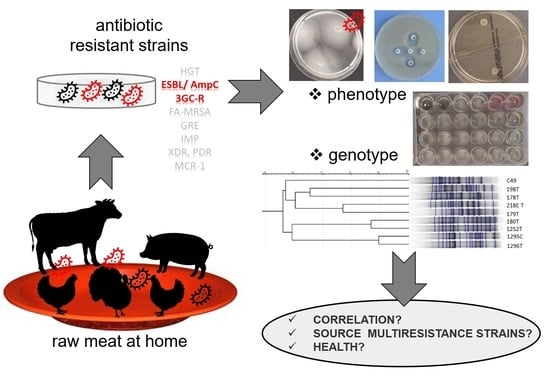
Raw Meat Contaminated with Cephalosporin-Resistant Enterobacterales as a Potential Source of Human Home Exposure to Multidrug-Resistant Bacteria
Abstract
:1. Introduction
2. Results
2.1. Most of the Third-Generation Cephalosporine-Resistant Strains Were Isolated from Turkey Meat
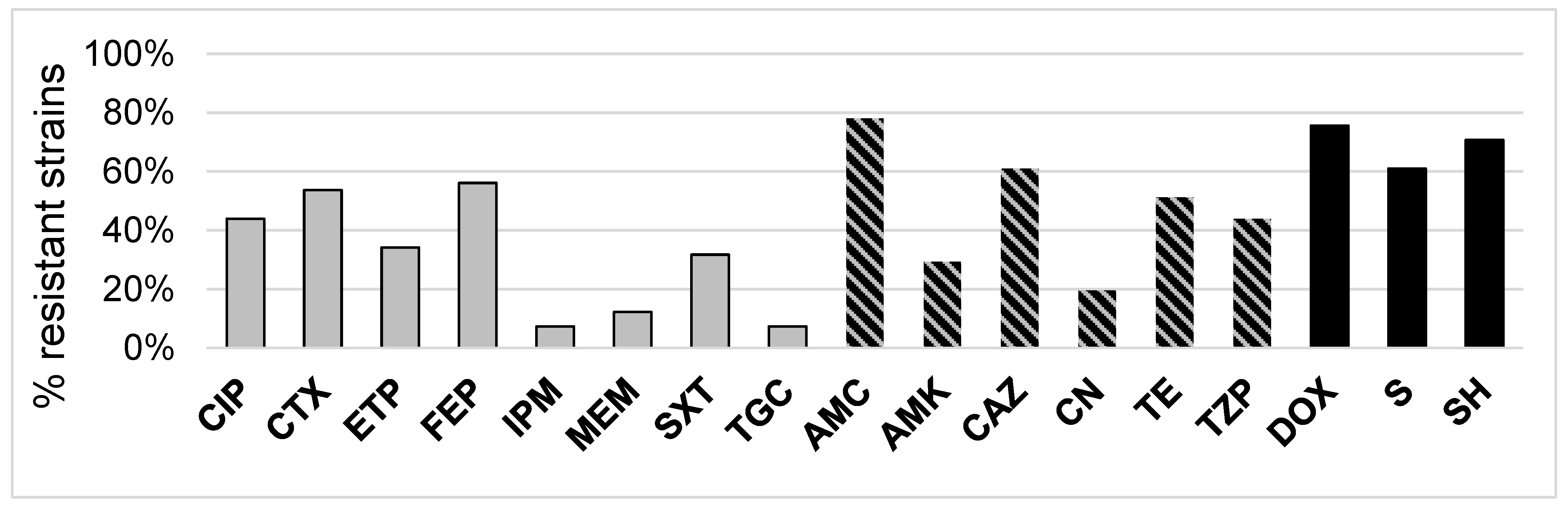
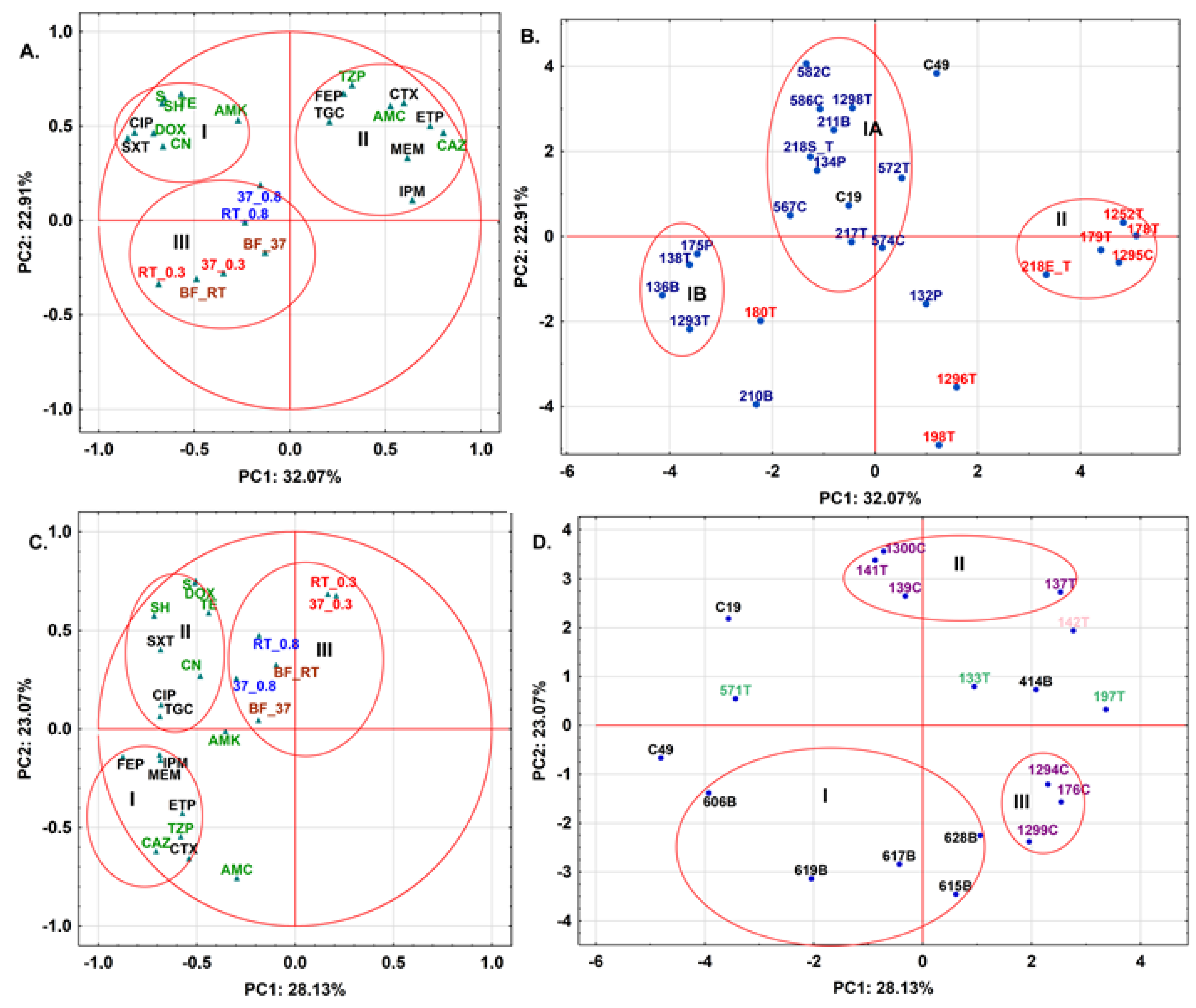
2.2. 3CG-R Strains Isolated from Meat Express Diversity in Phenotypic Traits
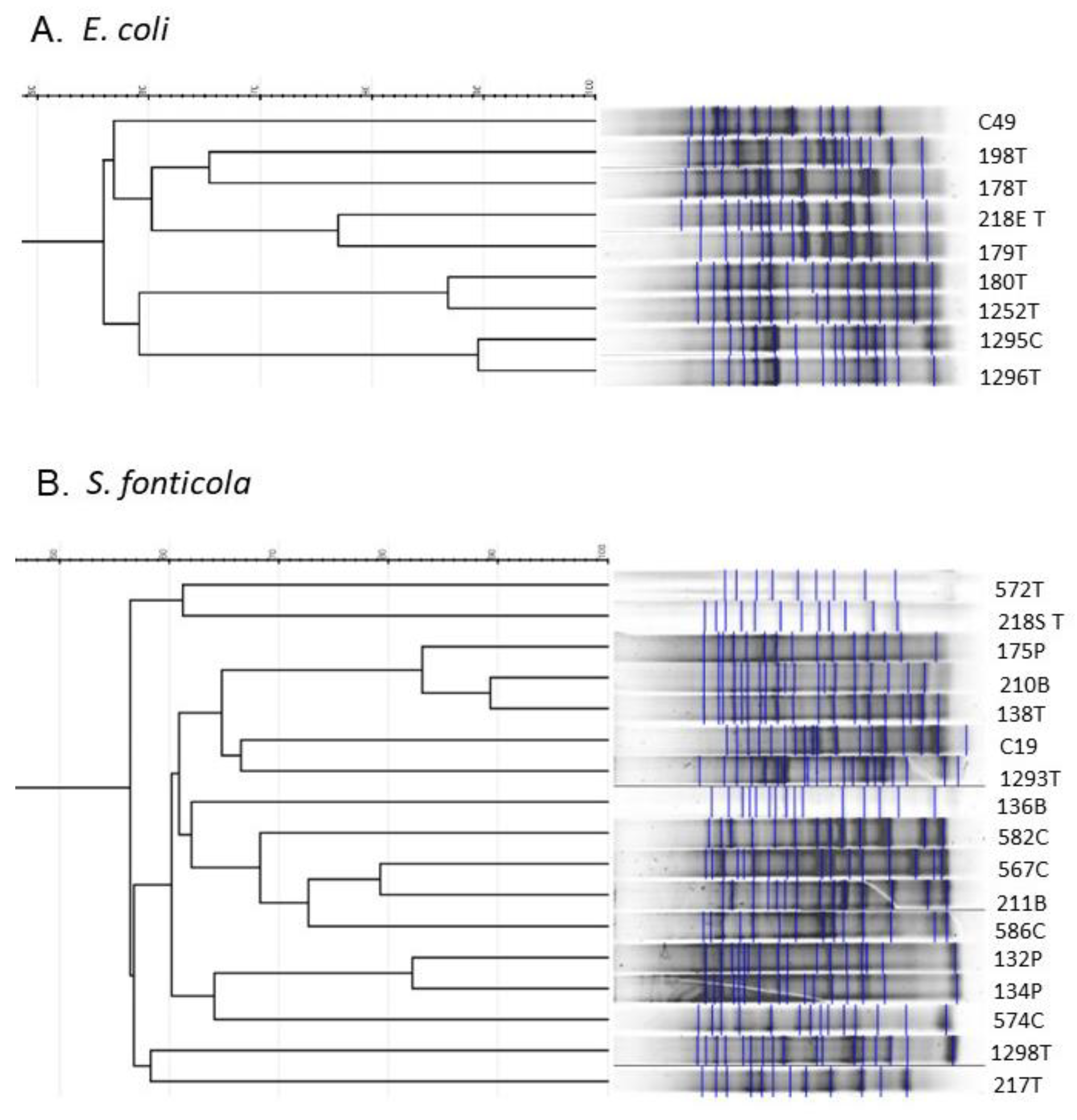
2.3. Repetitive Sequence Profiles Are Diverse among E. coli and S. fonticola 3CG-R Strains
2.4. Antibiotic Resistance Patterns May Be Explained by Meat Type for S. fonticola and En. cloacae
3. Discussion
4. Materials and Methods
4.1. Fresh Produce Samples
4.2. Isolation of Cephalosporin-Resistant Bacterial Strains
4.3. Antimicrobial Susceptibility Testing
4.4. Motility and Biofilm Formation
4.5. DNA Fingerprinting
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- The Third OIE Annual Report on the Use of Antimicrobial Agents Intended for Use in Animals; World Organisation for Animal Health (OIE): Paris, France, 2018; Available online: www.oie.int (accessed on 20 January 2022).
- EUCAST. Available online: http://www.eucast.org/expert_rules_and_intrinsic_resistance/ (accessed on 10 October 2021).
- ECDC/EFSA/EMA. First Joint Report on the Integrated Analysis of the Consumption of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Humans and Food-Producing Animals. EFSA J. 2015, 13, 4006. [Google Scholar] [CrossRef]
- Dandachi, I.; Chabou, S.; Daoud, Z.; Rolain, J.M. Prevalence and Emergence of Extended-Spectrum Cephalosporin-, Carbapenem- and Colistin-Resistant Gram Negative Bacteria of Animal Origin in the Mediterranean Basin. Front. Microbiol. 2018, 9, 2299. [Google Scholar] [CrossRef] [PubMed]
- Esteve-Palau, E.; Solande, G.; Sanchez, F.; Sorli, L.; Montero, M.; Guerri, R.; Villar, J.; Grau, S.; Horcajada, J.P. Clinical and economic impact of urinary tract infections caused by ESBL-producing Escherichia coli requiring hospitalization: A matched cohort study. J. Infect. 2015, 71, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States. 2013. Available online: https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf (accessed on 5 March 2022).
- Tanner, W.D.; VanDerslice, J.A.; Goel, R.K.; Leecaster, M.K.; Fisher, M.A.; Olstadt, J.; MGurley, C.M.; Morris, A.G.; Seely, K.A.; Chapman, L.; et al. Multi-state study of Enterobacteriaceae harboring extended-spectrum beta-lactamase and carbapenemase genes in U.S. drinking water. Sci. Rep. 2019, 9, 3938. [Google Scholar] [CrossRef] [Green Version]
- Rybak, B.; Wawrzyniak, N.; Wolska, L.; Potrykus, M. Escherichia coli and Serratia fonticola ESBLs as a potential source of antibiotics resistance dissemination in the Tricity water reservoirs. Biochim. Pol. 2021, 68, 437–448. [Google Scholar] [CrossRef]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial resist ance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef]
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance. 2014. Available online: https://www.who.int/drugresistance/documents/surveillancereport/en/ (accessed on 8 March 2022).
- World Health Organization. WHO Publishes List of Bacteria for Which New Antibiotics are Urgently Needed. 2017. Available online: http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/ (accessed on 27 February 2022).
- Pfeifer, Y.; Cullik, A.; Witte, W. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int. J. Med. Microbiol. 2010, 300, 371–379. [Google Scholar] [CrossRef]
- Bush, K.; Jacoby, G.A. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976. [Google Scholar] [CrossRef] [Green Version]
- Pulss, S.; Semmler, T.; Prenger-Berninghoff, E.; Bauerfeind, R.; Ewers, C. First report of an Escherichia coli strain from swine carrying an OXA-181 carbapenemase and the colistin resistance determinant MCR-1. Int. J. Antimicrob. Agents 2017, 50, 232–236. [Google Scholar] [CrossRef]
- Mathers, A.J.; Peirano, G.; Pitout, J.D. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 2015, 28, 565–591. [Google Scholar] [CrossRef] [Green Version]
- Bouchillon, S.K.; Badal, R.E.; Hoban, D.J.; Hawser, S.P. Antimicrobial susceptibility of inpatient urinary tract isolates of gram negative bacilli in the United States: Results from the study for monitoring antimicrobial resistance trends (SMART) program: 2009–2011. Clin. Ther. 2013, 35, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Kelly, A.M.; Mathema, B.; Larson, E.L. Carbapenem-resistant Enterobacteriaceae in the community: A scoping review. Int. J. Antimicrob. Agents 2017, 50, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Cekanova, L.; Kolar, M.; Chroma, M.; Sauer, P.; Sedlackova, M.; Koukalova, D. Prevalence of ESBL-positive bacteria in the community in the Czech Republic. Med. Sci. Monit. 2009, 15, BR202–BR206. [Google Scholar] [PubMed]
- Reuland, E.A.; Overdevest, I.T.; Al Naiemi, N.; Kalpoe, J.S.; Rijnsburger, M.C.; Raadsen, S.A.; Ligtenberg-Burgman, I.; van der Zwaluw, K.W.; Heck, M.; Savelkoul, P.H.; et al. Highprevalence of ESBL-producing Enterobacteriaceae carriage in Dutch community patients with gastrointestinal complaints. Clin. Microbiol. Infect. 2013, 19, 542–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bush, K. Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr. Opin. Microbiol. 2010, 13, 558–564. [Google Scholar] [CrossRef]
- Uyanik, T.; Gülel, G.T.; Alişarli, M. Characterization of extended-spectrum beta-lactamase-producing Enterobacterales from organic and conventional chicken meats. Lett. Appl. Microbiol. 2021, 72, 783–790. [Google Scholar] [CrossRef]
- Tanimoto, K.; Nomura, T.; Hashimoto, Y.; Hirakawa, H.; Watanabe, H.; Tomita, H. Isolation of Serratia fonticola producing FONA, a minor extended-spectrum β-lactamase (ESBL), from imported chicken meat in Japan. Jpn. J. Infect. Dis. 2021, 74, 79–81. [Google Scholar] [CrossRef]
- Yang, Y.Q.; Li, Y.X.; Lei, C.W.; Zhang, A.Y.; Wang, H.N. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2018, 73, 1791–1795. [Google Scholar] [CrossRef] [Green Version]
- Guo, S.; Aung, K.T.; Leekitcharoenphon, P.; Tay, M.Y.; Seow, K.L.; Zhong, Y.; Ching, L.; Møller Aarestrup, F.; Schlundt, J. Prevalence and genomic analysis of ESBL-producing Escherichia coli in retail raw meats in Singapore. J. Antimicrob. Chemoth. 2021, 76, 601–605. [Google Scholar] [CrossRef]
- Mezhoud, H.; Chantziaras, I.; Iguer-Ouada, M.; Moula, N.; Garmyn, A.; Martel, A.; Touati, A.; Smet, A.; Haesebrouck, F.; Boyen, F. Presence of antimicrobial resistance in coliform bacteria from hatching broiler eggs with emphasis on ESBL/AmpC-producing bacteria. Avian Pathol. 2016, 45, 493–500. [Google Scholar] [CrossRef] [Green Version]
- Badr, H.; Reda, R.M.; Hagag, N.M.; Kamel, E.; Elnomrosy, S.M.; Mansour, A.I.; Shahein, M.A.; Ali, S.F.; Ali, H.R. Multidrug-Resistant and Genetic Characterization of Extended-Spectrum Beta-Lactamase-Producing E. coli Recovered from Chickens and Humans in Egypt. Animals 2022, 31, 346. [Google Scholar] [CrossRef] [PubMed]
- Zurfluh, K.; Nuesch-Inderbinen, M.; Morach, M.; Berner, A.Z.; Hachler, H.; Stephan, R. Extended-spectrum-beta-lactamase-producing Enterobacteriaceae isolated from vegetables imported from the Dominican Republic, India, Thailand, and Vietnam. Appl. Environ. Microbiol. 2015, 81, 3115–3120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Rooij, M.M.; Hoek, G.; Schmitt, H.; Janse, I.; Swart, A.; Maassen, C.B.; Schalk, M.; van Leeuwenhoeklaan, A.; Heederik, D.J.J.; Wouters, I.M.; et al. Insights into Livestock-Related Microbial Concentrations in Air at Residential Level in a Livestock Dense Area. Environ. Sci. Technol. 2019, 53, 7746–7758. [Google Scholar] [CrossRef] [PubMed]
- Beyrouthy, R.; Robin, F.; Lessene, A.; Lacombat, I.; Dortet, L.; Naas, T.; Ponties, V.; Bonnet, R. MCR-1 and OXA-48 in vivo acquisition in KPC-producing Escherichia coli after colistin treatment. Antimicrob. Agents Chemother. 2017, 61, e02540-16. [Google Scholar] [CrossRef] [Green Version]
- McLellan, J.E.; Pitcher, J.I.; Ballard, S.A.; Grabsch, E.A.; Bell, J.M.; Barton, M.; Grayson, M.L. Superbugs in the supermarket? Assessing the rate of contamination with thirdgeneration cephalosporin-resistant gram negative bacteria in fresh Australian pork and chicken. Antimicrob. Resist. Infect. Control. 2018, 7, 30. [Google Scholar] [CrossRef] [Green Version]
- Müller, A.; Jansen, W.; Grabowski, N.T.; Monecke, S.; Ehricht, R.; Kehrenberg, C. ESBL- and AmpC-producing Escherichia coli from legally and illegally imported meat: Characterization of isolates brought into the EU from third countries. Int. J. Food Microbiol. 2018, 283, 52–58. [Google Scholar] [CrossRef]
- Palmeira, J.D.; Ferreira, H.; Madec, J.Y.; Haenni, M. Draft genome of a ST443 mcr-1- and blaCTX-M-2-carrying Escherichia coli from cattle in Brazil. J. Glob. Antimicrob. Resist. 2018, 13, 269–270. [Google Scholar] [CrossRef]
- Riley, L.W. Extraintestinal foodborne pathogens. Annu. Rev. Food Sci. Technol. 2020, 11, 275–294. [Google Scholar] [CrossRef] [Green Version]
- Cyoia, P.S.; Koga, V.L.; Nishio, E.K.; Houle, S.; Dozois, C.M.; de Brito, K.C.T.; de Brito, B.G.; Nakazato, G. Kobayashi, R.K.T. Distribution of ExPEC virulence factors, blaCTX-M, fosA3, and mcr-1 in Escherichia coli isolated from commercialized chicken carcasses. Front. Microbiol. 2018, 9, 3254. [Google Scholar] [CrossRef] [Green Version]
- Dominguez, J.E.; Faccone, D.; Tijet, N.; Gomez, S.; Corso, A.; Fernandez-Miyakawa, M.E.; Melano, R.G. Characterization of Escherichia coli carrying mcr-1-plasmids recovered from food animals from Argentina. Front. Cell Infect. Microbiol. 2019, 9, 41. [Google Scholar] [CrossRef] [Green Version]
- Davis, G.S.; Waits, K.; Nordstrom, L.; Weaver, B.; Aziz, M.; Gauld, L.; Grande, H.; Bigler, R.; Horwinski, J.; Porter, S.; et al. Intermingled Klebsiella pneumoniae populations between retail meats and human urinary tract infections. Clin. Infect. Dis. 2015, 61, 892–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasman, H.; Hammerum, A.M.; Hansen, F.; Hendriksen, R.S.; Olesen, B.; Agerso, Y.; Zankari, E.; Leekitcharoenphon, P.; Stegger, M.; Kaas, R.S.; et al. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Eurosurveillance 2015, 20, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 6, 161–168. [Google Scholar] [CrossRef]
- Köck, R.; Daniels-Haardt, I.; Becker, K.; Mellmann, A.; Friedrich, A.W.; Mevius, D.; Schwarz, S.; Jurke, A. Carbapenem-resistant Enterobacteriaceae in wildlife, food-producing, and companion animals: A systematic review. Clin. Microbiol. Infect. 2018, 24, 1241–1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roer, L.; Hansen, F.; Stegger, M.; Sönksen, U.W.; Hasman, H.; Hammerum, A.M. Novel mcr-3 variant, encoding mobile colistin resistance, in an ST131 Escherichia coli isolate from bloodstream infection, Denmark, 2014. Eurosurveillance 2017, 22, 31. [Google Scholar] [CrossRef]
- EMA/CVMP/CHMP/682198/2017. Committee for Medicinal Products for Veterinary Use (CVMP); Committee for Medicinal Products for Human Use (CHMP), Categorisation of Antibiotics in the European Union. 2019. Available online: www.ema.europa.eu (accessed on 25 February 2022).
- Clemente, L.; Manageiro, V.; Correia, I.; Amaro, A.; Albuquerque, T.; Themudo, P.; Ferreira, E.; Caniça, M. Revealing mcr-1-positive ESBL-producing Escherichia coli strains among Enterobacteriaceae from food-producing animals (bovine, swine and poultry) and meat (bovine and swine), Portugal, 2010–2015. Int. J. Food Microbiol. 2019, 296, 37–42. [Google Scholar] [CrossRef]
- Doi, Y.; Paterson, D.L.; Pascual, E.A.; López-Cerero, L.; Navarro, M.D.; Adams-Haduch, J.M.; Qureshi, Z.A.; Sidjabat, H.E.; Rodríguez-Baño, J. Extended-spectrum and CMY-type β-lactamasep-producing Escherichia coli in clinical samples and retial meat from Pittsburgh, USA and Sevile, Spain. Clin. Microbiol. Infect. 2009, 20, 33–38. [Google Scholar]
- Randall, L.P.; Lodge, M.P.; Elviss, N.C.; Lemma, F.L.; Hopkins, K.L.; Teale, C.J.; Woodford, N. Evaluation of meat, fruit and vegetables from retail stores in five United Kingdom regions as sources of extended-spectrum beta-lactamase (ESBL)-producing and carbapenem-resistant Escherichia coli. Int. J. Food Microbiol. 2017, 241, 283–290. [Google Scholar] [CrossRef]
- Kola, A.; Kohler, C.; Pfeifer, Y.; Schwab, F.; Kuhn, K.; Schulz, K.; Balau, V.; Breitbach, K.; Bast, A.; Witte, W.; et al. High prevalence of extended-spectrum-b-lactamase-producing Enterobacteriaceae in organic and conventional retail chicken meat, Germany. J. Antimicrob. Chemother. 2012, 67, 2631–2634. [Google Scholar] [CrossRef] [Green Version]
- Leverstein-van Hall, M.A.; Dierikx, C.M.; Stuart, J.C.; Voets, G.M.; van den Munckhof, M.P.; van Essen-Zandbergen, A.; Platteel, T.; Fluit, A.C.; van de Sande-Bruinsma, N.; Scharinga, J.; et al. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin. Microbiol. Infect. 2011, 17, 873–880. [Google Scholar] [CrossRef] [Green Version]
- Carattoli, A.; Villa, L.; Feudi, C.; Curcio, L.; Orsini, S.; Luppi, A.; Pezzotti, G.; Magistrali, C.F. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Eurosurveillance 2017, 22, 30589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borowiak, M.; Fischer, J.; Hammerl, J.A.; Hendriksen, R.S.; Szabo, I.; Malorny, B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi. B. J. Antimicrob. Chemother. 2017, 72, 3317–3324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camposa, C.B.; Fennerb, I.; Wieseb, N.; Lensingb, C.; Christnera, M.; Rohdea, H.; Aepfelbachera, M.; Fennerb, T.; Hentschke, M. Prevalence and genotypes of extended spectrum beta-lactamases in Enterobacteriaceae isolated from human stool and chicken meat in Hamburg, Germany. Int. J. Med. Microbiol. 2014, 304, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Casella, T.; Lelles Nogueira, M.C.; Saras, E.; Haenni, M. High prevalence of ESBLs in retail chicken meat despite reduced use of antimicrobials in chicken production, France. Int. J. Food Microbiol. 2017, 257, 271–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- von Tippelskirch, P.; Gölz, G.; Projahn, M.; Daehre, K.; Friese, A.; Roesler, U.; Alter, T.; Orquera, S. Prevalence and quantitative analysis of ESBL and AmpC beta-lactamase producing Enterobacteriaceae in broiler chicken during slaughter in Germany. Int. J. Food Microbiol. 2018, 281, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Dierikx, C.M.; van der Goot, J.A.; Smith, H.E.; Kant, A.; Mevius, D.J. Presence of ESBL/AmpC-producing Escherichia coli in the broiler production pyramid: A descriptive study. PLoS ONE 2013, 8, e79005. [Google Scholar] [CrossRef]
- Hering, J.; Fromke, C.; von Munchhausen, C.; Hartmann, M.; Schneider, B.; Friese, A.; Rosler, U.; Kreienbrock, L.; Hille, K. Cefotaxime-resistant Escherichia coli in broiler farms-a cross-sectional investigation in Germany. Prev. Vet. Med. 2016, 125, 154–157. [Google Scholar] [CrossRef]
- Schill, F.; Abdulmawjood, A.; Klein, G.; Reich, F. Prevalence and characterization of extended-spectrum beta-lactamase (ESBL) and AmpC beta-lactamase producing Enterobacteriaceae in fresh pork meat at processing level in Germany. Int. J. Food Microbiol. 2017, 257, 58–66. [Google Scholar] [CrossRef]
- Machado, E.; Coque, T.M.; Canton, R.; Sousa, J.C.; Peixe, L. Antibiotic resistance integrons and extended-spectrum β-lactamases among Enterobacteriaceae isolates recovered from chickens and swine in Portugal. J. Antimicrob. Chemother. 2008, 62, 296–302. [Google Scholar] [CrossRef] [Green Version]
- Geser, N.; Stephan, R.; Hachler, H. Occurrence and characteristics of extended spectrum beta-lactamase (ESBL) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. BMC Vet. Res. 2012, 8, 21. [Google Scholar] [CrossRef] [Green Version]
- Depoorter, P.; Persoons, D.; Uyttendaele, M.; Butaye, P.; De Zutter, L.; Dierick, K.; Herman, L.; Imberechts, H.; Van Huffel, X.; Dewulf, J. Assessment of human exposure to 3rd generation cephalosporin resistant E. coli (CREC) through consumption of broiler meat in Belgium. Int. J. Food Microbiol. 2012, 159, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Tschudin-Sutter, S.; Frei, R.; Stephan, R.; Hachler, H.; Nogarth, D.; Widmer, A.F. Extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae: A threat from the kitchen. Infect. Control. Hosp. Epidemiol. 2014, 35, 581–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz-Jimenez, D.; Garcia-Menino, I.; Fernandez, J.; Garcia, V.; Mora, A. Chicken and turkey meat: Consumer exposure to multidrug-resistant Enterobacteriaceae including mcr-carriers, uropathogenic E. coli and highrisk lineages such as ST131. Int. J. Food Microbiol. 2020, 331, 108750. [Google Scholar] [CrossRef] [PubMed]
- Zowawi, H.M.; Harris, P.N.; Roberts, M.J.; Tambyah, P.A.; Schembri, M.A.; Pezzani, M.D.; Williamson, D.A.; Paterson, D.L. The emerging threat of multidrug-resistant gram-negative bacteria in urology. Nat. Rev. Urol. 2015, 12, 570–584. [Google Scholar] [CrossRef]
- National Agricultural Support Center; Analysis and Strategy Office. Polish Foreign Trade in Meat Products in 2019, Warsaw. 2020. Available online: www.kowr.gov.pl (accessed on 15 February 2022).
- Polkowska, E. Wykorzystywanie Antybiotyków w Produkcji Zwierzęcej w Województwie Lubuskim, Najwyższa Izba Kontroli, Warszawa. Available online: www.nik.gov.pl (accessed on 15 February 2022).
- Krasucka, D.; Biernacki, B.; Szumiło, J.; Burmańczuk, A. Monitoring zużycia leków przeciwdrobnoustrojowych u bydła, trzody chlewnej i koni w Polsce w latach 2014–2016 na podstawie Programu Wieloletniego. Życie Weter. 2017, 92, 8. [Google Scholar]
- Granato, D.; Santos, J.S.; Escher, G.B.; Ferreira, B.L.; Maggio, R.M. Use of principal component analysis (PCA) and hierarchical cluster analysis (HCA) for multivariate association between bioactive compounds and functional properties in foods: A critical perspective. Trends Food Sci. Technol. 2018, 72, 83–90. [Google Scholar] [CrossRef]
- OECD. Meat Consumption; Organisation for Economic Co-operation and Development: Paris, France, 2022. [Google Scholar] [CrossRef]
- Central Statistical Office (GUS). Agriculture in 2019, Warsaw; Central Statistical Office (GUS): Warszawa, Poland, 2020. [Google Scholar]
- Data Sheet on Poultry Production in Europe, Agridata. Available online: https://ec.europa.eu/info/sites/default/files/food-farming-fisheries/farming/documents/poultry-meat-dashboard_en.pdf (accessed on 1 March 2022).
- Data Sheet on Pigs Production in EUROPE, Agridata. Available online: https://agridata.ec.europa.eu/Reports/Pigmeat_Dashboard.pdf (accessed on 3 March 2022).
- Bergšpica, I.; Kaprou, G.; Alexa, E.A.; Prieto, M.; Alvarez-Ordóñez, A. Extended spectrum β-lactamase (ESBL) producing Escherichia coli in pigs and pork meat in the European Union. Antibiotics 2020, 9, 678. [Google Scholar] [CrossRef]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2017/2018. EFSA J. 2020, 18, e06007. [Google Scholar]
- Melzer, M.; Petersen, I. Mortality following bacteraemic infection caused by extended spectrum beta-lactamase (ESBL) producing E. coli compared to non-ESBL producing E. coli. J. Infect. 2007, 55, 254–259. [Google Scholar] [CrossRef]
- MacKinnon, M.C.; McEwen, S.A.; Pearl, D.L.; Parfitt, E.C.; Pasquill, K.; Steele, L.; Laupland, K.B. Escherichia coli bloodstream infections in the western interior of British Columbia, Canada: A population-based cohort study. Epidemiol. Infect. 2021, 6, e195. [Google Scholar] [CrossRef]
- van den Bunt, G.; van Pelt, W.; Hidalgo, L.; Scharringa, J.; de Greeff, S.C.; Schürch, A.C.; Mughini-Gras, L.; Bonten, M.J.M.; Fluit, A.C. Prevalence, risk factors and genetic characterisation of extended-spectrum beta-lactamase and carbapenemase-producing Enterobacteriaceae (ESBL-E and CPE): A community-based cross-sectional study, the Netherlands, 2014 to 2016. Eurosurveillance 2019, 24, 1800594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaesbohrer, A.; Bakran-Lebl, K.; Irrgang, A.; Fischer, J.; Kämpf, P.; Schiffmann, A.; Werckenthin, C.; Busch, M.; Kreienbrock, L.; Hille, K. Diversity in prevalence and characteristics of ESBL/pAmpC producing E. coli in food in Germany. Vet. Microbiol. 2019, 233, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Parvin, M.; Talukder, S.; Ali, M.; Chowdhury, E.H.; Rahman, M.; Islam, M. Antimicrobial resistance pattern of Escherichia coli isolated from frozen chicken meat in Bangladesh. Pathogens 2020, 9, 420. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Moon, J.S.; Oh, D.H.; Chon, J.W.; Song, B.R.; Lim, J.S.; Heo, E.J.; Park, H.J.; Wee, S.H.; Sung, K. Genotypic characterization of ESBL-producing E. coli from imported meat in South Korea. Food Res. Int. 2018, 107, 158–164. [Google Scholar] [CrossRef]
- Rahman, S.U.; Ahmad, S.; Khan, I. Incidence of ESBL-producing-Escherichia coli in poultry farm environment and retail poultry meat. Pak. Vet. J. 2018, 39, 116–120. [Google Scholar] [CrossRef]
- Ojer-Usoz, E.; González, D.; Vitas, A.I.; Leiva, J.; García-Jalón, I.; Febles-Casquero, A.; de la Soledad Escolano, M. Prevalence of extended-spectrum β-lactamase-producing Enterobacteriaceae in meat products sold in Navarra, Spain. Meat Sci. 2013, 93, 316–321. [Google Scholar] [CrossRef]
- Huizinga, P.; Kluytmans-van den Bergh, M.; Rossen, J.W.; Willemsen, I.; Verhulst, C.; Savelkoul, P.H.; Friedrich, A.W.; García-Cobos, S.; Kluytmans, J. Decreasing prevalence of contamination with extended-spectrum beta-lactamase-producing Enterobacteriaceae (ESBL-E) in retail chicken meat in the Netherlands. PLoS ONE 2019, 14, e0226828. [Google Scholar] [CrossRef]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. EFSA J. 2021, 19, e06490. [Google Scholar]
- Evers, E.G.; Pielaat, A.; Smid, J.H.; van Duijkeren, E.; Vennemann, F.B.; Wijnands, L.M.; Chardon, J.E. Comparative exposure assessment of ESBL-producing Escherichia coli through meat consumption. PLoS ONE 2017, 12, e0169589. [Google Scholar] [CrossRef]
- Özpınar, H.; Tekiner, İ.H.; Sarıcı, B.; Çakmak, B.; Gökalp, F.; Özadam, A. Phenotypic Characterization of ESBL-and AmpC-Type Betalactamases in Enterobacteriaceae From Chicken Meat and Dairy Products. Ank. Üniversitesi Vet. Fakültesi Derg. 2017, 64, 267–272. [Google Scholar]
- Bello-López, J.M.; Cabrero-Martínez, O.A.; Ibáñez-Cervantes, G.; Hernández-Cortez, C.; Pelcastre-Rodríguez, L.I.; Gonzalez-Avila, L.U.; Castro-Escarpulli, G. Horizontal Gene Transfer and Its Association with Antibiotic Resistance in the Genus Aeromonas spp. Microorganisms 2019, 18, 363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giaouris, E.; Heir, E.; Hébraud, M.; Chorianopoulos, N.; Langsrud, S.; Møretrø, T.; Habimana, O.; Desvaux, M.; Renier, S.; Nychas, G.J. Attachment and biofilm formation by foodborne bacteria in meat processing environments: Causes, implications, role of bacterial interactions and control by alternative novel methods. Meat Sci. 2014, 97, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Ehuwa, O.; Jaiswal, A.K.; Jaiswal, S. Salmonella, Food Safety and Food Handling Practices. Foods 2021, 21, 907. [Google Scholar] [CrossRef]
- Jarlier, V.; Nicolas, M.H.; Fournier, G.; Philippon, A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: Hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 1988, 10, 867–878. [Google Scholar] [CrossRef]
- Versalovic, J.; de Bruijn, F.J.; Lupski, J.R. Repetitive Sequence-based PCR (rep-PCR) DNA Fingerprinting of Bacterial Genomes. In Bacterial Genomes 437–454; Springer: Boston, MA, USA, 1998. [Google Scholar]
- Heras, J.; Domínguez, C.; Mata, E.; Pascual, V.; Lozano, C.; Torres, C.; Zarazaga, M. GelJ—A tool for analyzing DNA fingerprint gel images. BMC Bioinform. 2015, 16, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
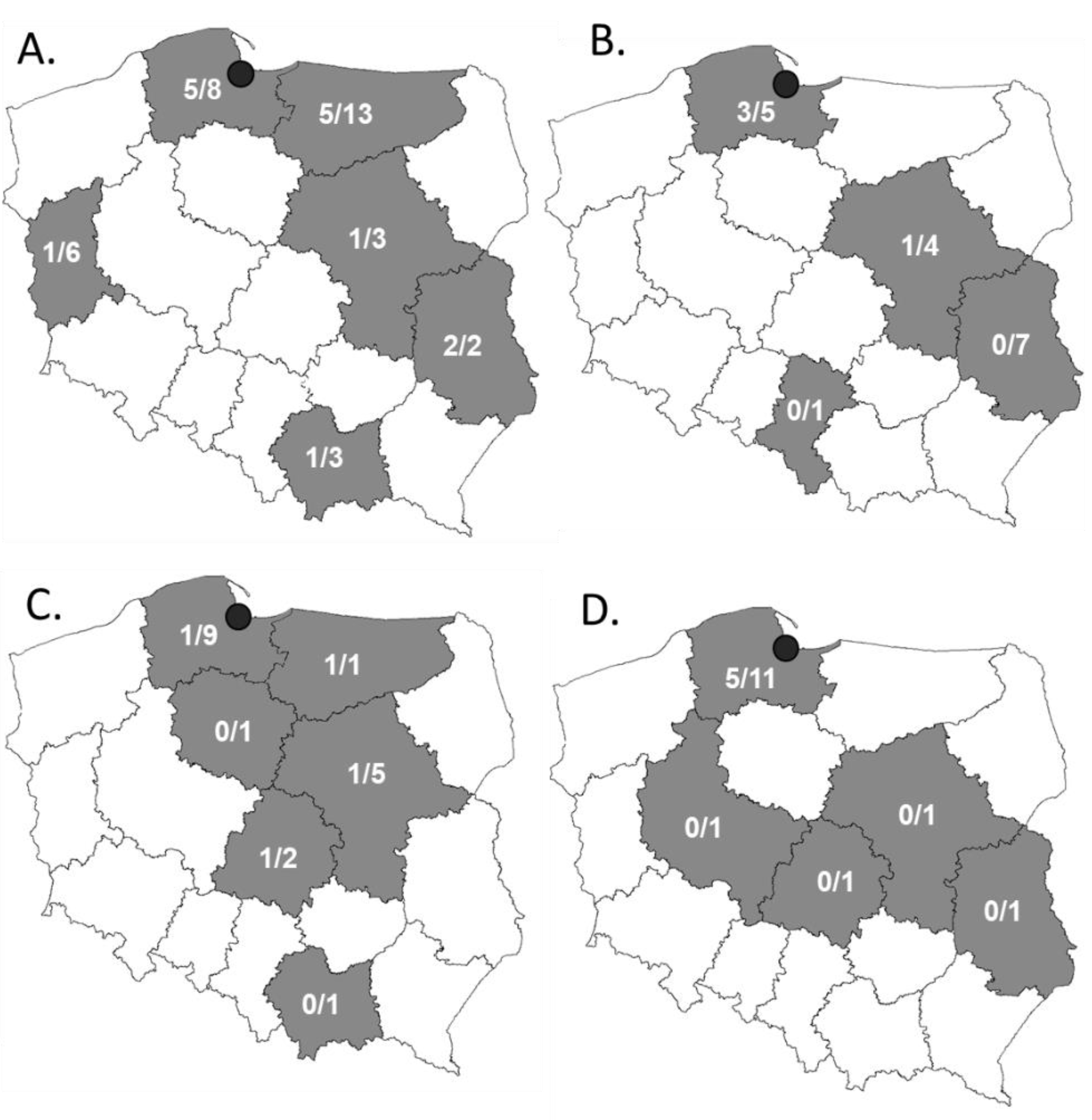

| Meat Type | Packaging | Summary | ||||
|---|---|---|---|---|---|---|
| Vacuum | Tray | |||||
| Samples | % | Samples | % | Samples | % | |
| turkey | 33 | 38.4% | 2 | 2.3% | 35 | 40.7% |
| chicken | 4 | 4.7% | 13 | 15.1% | 17 | 19.8% |
| pork | 15 | 17.4% | 4 | 4.7% | 19 | 22.1% |
| beef | 4 | 4.7% | 11 | 12.8% | 15 | 17.4% |
| SUM | 56 | 30 | 86 | |||
| Samples | Lactose + | Lactose − | E. coli ESBL | KESC ESBL | Mannitol + | CT-R | |||
|---|---|---|---|---|---|---|---|---|---|
| Samples | % | ||||||||
| Meat type | Turkey | 35 | 16.6 ± 27.5 | 27.3 ± 32.3 | 0.4 ± 0.8 | 6 ± 17.1 | 14.9 ± 25.5 | 15 | 43% |
| Chicken | 17 | 27 ± 40 | 25 ± 43.5 | 0.9 ± 2.4 | 9 ± 24.6 | 13.9 ± 23.5 | 5 | 33% | |
| Pork | 19 | 14.76 ± 29 | 15.2 ± 16.3 | 0.05 ± 0.2 | 1.9 ± 3.6 | 5.2 ± 7.2 | 4 | 24% | |
| Beef | 15 | 13.4 ± 24.8 | 15.8 ± 23.8 | 0.06 ± 0.2 | 5.8 ± 15.5 | 3.8 ± 6 | 4 | 21% | |
| Packaging | Vacuum | 56 | 23.4 ± 27.3 | 27.5 ± 26.9 | 0.7 ± 0.5 | 3.9 ± 19.3 | 15.1 ± 13.6 | 16 | 57% |
| Tray | 30 | 17.4 ± 33.9 | 21.3 ± 30.1 | 0.3 ± 1.9 | 5.6 ± 11.2 | 10.5 ± 27.7 | 12 | 43% | |
| Total | 86 | 17.4 ± 29.8 | 21.3 ± 27.9 | 0.3 ± 1.2 | 5.6 ± 16.9 | 10.5 ± 20 | 28 | nd | |
| Species | Meat Type | Packaging | Total | |||||
|---|---|---|---|---|---|---|---|---|
| Turkey (n = 35) | Chicken (n = 17) | Pork (n = 19) | Beef (n = 15) | Vacuum (n = 56) | Tray (n = 30) | Isolates | % | |
| En. cloacae | 2 | 1 | 2 | 2 | 4 | 3 | 7 | 17 |
| E. coli | 7 | 1 | 0 | 0 | 7 | 1 | 8 | 19.5 |
| K. pneumoniae | 5 | 0 | 1 | 0 | 3 | 3 | 6 | 14.6 |
| S. fonticola | 6 | 4 | 3 | 3 | 9 | 7 | 16 | 39 |
| S. liquefaciens | 2 | 0 | 1 | 0 | 3 | 0 | 3 | 7.3 |
| S. marcescens | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 2.4 |
| Total | 22 | 6 | 7 | 6 | 26 | 15 | 41 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rybak, B.; Potrykus, M.; Plenis, A.; Wolska, L. Raw Meat Contaminated with Cephalosporin-Resistant Enterobacterales as a Potential Source of Human Home Exposure to Multidrug-Resistant Bacteria. Molecules 2022, 27, 4151. https://doi.org/10.3390/molecules27134151
Rybak B, Potrykus M, Plenis A, Wolska L. Raw Meat Contaminated with Cephalosporin-Resistant Enterobacterales as a Potential Source of Human Home Exposure to Multidrug-Resistant Bacteria. Molecules. 2022; 27(13):4151. https://doi.org/10.3390/molecules27134151
Chicago/Turabian StyleRybak, Bartosz, Marta Potrykus, Alina Plenis, and Lidia Wolska. 2022. "Raw Meat Contaminated with Cephalosporin-Resistant Enterobacterales as a Potential Source of Human Home Exposure to Multidrug-Resistant Bacteria" Molecules 27, no. 13: 4151. https://doi.org/10.3390/molecules27134151
APA StyleRybak, B., Potrykus, M., Plenis, A., & Wolska, L. (2022). Raw Meat Contaminated with Cephalosporin-Resistant Enterobacterales as a Potential Source of Human Home Exposure to Multidrug-Resistant Bacteria. Molecules, 27(13), 4151. https://doi.org/10.3390/molecules27134151