The Effects of Charged Amino Acid Side-Chain Length on Diagonal Cross-Strand Interactions between Carboxylate- and Ammonium-Containing Residues in a β-Hairpin
Abstract
:1. Introduction
2. Results
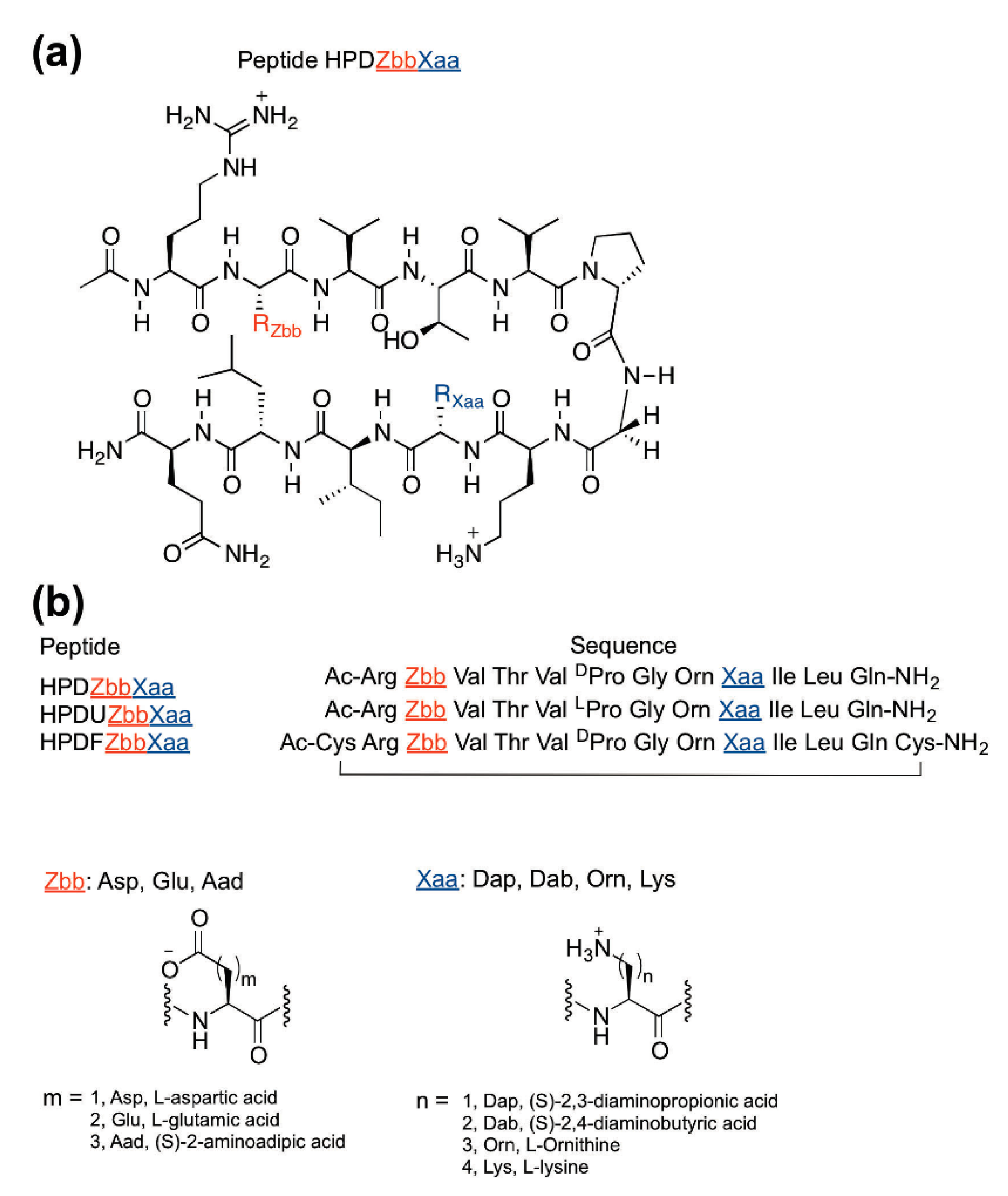
2.1. Peptide Design and Synthesis
2.2. Peptide Structure Characterization by NMR Spectroscopy
2.3. Fraction Folded Population and Folding Free Energy
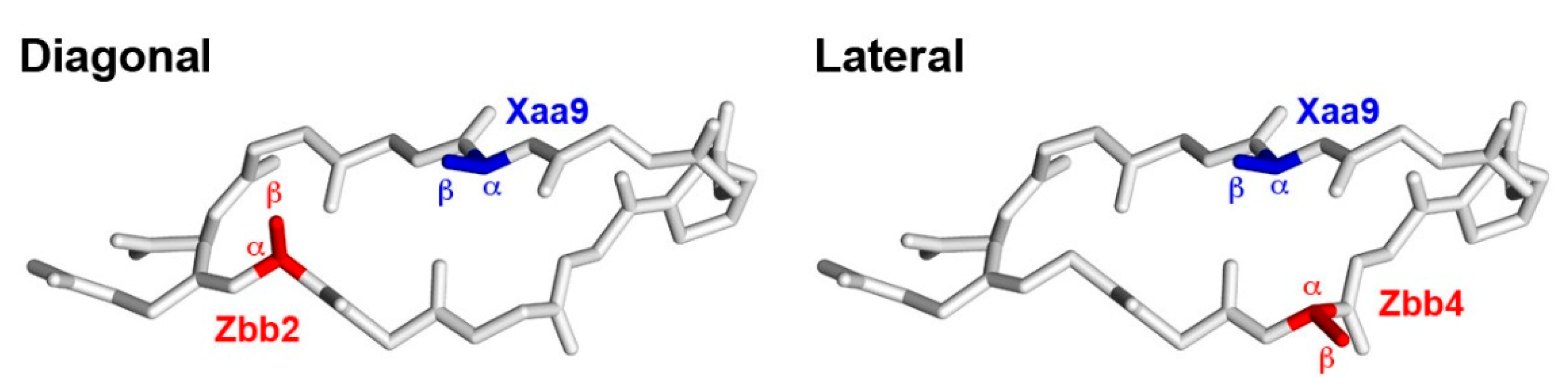
2.4. Diagonal Cross-Strand Zbb-Xaa Interactions
3. Discussion
4. Materials and Methods
4.1. Peptide Synthesis and Purification
4.2. Chemical Shift Deviation
4.3. Nuclear Magnetic Resonance Spectroscopy
4.4. 3JNHα Spin–Spin Coupling Constant
4.5. Interproton Distance Determination via NOE Integration
4.6. Fraction Folded Population and Folding Free Energy (ΔGfold)
4.7. Double Mutant Cycle Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Chou, P.Y.; Fasman, G.D. Conformational parameters for amino acids in helical, β-sheet, and random coil regions calculated from proteins. Biochemistry 1974, 13, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, V.; Serrano, L. Intrinsic secondary structure propensities of the amino acids, using statistical φ-ψ matrices: Comparison with experimental scales. Proteins 1994, 20, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Kuo, L.-H.; Li, J.-H.; Kuo, H.-T.; Hung, C.-Y.; Tsai, H.-Y.; Chiu, W.-C.; Wu, C.-H.; Wang, W.-R.; Yang, P.-A.; Yao, Y.-C.; et al. Effect of Charged Amino Acid Side Chain Length at Non-Hydrogen Bonded Strand Positions on β-Hairpin Stability. Biochemistry 2013, 52, 7785–7797. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.; Allsop, D. Amyloid deposition as the central event in the aetiology of Alzheimer’s disease. Trends Pharmacol. Sci. 1991, 12, 383–388. [Google Scholar] [CrossRef]
- Bartzokis, G.; Lu, P.H.; Mintz, J. Human brain myelination and amyloid beta deposition in Alzheimer’s disease. Alzheimers Dement. 2007, 3, 122–125. [Google Scholar] [CrossRef] [Green Version]
- Scherzinger, E.; Lurz, R.; Turmaine, M.; Mangiarini, L.; Hollenbach, B.; Hasenbank, R.; Bates, G.P.; Davies, S.W.; Lehrach, H.; Wanker, E.E. Huntingtin-Encoded Polyglutamine Expansions Form Amyloid-like Protein Aggregates In Vitro and In Vivo. Cell 1997, 90, 549–558. [Google Scholar] [CrossRef] [Green Version]
- Truant, R.; Atwal, R.S.; Desmond, C.; Munsie, L.; Tran, T. Huntington’s disease: Revisiting the aggregation hypothesis in polyglutamine neurodegenerative diseases. FEBS J. 2008, 275, 4252–4262. [Google Scholar] [CrossRef] [Green Version]
- Irvine, G.B.; El-Agnaf, O.M.; Shankar, G.M.; Walsh, D.M. Protein Aggregation in the Brain: The Molecular Basis for Alzheimer’s and Parkinson’s Diseases. Mol. Med. 2008, 14, 451–464. [Google Scholar] [CrossRef]
- Minor, D.L., Jr.; Kim, P.S. Measurement of the β-sheet-forming propensities of amino acids. Nature 1994, 367, 660–663. [Google Scholar] [CrossRef]
- Smith, C.K.; Withka, J.M.; Regan, L. A thermodynamic scale for the β-sheet forming tendencies of the amino acids. Biochemistry 1994, 33, 5510–5517. [Google Scholar] [CrossRef]
- Wouters, M.A.; Curmi, P.M.G. An analysis of side chain interactions and pair correlations within antiparallel β-sheets: The differences between backbone hydrogen-bonded and non-hydrogen-bonded residue pairs. Proteins 1995, 22, 119–131. [Google Scholar] [CrossRef]
- Cootes, A.P.; Curmi, P.M.; Cunningham, R.; Donnelly, C.; Torda, A.E. The dependence of amino acid pair correlations on structural environment. Proteins 1998, 32, 175–189. [Google Scholar] [CrossRef]
- Hutchinson, E.G.; Sessions, R.B.; Thornton, J.M.; Woolfson, D.N. Determinants of strand register in antiparallel β-sheets of proteins. Protein Sci. 1998, 7, 2287–2300. [Google Scholar] [CrossRef]
- Smith, C.K.; Regan, L. Guidelines for Protein Design: The Energetics of β Sheet Side Chain Interactions. Science 1995, 270, 980–982. [Google Scholar] [CrossRef]
- Merkel, J.S.; Sturtevant, J.M.; Regan, L. Sidechain interactions in parallel β sheets: The energetics of cross-strand pairings. Structure 1999, 7, 1333–1343. [Google Scholar] [CrossRef] [Green Version]
- Blasie, C.A.; Berg, J.M. Electrostatic Interactions across a β-Sheet. Biochemistry 1997, 36, 6218–6222. [Google Scholar] [CrossRef]
- Searle, M.S.; Griffiths-Jones, S.R.; Skinner-Smith, H. Energetics of Weak Interactions in a β-hairpin Peptide: Electrostatic and Hydrophobic Contributions to Stability from Lysine Salt Bridges. J. Am. Chem. Soc. 1999, 121, 11615–11620. [Google Scholar] [CrossRef]
- Russell, S.J.; Cochran, A.G. Designing Stable β-Hairpins: Energetic Contributions from Cross-Strand Residues. J. Am. Chem. Soc. 2000, 122, 12600–12601. [Google Scholar] [CrossRef]
- Syud, F.A.; Stanger, H.E.; Gellman, S.H. Interstrand Side Chain−Side Chain Interactions in a Designed β-Hairpin: Significance of Both Lateral and Diagonal Pairings. J. Am. Chem. Soc. 2001, 123, 8667–8677. [Google Scholar] [CrossRef]
- Ramírez-Alvarado, M.; Blanco, F.J.; Serrano, L. Elongation of the BH8 β-hairpin peptide: Electrostatic interactions in β-hairpin formation and stability. Protein Sci. 2001, 10, 1381–1392. [Google Scholar] [CrossRef]
- Tatko, C.D.; Waters, M.L. Selective Aromatic Interactions in β-Hairpin Peptides. J. Am. Chem. Soc. 2002, 124, 9372–9373. [Google Scholar] [CrossRef]
- Kiehna, S.E.; Waters, M.L. Sequence dependence of β-hairpin structure: Comparison of a salt bridge and an aromatic interaction. Protein Sci. 2003, 12, 2657–2667. [Google Scholar] [CrossRef]
- Ciani, B.; Jourdan, M.; Searle, M.S. Stabilization of β-Hairpin Peptides by Salt Bridges: Role of Preorganization in the Energetic Contribution of Weak Interactions. J. Am. Chem. Soc. 2003, 125, 9038–9047. [Google Scholar] [CrossRef]
- Tatko, C.D.; Waters, M.L. Comparison of C-H...π and hydrophobic interactions in a β-hairpin peptide: Impact on stability and specificity. J. Am. Chem. Soc. 2004, 126, 2028–2034. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.-T.; Fang, C.-J.; Tsai, H.-Y.; Yang, M.-F.; Chang, H.-C.; Liu, S.-L.; Kuo, L.-H.; Wang, W.-R.; Yang, P.-A.; Huang, S.-J.; et al. Effect of Charged Amino Acid Side Chain Length on Lateral Cross-Strand Interactions between Carboxylate-Containing Residues and Lysine Analogues in a β-Hairpin. Biochemistry 2013, 52, 9212–9222. [Google Scholar] [CrossRef]
- Kuo, H.-T.; Liu, S.-L.; Chiu, W.-C.; Fang, C.-J.; Chang, H.-C.; Wang, W.-R.; Yang, P.-A.; Li, J.-H.; Huang, S.-J.; Huang, S.-L.; et al. Effect of charged amino acid side chain length on lateral cross-strand interactions between carboxylate- and guanidinium-containing residues in a β-hairpin. Amino Acids 2015, 47, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-H.; Wong, T.W.; Yu, C.-H.; Chang, J.-Y.; Huang, S.-J.; Huang, S.-L.; Cheng, R.P. Swapping the Positions in a Cross-Strand Lateral Ion-Pairing Interaction between Ammonium- and Carboxylate-Containing Residues in a β-Hairpin. Molecules 2021, 26, 1346. [Google Scholar] [CrossRef] [PubMed]
- Chothia, C. Conformation of twisted β-pleated sheets in proteins. J. Mol. Biol. 1973, 75, 295–302. [Google Scholar] [CrossRef]
- Tatko, C.D.; Waters, M.L. The geometry and efficacy of cation-π interactions in a diagonal position of a designed β-hairpin. Protein Sci. 2003, 12, 2443–2452. [Google Scholar] [CrossRef]
- Hughes, R.M.; Waters, M.L. Influence of N-methylation on a cation-π interaction produces a remarkably stable β-hairpin peptide. J. Am. Chem. Soc. 2005, 127, 6518–6519. [Google Scholar] [CrossRef]
- Hughes, R.M.; Waters, M.L. Arginine methylation in a β-hairpin peptide: Implications for Arg-π interactions, ΔCpo, and the cold denatured state. J. Am. Chem. Soc. 2006, 128, 12735–12742. [Google Scholar] [CrossRef]
- Hughes, R.M.; Benshoff, M.L.; Waters, M.L. Effects of chain length and N-methylation on a cation-π interaction in a β-hairpin peptide. Chem. Eur. J. 2007, 13, 5753–5764. [Google Scholar] [CrossRef]
- Kiehna, S.E.; Laughrey, Z.R.; Waters, M.L. Evaluation of a carbohydrate–π interaction in a peptide model system. Chem. Commun. 2007, 4026–4028. [Google Scholar] [CrossRef]
- Laughrey, Z.R.; Kiehna, S.E.; Riemen, A.J.; Waters, M.L. Carbohydrate-π interactions: What are they worth? J. Am. Chem. Soc. 2008, 130, 14625–14633. [Google Scholar] [CrossRef] [Green Version]
- Hughes, R.M.; Waters, M.L. Effects of lysine acetylation in a β-hairpin peptide: Comparison of an amide-π and a cation-π interaction. J. Am. Chem. Soc. 2006, 128, 13586–13591. [Google Scholar] [CrossRef]
- Stanger, H.E.; Gellman, S.H. Rules for Antiparallel β-Sheet Design: D-Pro-Gly Is Superior to L-Asn-Gly for β-Hairpin Nucleation1. J. Am. Chem. Soc. 1998, 120, 4236–4237. [Google Scholar] [CrossRef]
- Syud, F.A.; Espinosa, J.F.; Gellman, S.H. NMR-Based Quantification of β-Sheet Populations in Aqueous Solution through Use of Reference Peptides for the Folded and Unfolded States. J. Am. Chem. Soc. 1999, 121, 11577–11578. [Google Scholar] [CrossRef]
- Chang, J.-Y.; Li, N.-Z.; Wang, W.-M.; Liu, C.-T.; Yu, C.-H.; Chen, Y.-C.; Lu, D.; Lin, P.-H.; Huang, C.-H.; Kono, O.; et al. Longer charged amino acids favor β-strand formation in hairpin peptides. J. Pept. Sci. 2021, 27, e3333. [Google Scholar] [CrossRef]
- Atherton, E.; Fox, H.; Harkiss, D.; Logan, C.J.; Sheppard, R.C.; Williams, B.J. A mild procedure for solid phase peptide synthesis: Use of fluorenylmethoxycarbonylamino-acids. J. Chem. Soc. Chem. Commun. 1978, 537–539. [Google Scholar] [CrossRef]
- Fields, G.B.; Noble, R.L. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int. J. Pept. Protein Res. 1990, 35, 161–214. [Google Scholar] [CrossRef]
- Volkmer-Engert, R.; Landgraf, C.; Schneider-Mergener, J. Charcoal surface-assisted catalysis of intramolecular disulfide bond formation in peptides. J. Pept. Res. 1998, 51, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Wüthrich, K. NMR of Proteins and Nucleic Acids; John Wiley & Sons: New York, NY, USA, 1986. [Google Scholar]
- Ramirez-Alvarado, M.; Blanco, F.J.; Serrano, L. De novo design and structural analysis of a model β-hairpin peptide system. Nat. Struct. Biol. 1996, 3, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Russell, S.J.; Blandl, T.; Skelton, N.J.; Cochran, A.G. Stability of Cyclic β-Hairpins: Asymmetric Contributions from Side Chains of a Hydrogen-Bonded Cross-Strand Residue Pair. J. Am. Chem. Soc. 2003, 125, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Dyson, H.; Wright, P.E. Chemical shift dispersion and secondary structure prediction in unfolded and partly folded proteins. FEBS Lett. 1997, 419, 285–289. [Google Scholar] [CrossRef] [Green Version]
- Dalgarno, D.C.; Levine, B.A.; Williams, R.J.P. Structural information from NMR secondary chemical shifts of peptide α C-H protons in proteins. Biosci. Rep. 1983, 3, 443–452. [Google Scholar] [CrossRef]
- Wishart, D.S.; Sykes, B.D.; Richards, F.M. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 1991, 222, 311–333. [Google Scholar] [CrossRef]
- Kim, Y.; Prestegard, J.H. Measurement of vicinal couplings from cross peaks in COSY spectra. J. Magn. Reson. 1989, 84, 9–13. [Google Scholar] [CrossRef]
- Pardi, A.; Billeter, M.; Wuthrich, K. Calibration of the angular dependence of the amide proton-Cα proton coupling constants, 3JNHα, in a globular protein. Use of 3JNHα for identification of helical secondary structure. J. Mol. Biol. 1984, 180, 741–751. [Google Scholar] [CrossRef]
- Horovitz, A. Double-mutant cycles: A powerful tool for analyzing protein structure and function. Fold. Des. 1996, 1, R121–R126. [Google Scholar] [CrossRef] [Green Version]
- Stanger, H.E.; Syud, F.A.; Espinosa, J.F.; Giriat, I.; Muir, T.; Gellman, S.H. Length-dependent stability and strand length limits in antiparallel β-sheet secondary structure. Proc. Natl. Acad. Sci. USA 2001, 98, 12015–12020. [Google Scholar] [CrossRef] [Green Version]
- Bax, A.; Davis, D.G. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 1985, 65, 355–360. [Google Scholar] [CrossRef]
- Bothner-by, A.A.; Stephens, R.L.; Lee, J.; Warren, C.D.; Jeanloz, R.W. Structure determination of a tetrasaccharide: Transient nuclear Overhauser effects in the rotating frame. J. Am. Chem. Soc. 1984, 106, 811–813. [Google Scholar] [CrossRef]
- Aue, W.P.; Bartholdi, E.; Ernst, R.R. Two-dimensional spectroscopy. Application to nuclear magnetic resonance. J. Chem. Phys. 1976, 64, 2229–2246. [Google Scholar] [CrossRef] [Green Version]
- Piotto, M.; Saudek, V.; Sklenar, V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 1992, 2, 661–665. [Google Scholar] [CrossRef]
- Butterfield, S.M.; Waters, M.L. A Designed β-Hairpin Peptide for Molecular Recognition of ATP in Water. J. Am. Chem. Soc. 2003, 125, 9580–9581. [Google Scholar] [CrossRef]
- Butterfield, S.M.; Cooper, W.J.; Waters, M.L. Minimalist Protein Design: A β-Hairpin Peptide That Binds ssDNA. J. Am. Chem. Soc. 2005, 127, 24–25. [Google Scholar] [CrossRef]
- Stewart, A.L.; Waters, M.L. Structural Effects on ss- and dsDNA Recognition by a β-Hairpin Peptide. ChemBioChem 2009, 10, 539–544. [Google Scholar] [CrossRef]
- Hamy, F.; Felder, E.R.; Heizmann, G.; Lazdins, J.; Aboul-Ela, F.; Varani, G.; Karn, J.; Klimkait, T. An inhibitor of the Tat/TAR RNA interaction that effectively supresses HIV-1 replication. Proc. Natl. Acad. Sci. USA 1997, 94, 3548–3553. [Google Scholar] [CrossRef] [Green Version]
- Runyon, S.T.; Puglisi, J.D. Design of a Cyclic Peptide that Targets a Viral RNA. J. Am. Chem. Soc. 2003, 125, 15704–15705. [Google Scholar] [CrossRef]
- Athanassiou, Z.; Dias, R.L.A.; Moehle, K.; Dobson, N.; Varani, G.; Robinson, J.A. Structural mimicry of retroviral Tat proteins by constrained β-hairpin peptidomimetics: Ligands with high affinity and seleectivity for viral TAR RNA regulatory elements. J. Am. Chem. Soc. 2004, 126, 6906–6913. [Google Scholar] [CrossRef]
- Moehle, K.; Athanassiou, Z.; Patora, K.; Davidson, A.; Varani, G.; Robinson, J.A. Design of β-Hairpin Peptidomimetics That Inhibit Binding of α-Helical HIV-1 Rev Protein to the Rev Response Element RNA. Angew. Chem. Int. Ed. 2007, 46, 9101–9104. [Google Scholar] [CrossRef] [Green Version]
- Davidson, A.; Leeper, T.C.; Athanassiou, Z.; Patora-Komisarska, K.; Karn, J.; Robinson, J.A.; Varani, G. Simultaneous recognition of HIV-1 TAR RNA bulge and loop sequences by cyclic peptide mimics of Tat protein. Proc. Natl. Acad. Sci. USA 2009, 106, 11931–11936. [Google Scholar] [CrossRef] [Green Version]
- Cline, L.L.; Waters, M.L. Design of a β-hairpin peptide-intercalator conjugate for simultaneous recognition of single stranded and double straded regions of RNA. Org. Biomol. Chem. 2009, 7, 4622–4630. [Google Scholar] [CrossRef]
- Drakopoulou, E.; Zinn-Justin, S.; Guenneugues, M.; Gilquin, B.; Ménez, A.; Vita, C. Changing the structural contect of a funtional β-hairpin. J. Biol. Chem. 1996, 271, 11979–11987. [Google Scholar] [CrossRef] [Green Version]
- González-Navarro, H.; Mora, P.; Pastor, M.; Serrano, L.; Mingarro, I.; Pérez-Payá, E. Identification of peptides that neutralize bacterial endotoxins using β-hairpin conformationally restricted libraries. Mol. Divers. 2000, 5, 117–126. [Google Scholar] [CrossRef]
- Lai, J.R.; Huck, B.R.; Weisblum, B.; Gellman, S.H. Design of Non-Cysteine-Containing Antimicrobial β-Hairpins: Structure−Activity Relationship Studies with Linear Protegrin-1 Analogues. Biochemistry 2002, 41, 12835–12842. [Google Scholar] [CrossRef]
- Schneider, J.P.; Pochan, D.J.; Ozbas, B.; Rajagopal, K.; Pakstis, L.; Kretsinger, J. Responsive Hydrogels from the Intramolecular Folding and Self-Assembly of a Designed Peptide. J. Am. Chem. Soc. 2002, 124, 15030–15037. [Google Scholar] [CrossRef]
- Pochan, D.J.; Schneider, J.P.; Kretsinger, J.; Ozbas, B.; Rajagopal, K.; Haines, L. Thermally Reversible Hydrogels via Intramolecular Folding and Consequent Self-Assembly of a de Novo Designed Peptide. J. Am. Chem. Soc. 2003, 125, 11802–11803. [Google Scholar] [CrossRef] [PubMed]
- Salick, D.A.; Kretsinger, J.K.; Pochan, D.J.; Schneider, J.P. Inherent Antibacterial Activity of a Peptide-Based β-Hairpin Hydrogel. J. Am. Chem. Soc. 2007, 129, 14793–14799. [Google Scholar] [CrossRef] [Green Version]
- Haines-Butterick, L.; Rajagopal, K.; Branco, M.; Salick, D.; Rughani, R.; Pilarz, M.; Lamm, M.S.; Pochan, D.J.; Schneider, J.P. Controlling hydrogelation kinetics by peptide design for three-dimensional encapsulation and injectable delivery of cells. Proc. Natl. Acad. Sci. USA 2007, 104, 7791–7796. [Google Scholar] [CrossRef] [Green Version]
- Nagarkar, R.P.; Hule, R.A.; Pochan, D.J.; Schneider, J.P. De Novo Design of Strand-Swapped β-Hairpin Hydrogels. J. Am. Chem. Soc. 2008, 130, 4466–4474. [Google Scholar] [CrossRef] [PubMed]
- Medina, S.H.; Li, S.; Howard, O.M.Z.; Dunlap, M.; Trivett, A.; Schneider, J.P.; Oppenheim, J.J. Enhanced immunostimulatory effects of DNA-encapsulated peptide hydrogels. Biomaterials 2015, 53, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Nagy-Smith, K.; Moore, E.; Schneider, J.; Tycko, R. Molecular structure of monomorphic peptide fibrils within a kinetically trapped hydrogel network. Proc. Natl. Acad. Sci. USA 2015, 112, 9816–9821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, Y.; Schneider, J.P. Fragmentation of Injectable Bioadhesive Hydrogels Affords Chemotherapeutic Macromolecules. Biomacromolecules 2016, 17, 2634–2641. [Google Scholar] [CrossRef]
- Medina, S.H.; Schneider, J.P. Cancer cell surface induced peptide folding allows intracellular translocation of drug. J. Control. Release 2015, 209, 317–326. [Google Scholar] [CrossRef] [Green Version]
- Medina, S.H.; Miller, S.E.; Keim, A.I.; Gorka, A.P.; Schnermann, M.J.; Schneider, J.P. An Intrinsically Disordered Peptide Facilitates Non-Endosomal Cell Entry. Angew. Chem. Int. Ed. 2016, 55, 3369–3372. [Google Scholar] [CrossRef]
- Cheng, Y.-S.; Chen, Z.-T.; Liao, T.-Y.; Lin, C.; Shen, H.C.-H.; Wang, Y.-H.; Chang, C.-W.; Liu, R.-S.; Chen, R.P.-Y.; Tu, P.-H. An intranasally delivered peptide drug ameliorates cognitive decline in Alzheimer transgenic mice. EMBO Mol. Med. 2017, 9, 703–715. [Google Scholar] [CrossRef]
- Watkins, A.M.; Arora, P.S. Anatomy of β-strands at protien-protein interfaces. ACS Chem. Biol. 2014, 9, 1747–1754. [Google Scholar] [CrossRef]
- Blosser, S.L.; Sawyer, N.; Maksimovic, I.; Ghosh, B.; Arora, P.S. Covalent and noncovalent targeting of the Tcf4/β-catenin strand interface with β-hairpin mimics. ACS Chem. Biol. 2021, 16, 1518–1525. [Google Scholar] [CrossRef]

| Peptide | δHN Range 1 (ppm) | Average δHN (ppm) | δHα Range 1 (ppm) | Average δHα (ppm) |
|---|---|---|---|---|
| HPDFAspDap | 1.145 | 8.697 ± 0.336 | 1.428 | 4.664 ± 0.426 |
| HPDAspDap | 0.728 | 8.406 ± 0.195 | 1.009 | 4.374 ± 0.276 |
| HPDUAspDap | 0.769 | 8.366 ± 0.196 | 0.782 | 4.351 ± 0.209 |
| HPDFAspDab | 1.169 | 8.662 ± 0.339 | 1.407 | 4.649 ± 0.406 |
| HPDAspDab | 0.665 | 8.386 ± 0.171 | 0.902 | 4.368 ± 0.265 |
| HPDUAspDab | 0.570 | 8.378 ± 0.153 | 0.693 | 4.318 ± 0.177 |
| HPDFAspOrn | 1.124 | 8.637 ± 0.333 | 1.397 | 4.622 ± 0.391 |
| HPDAspOrn | 0.530 | 8.346 ± 0.150 | 0.895 | 4.338 ± 0.250 |
| HPDUAspOrn | 0.429 | 8.317 ± 0.133 | 0.723 | 4.279 ± 0.193 |
| HPDFAspLys | 1.108 | 8.625 ± 0.329 | 1.373 | 4.608 ± 0.384 |
| HPDAspLys | 0.454 | 8.326 ± 0.135 | 0.878 | 4.328 ± 0.244 |
| HPDUAspLys | 0.432 | 8.302 ± 0.127 | 0.718 | 4.276 ± 0.192 |
| HPDFGluDap | 1.119 | 8.740 ± 0.330 | 1.506 | 4.673 ± 0.421 |
| HPDGluDap | 0.795 | 8.482 ± 0.206 | 1.135 | 4.404 ± 0.294 |
| HPDUGluDap | 0.574 | 8.398 ± 0.172 | 0.772 | 4.326 ± 0.187 |
| HPDFGluDab | 1.192 | 8.706 ± 0.337 | 1.467 | 4.653 ± 0.404 |
| HPDGluDab | 0.626 | 8.417 ± 0.179 | 1.008 | 4.398 ± 0.278 |
| HPDUGluDab | 0.373 | 8.368 ± 0.126 | 0.514 | 4.299 ± 0.148 |
| HPDFGluOrn | 1.166 | 8.695 ± 0.343 | 1.457 | 4.640 ± 0.394 |
| HPDGluOrn | 0.584 | 8.427 ± 0.176 | 0.977 | 4.369 ± 0.258 |
| HPDUGluOrn | 0.324 | 8.348 ± 0.106 | 0.494 | 4.289 ± 0.140 |
| HPDFGluLys | 1.217 | 8.686 ± 0.360 | 1.452 | 4.630 ± 0.390 |
| HPDGluLys | 0.618 | 8.427 ± 0.179 | 0.959 | 4.395 ± 0.237 |
| HPDUGluLys | 0.355 | 8.350 ± 0.102 | 0.584 | 4.278 ± 0.165 |
| HPDFAadDap | 1.135 | 8.738 ± 0.335 | 1.536 | 4.681 ± 0.424 |
| HPDAadDap | 0.793 | 8.476 ± 0.211 | 1.140 | 4.410 ± 0.296 |
| HPDUAadDap | 0.587 | 8.298 ± 0.169 | 0.782 | 4.244 ± 0.190 |
| HPDFAadDab | 1.183 | 8.702 ± 0.344 | 1.458 | 4.654 ± 0.402 |
| HPDAadDab | 0.675 | 8.445 ± 0.192 | 0.958 | 4.390 ± 0.268 |
| HPDUAadDab | 0.367 | 8.355 ± 0.116 | 0.515 | 4.300 ± 0.148 |
| HPDFAadOrn | 1.182 | 8.693 ± 0.340 | 1.442 | 4.638 ± 0.390 |
| HPDAadOrn | 0.535 | 8.405 ± 0.164 | 0.906 | 4.359 ± 0.247 |
| HPDUAadOrn | 0.273 | 8.333 ± 0.092 | 0.513 | 4.261 ± 0.163 |
| HPDFAadLys | 1.207 | 8.687 ± 0.357 | 1.455 | 4.629 ± 0.387 |
| HPDAadLys | 0.599 | 8.399 ± 0.172 | 0.927 | 4.360 ± 0.249 |
| HPDUAadLys | 0.276 | 8.317 ± 0.084 | 0.530 | 4.255 ± 0.165 |
| Zbb | Xaa | HPDFZbbXaa | HPDZbbXaa | HPDUZbbXaa |
|---|---|---|---|---|
| Asp | Dap | 11 ± 0.7 | 9.8 ± 1.0 | 10 ± 1.2 |
| Asp | Dab | 11 ± 1.6 | 9.4 ± 1.5 | 8.3 ± 2.2 |
| Asp | Orn | 12 ± 0.7 | 10 ± 1.0 | 9.7 ± 0.8 |
| Asp | Lys | 12 ± 0.6 | 10 ± 0.8 | 10 ± 1.0 |
| Glu | Dap | 10 ± 0.9 | 9.9 ± 0.9 | 9.6 ± 1.3 |
| Glu | Dab | 10 ± 1.1 | 9.5 ± 2.1 | 9.2 ± 1.6 |
| Glu | Orn | 11 ± 0.6 | 10 ± 0.6 | 10 ± 0.6 |
| Glu | Lys | 11 ± 1.1 | 9.6 ± 0.9 | 9.2 ± 1.0 |
| Aad | Dap | 11 ± 0.9 | 9.7 ± 1.4 | 9.9 ± 1.0 |
| Aad | Dab | 10 ± 1.0 | 9.4 ± 1.7 | 9.8 ± 1.0 |
| Aad | Orn | 12 ± 0.7 | 10 ± 0.7 | 9.3 ± 0.5 |
| Aad | Lys | 11 ± 0.5 | 9.9 ± 0.5 | 9.6 ± 0.7 |
| Xaa9 | Zbb2 | ||
|---|---|---|---|
| Asp | Glu | Aad | |
| Dap | 19 ± 4 | 38 ± 4 | 47 ± 4 |
| Dab | 25 ± 4 | 42 ± 3 | 40 ± 3 |
| Orn | 17 ± 4 | 36 ± 3 | 33 ± 3 |
| Lys | 15 ± 5 | 36 ± 4 | 36 ± 3 |
| Xaa9 | Zbb2 | ||
|---|---|---|---|
| Asp | Glu | Aad | |
| Dap | 0.864 ± 0.177 | 0.303 ± 0.091 | 0.062 ± 0.085 |
| Dab | 0.665 ± 0.143 | 0.185 ± 0.065 | 0.231 ± 0.070 |
| Orn | 0.975 ± 0.188 | 0.347 ± 0.075 | 0.424 ± 0.077 |
| Lys | 1.031 ± 0.231 | 0.333 ± 0.107 | 0.345 ± 0.086 |
| Xaa9 | Zbb2 | ||
|---|---|---|---|
| Asp | Glu | Aad | |
| Dap | −0.434 ± 0.204 | −0.412 ± 0.043 | −0.425 ± 0.077 |
| Dab | −0.794 ± 0.139 | −0.585 ± 0.058 | −0.284 ± 0.030 |
| Orn | −0.375 ± 0.081 | −0.291 ± 0.048 | −0.023 ± 0.040 |
| Lys | −0.206 ± 0.018 | −0.258 ± 0.026 | −0.020 ± 0.045 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, J.-Y.; Pan, Y.-J.; Huang, P.-Y.; Sun, Y.-T.; Yu, C.-H.; Ning, Z.-J.; Huang, S.-L.; Huang, S.-J.; Cheng, R.P. The Effects of Charged Amino Acid Side-Chain Length on Diagonal Cross-Strand Interactions between Carboxylate- and Ammonium-Containing Residues in a β-Hairpin. Molecules 2022, 27, 4172. https://doi.org/10.3390/molecules27134172
Chang J-Y, Pan Y-J, Huang P-Y, Sun Y-T, Yu C-H, Ning Z-J, Huang S-L, Huang S-J, Cheng RP. The Effects of Charged Amino Acid Side-Chain Length on Diagonal Cross-Strand Interactions between Carboxylate- and Ammonium-Containing Residues in a β-Hairpin. Molecules. 2022; 27(13):4172. https://doi.org/10.3390/molecules27134172
Chicago/Turabian StyleChang, Jing-Yuan, Yen-Jin Pan, Pei-Yu Huang, Yi-Ting Sun, Chen-Hsu Yu, Zhi-Jun Ning, Shou-Ling Huang, Shing-Jong Huang, and Richard P. Cheng. 2022. "The Effects of Charged Amino Acid Side-Chain Length on Diagonal Cross-Strand Interactions between Carboxylate- and Ammonium-Containing Residues in a β-Hairpin" Molecules 27, no. 13: 4172. https://doi.org/10.3390/molecules27134172
APA StyleChang, J.-Y., Pan, Y.-J., Huang, P.-Y., Sun, Y.-T., Yu, C.-H., Ning, Z.-J., Huang, S.-L., Huang, S.-J., & Cheng, R. P. (2022). The Effects of Charged Amino Acid Side-Chain Length on Diagonal Cross-Strand Interactions between Carboxylate- and Ammonium-Containing Residues in a β-Hairpin. Molecules, 27(13), 4172. https://doi.org/10.3390/molecules27134172







