Zebrafish—An Optimal Model in Experimental Oncology
Abstract
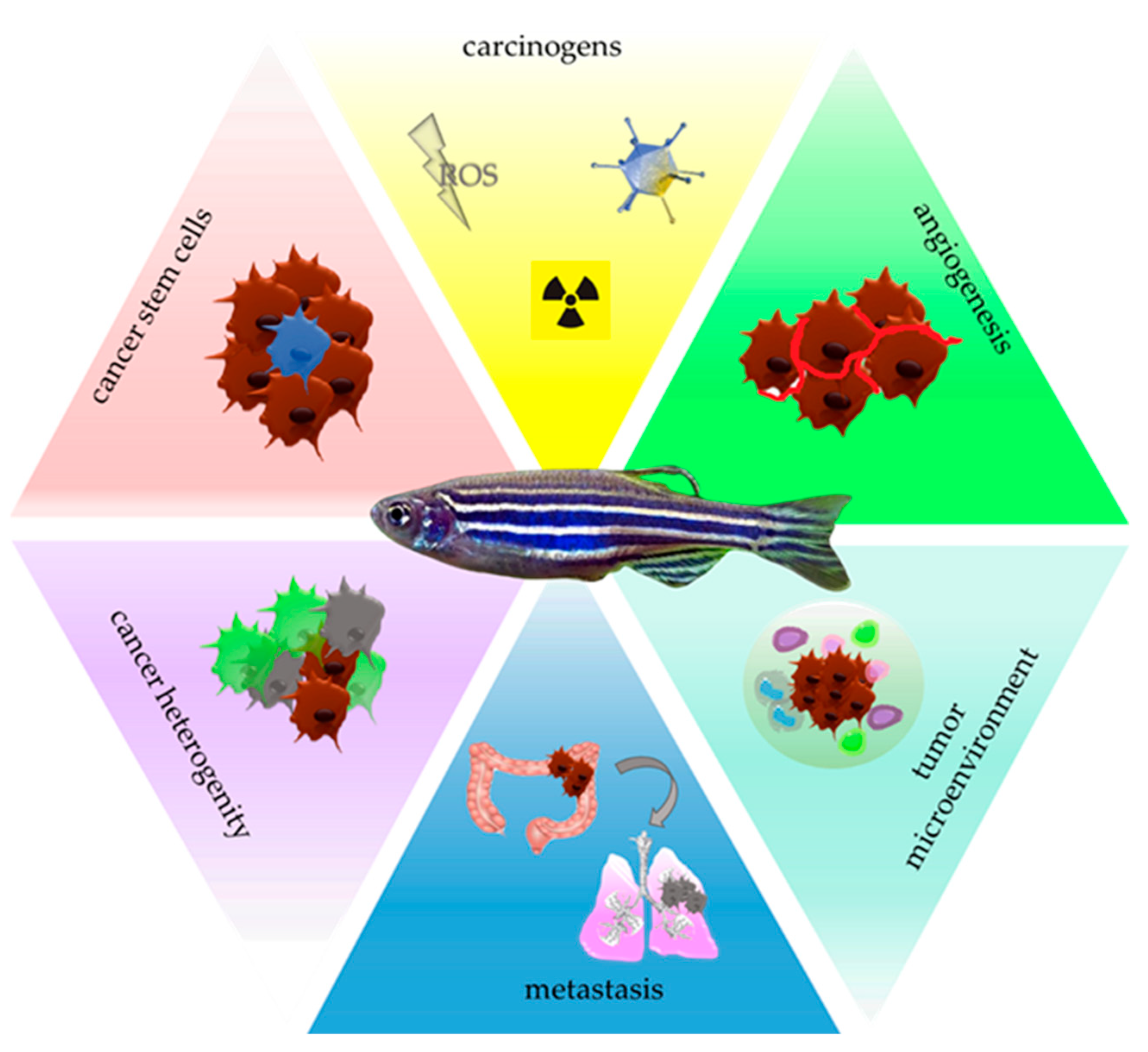
:1. Introduction
2. Zebrafish—A Tool for Carcinogens Screening
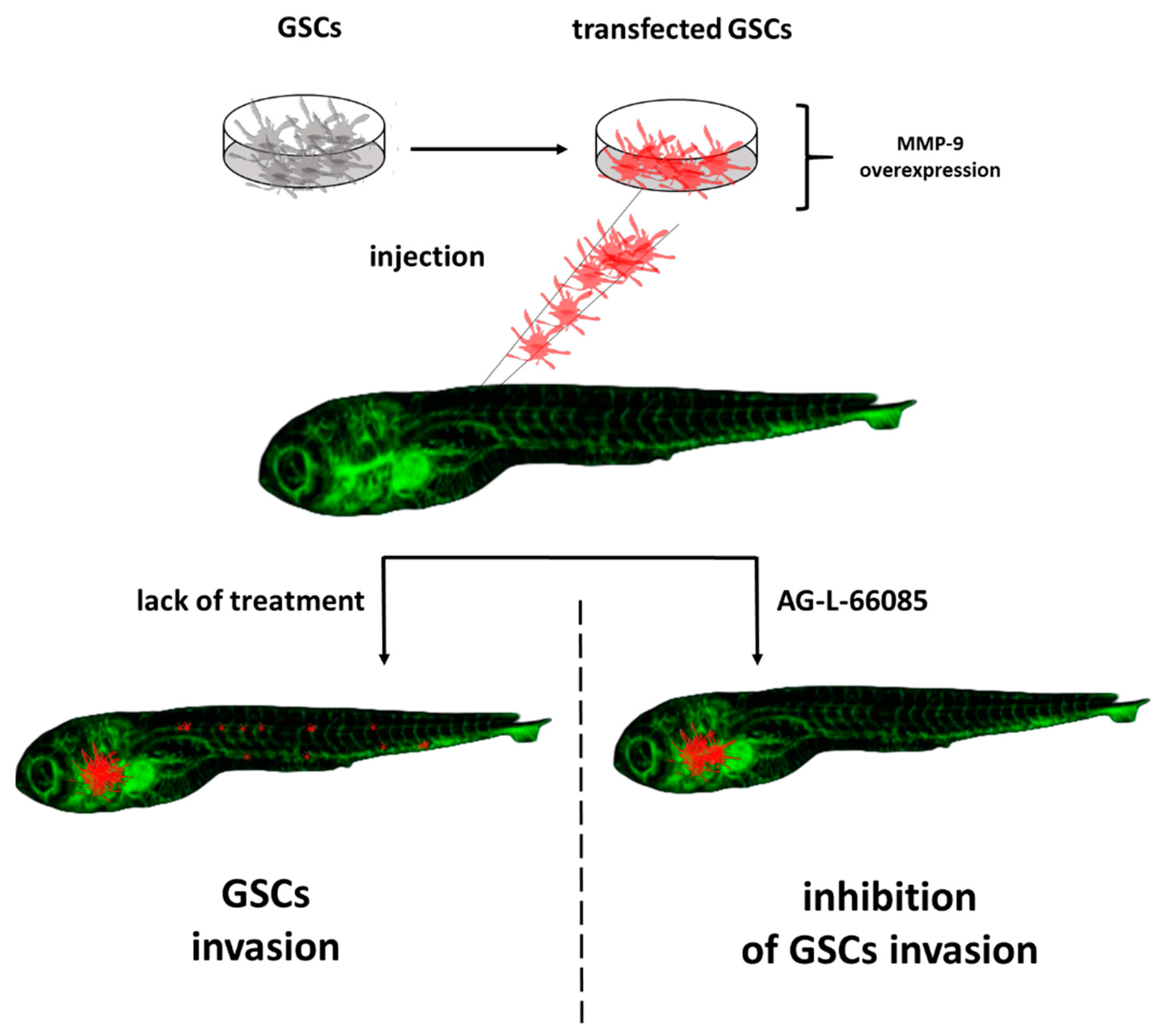
3. Zebrafish—A Tool for Studying Cancer Stem Cells
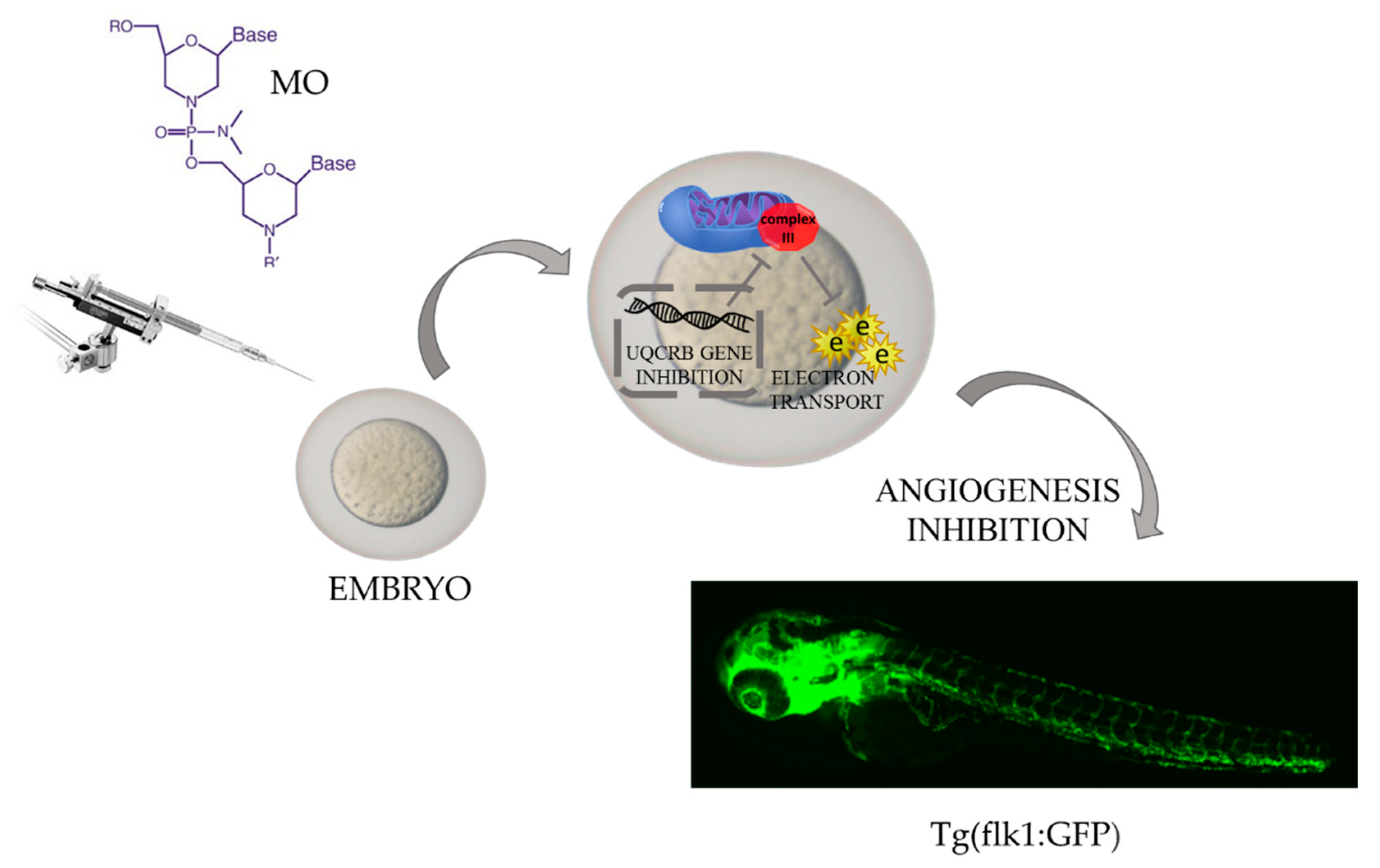
4. Zebrafish—A Tool for Studying Angiogenesis
5. Zebrafish—A Tool for Investigating Circulating Tumor Cells
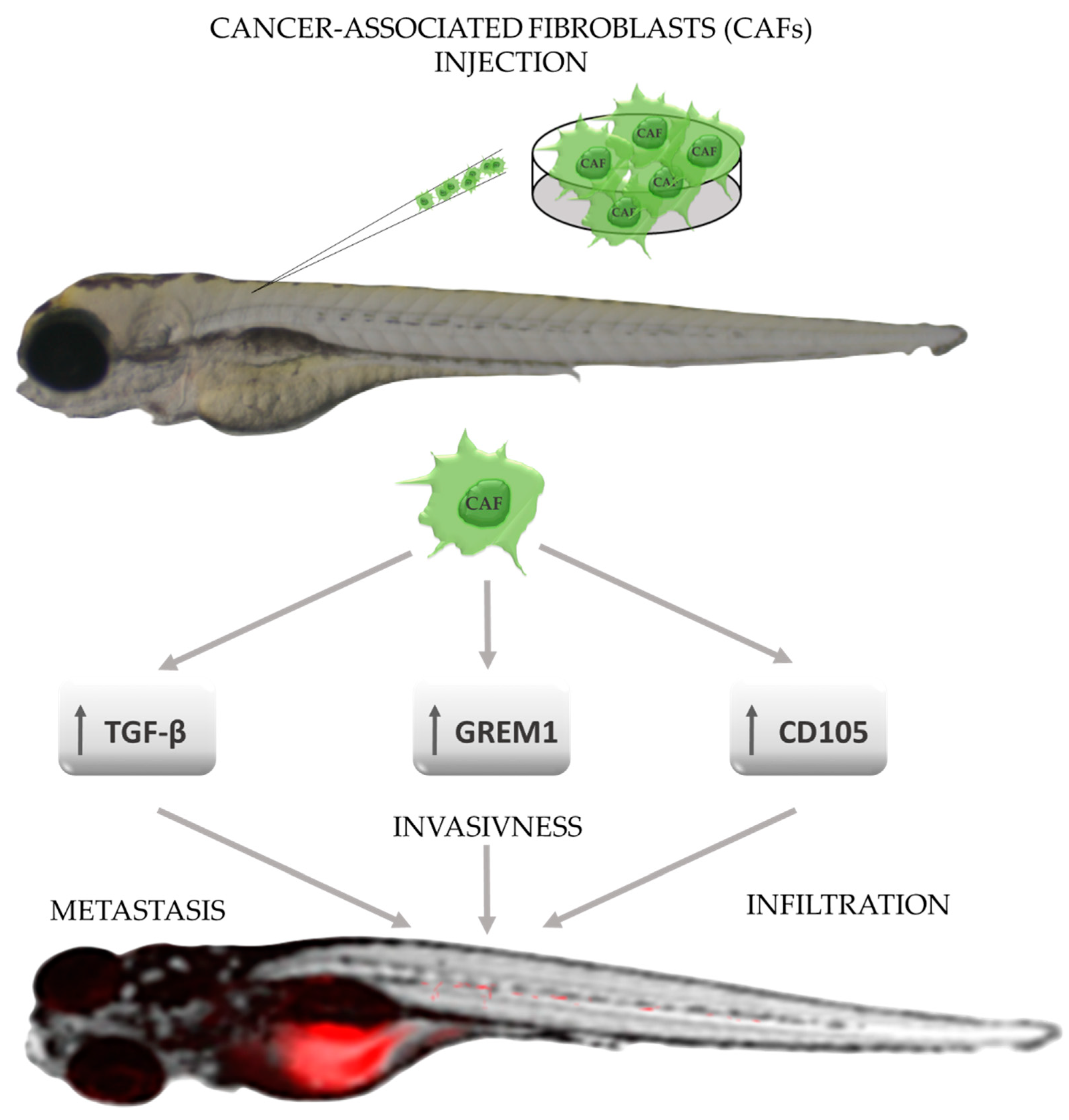
6. Zebrafish—A Tool for Studying the Tumor Microenvironment
7. Zebrafish—A Tool for Facing Other Challenges of Modern Oncology
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, J.T. Human-zebrafish non-coding conserved elements act in vivo to regulate transcription. Nucleic Acids Res. 2005, 33, 5437–5445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giardoglou, P.; Beis, D. On Zebrafish Disease Models and Matters of the Heart. Biomedicines 2019, 7, 15. [Google Scholar] [CrossRef] [Green Version]
- Outtandy, P.; Russell, C.; Kleta, R.; Bockenhauer, D. Zebrafish as a model for kidney function and disease. Pediatr. Nephrol. 2019, 34, 751–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubińska-Magiera, M.; Daczewska, M.; Lewicka, A.; Migocka-Patrzałek, M.; Niedbalska-Tarnowska, J.; Jagla, K. Zebrafish: A Model for the Study of Toxicants Affecting Muscle Development and Function. Int. J. Mol. Sci. 2016, 17, 1941. [Google Scholar] [CrossRef]
- Cox, A.G.; Goessling, W. The lure of zebrafish in liver research: Regulation of hepatic growth in development and regeneration. Curr. Opin. Genet. Dev. 2015, 32, 153–161. [Google Scholar] [CrossRef] [Green Version]
- Santoriello, C.; Zon, L.I. Hooked! Modeling human disease in zebrafish. J. Clin. Investig. 2012, 122, 2337–2343. [Google Scholar] [CrossRef] [Green Version]
- Meeker, N.D.; Trede, N.S. Immunology and zebrafish: Spawning new models of human disease. Dev. Comp. Immunol. 2008, 32, 745–757. [Google Scholar] [CrossRef]
- Smith, M.T.; Guyton, K.Z.; Gibbons, C.F.; Fritz, J.; Portier, C.; Rusyn, I.; DeMarini, D.; Caldwell, J.C.; Kavlock, R.J.; Lambert, P.F.; et al. Key Characteristics of Carcinogens as a Basis for Organizing Data on Mechanisms of Carcinogenesis. Environ. Health Perspect. 2016, 124, 713–721. [Google Scholar] [CrossRef] [Green Version]
- Pei, D.-S.; Strauss, P.R. Zebrafish as a model system to study DNA damage and repair. Mutat. Res. Mol. Mech. Mutagen. 2013, 743–744, 151–159. [Google Scholar] [CrossRef]
- Araldi, R.P.; de Melo, T.C.; Mendes, T.B.; de Sá Júnior, P.L.; Nozima, B.H.N.; Ito, E.T.; de Carvalho, R.F.; de Souza, E.B.; de Cassia Stocco, R. Using the comet and micronucleus assays for genotoxicity studies: A review. Biomed. Pharmacother. 2015, 72, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Le Bihanic, F.; Di Bucchianico, S.; Karlsson, H.L.; Dreij, K. In vivo micronucleus screening in zebrafish by flow cytometry. Mutagenesis 2016, 31, 643–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Šrut, M.; Štambuk, A.; Bourdineaud, J.-P.; Klobučar, G.I.V. Zebrafish genome instability after exposure to model genotoxicants. Ecotoxicology 2015, 24, 887–902. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Huang, C.; Bai, C.; Du, C.; Liao, J.; Dong, Q. In vivo DNA mismatch repair measurement in zebrafish embryos and its use in screening of environmental carcinogens. J. Hazard. Mater. 2016, 302, 296–303. [Google Scholar] [CrossRef]
- Mourabit, S.; Fitzgerald, J.A.; Ellis, R.; Takesono, A.; Porteus, C.S.; Trznadel, M.; Metz, J.; Winter, M.J.; Kudoh, T.; Tyler, C.R. New insights into organ-specific oxidative stress mechanisms using a novel biosensor zebrafish. Environ. Int. 2019, 133, 105138. [Google Scholar] [CrossRef]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gamble, J.; Elson, D.; Greenwood, J.; Tanguay, R.; Kolluri, S. The Zebrafish Xenograft Models for Investigating Cancer and Cancer Therapeutics. Biology 2021, 10, 252. [Google Scholar] [CrossRef]
- Cornet, C.; Dyballa, S.; Terriente, J.; Di Giacomo, V. ZeOncoTest: Refining and Automating the Zebrafish Xenograft Model for Drug Discovery in Cancer. Pharmaceuticals 2019, 13, 1. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Salim, L.; Yan, C.; Gong, Z. Rapid Analysis of Effects of Environmental Toxicants on Tumorigenesis and Inflammation Using a Transgenic Zebrafish Model for Liver Cancer. Mar. Biotechnol. 2019, 21, 396–405. [Google Scholar] [CrossRef]
- Rodgers, K.M.; Udesky, J.O.; Rudel, R.A.; Brody, J.G. Environmental chemicals and breast cancer: An updated review of epidemiological literature informed by biological mechanisms. Environ. Res. 2018, 160, 152–182. [Google Scholar] [CrossRef]
- Wielsøe, M.; Kern, P.; Bonefeld-Jørgensen, E.C. Serum levels of environmental pollutants is a risk factor for breast cancer in Inuit: A case control study. Environ. Health 2017, 16, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rattray, N.J.W.; Charkoftaki, G.; Rattray, Z.; Hansen, J.E.; Vasiliou, V.; Johnson, C.H. Environmental Influences in the Etiology of Colorectal Cancer: The Premise of Metabolomics. Curr. Pharmacol. Rep. 2017, 3, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Sohrabi, M.; Gholami, A.; Azar, M.H.; Yaghoobi, M.; Shahi, M.M.; Shirmardi, S.; Nikkhah, M.; Kohi, Z.; Salehpour, D.; Khoonsari, M.R.; et al. Trace Element and Heavy Metal Levels in Colorectal Cancer: Comparison Between Cancerous and Non-cancerous Tissues. Biol. Trace Element Res. 2017, 183, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Shankar, A.; Dubey, A.; Saini, D.; Singh, M.; Prasad, C.P.; Roy, S.; Bharati, S.J.; Rinki, M.; Singh, N.; Seth, T.; et al. Environmental and occupational determinants of lung cancer. Transl. Lung Cancer Res. 2019, 8, S31–S49. [Google Scholar] [CrossRef]
- Xu, X.; Nie, S.; Ding, H.; Hou, F.F. Environmental pollution and kidney diseases. Nat. Rev. Nephrol. 2018, 14, 313–324. [Google Scholar] [CrossRef]
- Scelo, G.; Larose, T.L. Epidemiology and Risk Factors for Kidney Cancer. J. Clin. Oncol. 2018, 36, 3574–3581. [Google Scholar] [CrossRef]
- Yuan, W.; Yang, N.; Li, X. Advances in Understanding How Heavy Metal Pollution Triggers Gastric Cancer. BioMed Res. Int. 2016, 2016, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Georgescu, S.R.; Mitran, C.I.; Mitran, M.I.; Caruntu, C.; Sarbu, M.I.; Matei, C.; Nicolae, I.; Tocut, M.; Popa, M.I.; Tampa, M. New Insights in the Pathogenesis of HPV Infection and the Associated Carcinogenic Processes: The Role of Chronic Inflammation and Oxidative Stress. J. Immunol. Res. 2018, 2018, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, A.; Ray, R.B.; Ray, R. Oncogenic Potential of Hepatitis C Virus Proteins. Viruses 2010, 2, 2108–2133. [Google Scholar] [CrossRef]
- De Mitri, M.S.; Cassini, R.; Bernardi, M. Hepatitis B virus-related hepatocarcinogenesis: Molecular oncogenic potential of clear or occult infections. Eur. J. Cancer 2010, 46, 2178–2186. [Google Scholar] [CrossRef]
- Rickinson, A. Co-infections, inflammation and oncogenesis: Future directions for EBV research. Semin. Cancer Biol. 2014, 26, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Chen, J.-R.; Hsu, C.-H.; Li, Y.-H.; Chen, Y.-M.; Lin, C.-Y.; Huang, S.-J.; Chang, Z.-K.; Chen, Y.-C.; Lin, C.-H.; et al. A zebrafish model of intrahepatic cholangiocarcinoma by dual expression of hepatitis B virus X and hepatitis C virus core protein in liver. Hepatology 2012, 56, 2268–2276. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Pestell, T.G.; Lisanti, M.P.; Pestell, R.G. Cancer stem cells. Int. J. Biochem. Cell Biol. 2012, 44, 2144–2151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Najafi, M.; Farhood, B.; Mortezaee, K. Cancer stem cells (CSCs) in cancer progression and therapy. J. Cell. Physiol. 2019, 234, 8381–8395. [Google Scholar] [CrossRef] [PubMed]
- Kretzschmar, K.; Watt, F.M. Lineage Tracing. Cell 2012, 148, 33–45. [Google Scholar] [CrossRef] [Green Version]
- Jones, M.G.; Khodaverdian, A.; Quinn, J.J.; Chan, M.M.; Hussmann, J.A.; Wang, R.; Xu, C.; Weissman, J.S.; Yosef, N. Inference of single-cell phylogenies from lineage tracing data using Cassiopeia. Genome Biol. 2020, 21, 92. [Google Scholar] [CrossRef] [Green Version]
- Rycaj, K.; Tang, D.G. Cell-of-Origin of Cancer versus Cancer Stem Cells: Assays and Interpretations. Cancer Res. 2015, 75, 4003–4011. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Groenewoud, A.; Tulotta, C.; Zoni, E.; Julio, M.K.-D.; van der Horst, G.; van der Pluijm, G.; Snaar-Jagalska, B.E. A zebrafish xenograft model for studying human cancer stem cells in distant metastasis and therapy response. In Methods In Cell Biology; Elsevier: Amsterdam, The Netherlands, 2017; Volume 138, pp. 471–496. [Google Scholar] [CrossRef]
- Zhang, B.; Shimada, Y.; Kuroyanagi, J.; Nishimura, Y.; Umemoto, N.; Nomoto, T.; Shintou, T.; Miyazaki, T.; Tanaka, T. Zebrafish xenotransplantation model for cancer stem-like cell study and high-throughput screening of inhibitors. Tumor Biol. 2014, 35, 11861–11869. [Google Scholar] [CrossRef]
- Yang, Y.; Hao, E.; Pan, X.; Tan, D.; Du, Z.; Xie, J.; Hou, X.; Deng, J.; Wei, K. Gomisin M2 from Baizuan suppresses breast cancer stem cell proliferation in a zebrafish xenograft model. Aging 2019, 11, 8347–8361. [Google Scholar] [CrossRef]
- Yang, X.-J.; Cui, W.; Gu, A.; Xu, C.; Yu, S.-C.; Li, T.-T.; Cui, Y.-H.; Zhang, X.; Bian, X.-W. A Novel Zebrafish Xenotransplantation Model for Study of Glioma Stem Cell Invasion. PLoS ONE 2013, 8, e61801. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.-B.; Tang, Y.-L.; Liang, X.-H. Targeting VEGF pathway to normalize the vasculature: An emerging insight in cancer therapy. OncoTargets Ther. 2018, 11, 6901–6909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.; Chen, T.; Ding, Z.; Wang, Y.; Wei, Y.; Wei, X. Inhibition of FGF-FGFR and VEGF-VEGFR signalling in cancer treatment. Cell Prolif. 2021, 54, e13009. [Google Scholar] [CrossRef] [PubMed]
- Omorphos, N.P.; Gao, C.; Tan, S.S.; Sangha, M.S. Understanding angiogenesis and the role of angiogenic growth factors in the vascularisation of engineered tissues. Mol. Biol. Rep. 2021, 48, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Parmar, D.; Apte, M. Angiopoietin inhibitors: A review on targeting tumor angiogenesis. Eur. J. Pharmacol. 2021, 899, 174021. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.; Singh, R.K. Chemokines orchestrate tumor cells and the microenvironment to achieve metastatic heterogeneity. Cancer Metastasis Rev. 2021, 40, 447–476. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Fabián, S.; Arreola, R.; Becerril-Villanueva, E.; Torres-Romero, J.C.; Arana-Argáez, V.; Lara-Riegos, J.; Ramírez-Camacho, M.A.; Alvarez-Sánchez, M.E. Role of Matrix Metalloproteinases in Angiogenesis and Cancer. Front. Oncol. 2019, 9, 1370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharif, P.M.; Jabbari, P.; Razi, S.; Keshavarz-Fathi, M.; Rezaei, N. Importance of TNF-alpha and its alterations in the development of cancers. Cytokine 2020, 130, 155066. [Google Scholar] [CrossRef]
- Lugano, R.; Ramachandran, M.; Dimberg, A. Tumor angiogenesis: Causes, consequences, challenges and opportunities. Cell. Mol. Life Sci. 2020, 77, 1745–1770. [Google Scholar] [CrossRef] [Green Version]
- Muz, B.; de la Puente, P.; Azab, F.; Azab, A.K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia 2015, 3, 83–92. [Google Scholar] [CrossRef] [Green Version]
- Mortara, L.; Benest, A.V.; Bates, D.O.; Noonan, D.M. Can the co-dependence of the immune system and angiogenesis facilitate pharmacological targeting of tumours? Curr. Opin. Pharmacol. 2017, 35, 66–74. [Google Scholar] [CrossRef]
- Guerra, J.; Tobia, C.; Presta, M.; Barbieri, A. Zebrafish embryo as an experimental model to study tumor angiogenesis. In Tumor Vascularization; Elsevier: Amsterdam, The Netherlands, 2020; pp. 129–145. [Google Scholar] [CrossRef]
- Tobia, C.; Gariano, G.; De Sena, G.; Presta, M. Zebrafish embryo as a tool to study tumor/endothelial cell cross-talk. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2013, 1832, 1371–1377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.-J.; Chen, G.-L.; Yu, S.-C.; Xu, C.; Xin, Y.-H.; Li, T.-T.; Shi, Y.; Gu, A.; Duan, J.-J.; Qian, C.; et al. TGF-β1 enhances tumor-induced angiogenesis via JNK pathway and macrophage infiltration in an improved zebrafish embryo/xenograft glioma model. Int. Immunopharmacol. 2012, 15, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Britto, D.D.; Wyroba, B.; Chen, W.; Lockwood, R.A.; Tran, K.B.; Shepherd, P.R.; Hall, C.; Crosier, K.E.; Crosier, P.S.; Astin, J.W. Macrophages enhance Vegfa-driven angiogenesis in an embryonic zebrafish tumour xenograft model. Dis. Model. Mech. 2018, 11, dmm035998. [Google Scholar] [CrossRef] [Green Version]
- Duan, J.; Hu, H.; Feng, L.; Yang, X.; Sun, Z. Silica nanoparticles inhibit macrophage activity and angiogenesis via VEGFR2-mediated MAPK signaling pathway in zebrafish embryos. Chemosphere 2017, 183, 483–490. [Google Scholar] [CrossRef] [PubMed]
- Sie, Z.-L.; Li, R.-Y.; Sampurna, B.P.; Hsu, P.-J.; Liu, S.-C.; Wang, H.-D.; Huang, C.-L.; Yuh, C.-H. WNK1 Kinase Stimulates Angiogenesis to Promote Tumor Growth and Metastasis. Cancers 2020, 12, 575. [Google Scholar] [CrossRef] [Green Version]
- Basnet, R.; Zizioli, D.; Muscò, A.; Finazzi, D.; Sigala, S.; Rossini, E.; Tobia, C.; Guerra, J.; Presta, M.; Memo, M. Caffeine Inhibits Direct and Indirect Angiogenesis in Zebrafish Embryos. Int. J. Mol. Sci. 2021, 22, 4856. [Google Scholar] [CrossRef]
- Letrado, P.; de Miguel, I.; Lamberto, I.; Díez-Martínez, R.; Oyarzabal, J. Zebrafish: Speeding Up the Cancer Drug Discovery Process. Cancer Res. 2018, 78, 6048–6058. [Google Scholar] [CrossRef] [Green Version]
- Kobar, K.; Collett, K.; Prykhozhij, S.V.; Berman, J.N. Zebrafish Cancer Predisposition Models. Front. Cell Dev. Biol. 2021, 9, 660069. [Google Scholar] [CrossRef]
- Gauert, A.; Olk, N.; Pimentel-Gutiérrez, H.; Astrahantseff, K.; Jensen, L.D.; Cao, Y.; Eggert, A.; Eckert, C.; Hagemann, A.I. Fast, In Vivo Model for Drug-Response Prediction in Patients with B-Cell Precursor Acute Lymphoblastic Leukemia. Cancers 2020, 12, 1883. [Google Scholar] [CrossRef]
- Cho, Y.S.; Jung, H.J.; Seok, S.H.; Payumo, A.Y.; Chen, J.K.; Kwon, H.J. Functional inhibition of UQCRB suppresses angiogenesis in zebrafish. Biochem. Biophys. Res. Commun. 2013, 433, 396–400. [Google Scholar] [CrossRef] [Green Version]
- Hollenbach, M.; Stoll, S.J.; Jörgens, K.; Seufferlein, T.; Kroll, J. Different Regulation of Physiological and Tumor Angiogenesis in Zebrafish by Protein Kinase D1 (PKD1). PLoS ONE 2013, 8, e68033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Attri, K.S.; Mehla, K.; Singh, P.K. Evaluation of Macrophage Polarization in Pancreatic Cancer Microenvironment Under Hypoxia. In Methods in Molecular Biology; Huang, L.E., Ed.; Springer: New York, NY, USA, 2018; Volume 1742, pp. 265–276. [Google Scholar] [CrossRef]
- Triner, D.; Shah, Y.M. Hypoxia-inducible factors: A central link between inflammation and cancer. J. Clin. Investig. 2016, 126, 3689–3698. [Google Scholar] [CrossRef] [PubMed]
- Daniel, S.K.; Sullivan, K.M.; Labadie, K.P.; Pillarisetty, V.G. Hypoxia as a barrier to immunotherapy in pancreatic adenocarcinoma. Clin. Transl. Med. 2019, 8, 10. [Google Scholar] [CrossRef] [PubMed]
- Al Tameemi, W.; Dale, T.P.; Al-Jumaily, R.M.K.; Forsyth, N.R. Hypoxia-Modified Cancer Cell Metabolism. Front. Cell Dev. Biol. 2019, 7, 4. [Google Scholar] [CrossRef] [Green Version]
- Elks, P.M.; Renshaw, S.A.; Meijer, A.H.; Walmsley, S.R.; van Eeden, F.J. Exploring the HIFs, buts and maybes of hypoxia signalling in disease: Lessons from zebrafish models. Dis. Model. Mech. 2015, 8, 1349–1360. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Wu, C.; Yang, C.; Li, G.; Han, Q.; Li, S.; Lee, S.M.-Y.; Leung, C.-H.; Ma, D.-L. A dual-functional luminescent probe for imaging H2S in living zebrafish and discrimination hypoxic cells from normoxic cells. Sens. Actuators B Chem. 2018, 255, 1953–1959. [Google Scholar] [CrossRef]
- Marchi, D.; Santhakumar, K.; Markham, E.; Li, N.; Storbeck, K.-H.; Krone, N.; Cunliffe, V.T.; van Eeden, F.J.M. Bidirectional crosstalk between Hypoxia-Inducible Factor and glucocorticoid signalling in zebrafish larvae. PLoS Genet. 2020, 16, e1008757. [Google Scholar] [CrossRef]
- Lewis, A.; Elks, P.M. Hypoxia Induces Macrophage tnfa Expression via Cyclooxygenase and Prostaglandin E2 in vivo. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Zhao, Y.; Huang, X.; Ding, T.W.; Gong, Z. Enhanced angiogenesis, hypoxia and neutrophil recruitment during Myc-induced liver tumorigenesis in zebrafish. Sci. Rep. 2016, 6, 31952. [Google Scholar] [CrossRef] [Green Version]
- Follain, G.; Osmani, N.; Fuchs, C.; Allio, G.; Harlepp, S.; Goetz, J.G. Using the zebrafish embryo to dissect the early steps of the metastasis cascade. In Cell Migration. Methods in Molecular Biology; Gautreau, A., Ed.; Springer: New York, NY, USA, 2018; Volume 1749, pp. 195–211. [Google Scholar] [CrossRef]
- Follain, G.; Osmani, N.; Azevedo, A.S.; Allio, G.; Mercier, L.; Karreman, M.A.; Solecki, G.; Leòn, M.J.G.; Lefebvre, O.; Fekonja, N.; et al. Hemodynamic Forces Tune the Arrest, Adhesion, and Extravasation of Circulating Tumor Cells. Dev. Cell 2018, 45, 33–52.e12. [Google Scholar] [CrossRef] [Green Version]
- Allen, T.A.; Asad, D.; Amu, E.; Hensley, M.T.; Cores, J.; Vandergriff, A.; Tang, J.; Dinh, P.-U.; Shen, D.; Qiao, L.; et al. Circulating tumor cells exit circulation while maintaining multicellularity augmenting metastatic potential. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, Y.; Fang, F.; Zhang, Q. Circulating tumor cell clusters: What we know and what we expect (Review). Int. J. Oncol. 2016, 49, 2206–2216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, T.; Amu, E.; Asad, D.; Cheng, K. Abstract 90: Metastatic melanoma and cervical tumor cell clusters can exit blood vessels through angiopellosis augmenting tumor formation ability. Tumor Biol. Am. Assoc. Cancer Res. 2018, 78, 90. [Google Scholar] [CrossRef]
- Martínez-Pena, I.; Hurtado, P.; Carmona-Ule, N.; Abuín, C.; Dávila-Ibáñez, A.B.; Sánchez, L.; Abal, M.; Chaachou, A.; Hernández-Losa, J.; Cajal, S.R.Y.; et al. Dissecting Breast Cancer Circulating Tumor Cells Competence via Modelling Metastasis in Zebrafish. Int. J. Mol. Sci. 2021, 22, 9279. [Google Scholar] [CrossRef]
- Amintas, S.; Bedel, A.; Moreau-Gaudry, F.; Boutin, J.; Buscail, L.; Merlio, J.-P.; Vendrely, V.; Dabernat, S.; Buscail, E. Circulating Tumor Cell Clusters: United We Stand Divided We Fall. Int. J. Mol. Sci. 2020, 21, 2653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fieuws, C.; Vierstraete, J.; Van De Vijver, K.; Denys, H.; Claes, K.B. Abstract B25: Isolation and engraftment of circulating tumor cells into zebrafish embryos to predict tumor response of ovarian cancer patients. Clin. Poster Present. Proffered Abstracts. Am. Assoc. Clin. Cancer Res. 2020, 26, B25. [Google Scholar] [CrossRef]
- Hanahan, D.; Coussens, L.M. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef] [Green Version]
- Ishii, G.; Ochiai, A.; Neri, S. Phenotypic and functional heterogeneity of cancer-associated fibroblast within the tumor microenvironment. Adv. Drug Deliv. Rev. 2016, 99, 186–196. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, Y.; Lim, S.; Hosaka, K.; Yang, Y.; Pavlova, T.; Alkasalias, T.; Hartman, J.; Jensen, L.; Xing, X.; et al. A Zebrafish Model Discovers a Novel Mechanism of Stromal Fibroblast-Mediated Cancer Metastasis. Clin. Cancer Res. 2017, 23, 4769–4779. [Google Scholar] [CrossRef] [Green Version]
- Sun, D.-Y.; Wu, J.-Q.; He, Z.-H.; He, M.-F.; Sun, H.-B. Cancer-associated fibroblast regulate proliferation and migration of prostate cancer cells through TGF-β signaling pathway. Life Sci. 2019, 235, 116791. [Google Scholar] [CrossRef]
- Ren, J.; Smid, M.; Iaria, J.; Salvatori, D.C.F.; van Dam, H.; Zhu, H.J.; Martens, J.W.M.; Dijke, P.T. Cancer-associated fibroblast-derived Gremlin 1 promotes breast cancer progression. Breast Cancer Res. 2019, 21, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paauwe, M.; Schoonderwoerd, M.J.; Helderman, R.F.; Harryvan, T.J.; Groenewoud, A.; van Pelt, G.W.; Bor, R.; Hemmer, D.M.; Versteeg, H.H.; Snaar-Jagalska, B.E.; et al. Endoglin Expression on Cancer-Associated Fibroblasts Regulates Invasion and Stimulates Colorectal Cancer Metastasis. Clin. Cancer Res. 2018, 24, 6331–6344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elliot, A.; Myllymäki, H.; Feng, Y. Inflammatory Responses during Tumour Initiation: From Zebrafish Transgenic Models of Cancer to Evidence from Mouse and Man. Cells 2020, 9, 1018. [Google Scholar] [CrossRef] [Green Version]
- De Oliveira, S.; Houseright, R.A.; Graves, A.L.; Golenberg, N.; Korte, B.G.; Miskolci, V.; Huttenlocher, A. High Cholesterol Diet Modulates Macrophage Polarization and Liver Inflammation during Early Hepatocellular Carcinoma Progression in Zebrafish. Cancer Biol. 2018. [Google Scholar] [CrossRef]
- Wikberg, M.L.; Ling, A.; Li, X.; Öberg, Å.; Edin, S.; Palmqvist, R. Neutrophil infiltration is a favorable prognostic factor in early stages of colon cancer. Hum. Pathol. 2017, 68, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Hu, P.; Donskov, F.; Wang, G.; Liu, Q.; Du, J. Tumor-Associated Neutrophils as a New Prognostic Factor in Cancer: A Systematic Review and Meta-Analysis. PLoS ONE 2014, 9, e98259. [Google Scholar] [CrossRef] [Green Version]
- Geng, S.-K.; Fu, S.-M.; Ma, S.-H.; Fu, Y.-P.; Zhang, H.-W. Tumor infiltrating neutrophil might play a major role in predicting the clinical outcome of breast cancer patients treated with neoadjuvant chemotherapy. BMC Cancer 2021, 21, 68. [Google Scholar] [CrossRef]
- Zielińska, K.A.; Katanaev, V.L. The Signaling Duo CXCL12 and CXCR4: Chemokine Fuel for Breast Cancer Tumorigenesis. Cancers 2020, 12, 3071. [Google Scholar] [CrossRef]
- Mortezaee, K. CXCL12/CXCR4 axis in the microenvironment of solid tumors: A critical mediator of metastasis. Life Sci. 2020, 249, 117534. [Google Scholar] [CrossRef]
- Tulotta, C.; Stefanescu, C.; Chen, Q.; Torraca, V.; Meijer, A.H.; Snaar-Jagalska, B.E. CXCR4 signaling regulates metastatic onset by controlling neutrophil motility and response to malignant cells. Sci. Rep. 2019, 9, 2399. [Google Scholar] [CrossRef] [Green Version]
- Mehla, K.; Singh, P.K. Metabolic Regulation of Macrophage Polarization in Cancer. Trends Cancer 2019, 5, 822–834. [Google Scholar] [CrossRef] [PubMed]
- Feng, R.; Morine, Y.; Ikemoto, T.; Imura, S.; Iwahashi, S.; Saito, Y.; Shimada, M. Nrf2 activation drive macrophages polarization and cancer cell epithelial-mesenchymal transition during interaction. Cell Commun. Signal. 2018, 16, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen-Chi, M.; Laplace-Builhe, B.; Travnickova, J.; Luz-Crawford, P.; Tejedor, G.; Phan, Q.T.; Duroux-Richard, I.; Levraud, J.-P.; Kissa, K.; Lutfalla, G.; et al. Identification of polarized macrophage subsets in zebrafish. eLife 2015, 4, e07288. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cao, Z.; Zhang, X.-M.; Nakamura, M.; Sun, M.; Hartman, J.; Harris, R.A.; Sun, Y.; Cao, Y. Novel Mechanism of Macrophage-Mediated Metastasis Revealed in a Zebrafish Model of Tumor Development. Cancer Res. 2015, 75, 306–315. [Google Scholar] [CrossRef] [Green Version]
- Martins, R.R.; Ellis, P.S.; Macdonald, R.B.; Richardson, R.J.; Henriques, C.M. Resident Immunity in Tissue Repair and Maintenance: The Zebrafish Model Coming of Age. Front. Cell Dev. Biol. 2019, 7. [Google Scholar] [CrossRef]
- Sullivan, C.; Matty, M.; Jurczyszak, D.; Gabor, K.; Millard, P.; Tobin, D.; Kim, C. Infectious disease models in zebrafish. In Methods in Cell Biology; Elsevier: Amsterdam, The Netherlands, 2017; Volume 138, pp. 101–136. [Google Scholar] [CrossRef]
- Zhang, J.; Späth, S.S.; Marjani, S.L.; Zhang, W.; Pan, X. Characterization of cancer genomic heterogeneity by next-generation sequencing advances precision medicine in cancer treatment. Precis. Clin. Med. 2018, 1, 29–48. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Hernandez, M.O.; Zhao, Y.; Mehta, M.; Tran, B.; Kelly, M.; Rae, Z.; Hernandez, J.M.; Davis, J.L.; Martin, S.P.; et al. Tumor Cell Biodiversity Drives Microenvironmental Reprogramming in Liver Cancer. Cancer Cell 2019, 36, 418–430.e6. [Google Scholar] [CrossRef] [Green Version]
- Ignatius, M.S.; Chen, E.; Elpek, N.M.; Fuller, A.Z.; Tenente, I.M.; Clagg, R.; Liu, S.; Blackburn, J.; Linardic, C.M.; Rosenberg, A.E.; et al. In Vivo Imaging of Tumor-Propagating Cells, Regional Tumor Heterogeneity, and Dynamic Cell Movements in Embryonal Rhabdomyosarcoma. Cancer Cell 2012, 21, 680–693. [Google Scholar] [CrossRef] [Green Version]
- Tang, Q.; Moore, J.C.; Ignatius, M.S.; Tenente, I.M.; Hayes, M.N.; Garcia, E.G.; Yordán, N.T.; Bourque, C.; He, S.N.; Blackburn, J.S.; et al. Imaging tumour cell heterogeneity following cell transplantation into optically clear immune-deficient zebrafish. Nat. Commun. 2016, 7, 10358. [Google Scholar] [CrossRef]
- He, S.; Jing, C.-B.; Look, A. Zebrafish models of leukemia. In Methods in Cell Biology; Elsevier: Amsterdam, The Netherlands, 2017; Volume 138, pp. 563–592. [Google Scholar] [CrossRef]
- Borga, C.; Park, G.; Foster, C.; Burroughs-Garcia, J.; Marchesin, M.; Shah, R.; Hasan, A.; Ahmed, S.T.; Bresolin, S.; Batchelor, L.; et al. Simultaneous B and T cell acute lymphoblastic leukemias in zebrafish driven by transgenic MYC: Implications for oncogenesis and lymphopoiesis. Leukemia 2018, 33, 333–347. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Yin, Y.; Yi, Y.; Cheng, Z.; Kuang, W.; Li, R.; Zhong, H.; Cui, Y.; Yuan, L.; Gong, F.; et al. Targeting lactate dehydrogenase A (LDHA) exerts antileukemic effects on T-cell acute lymphoblastic leukemia. Cancer Commun. 2020, 40, 501–517. [Google Scholar] [CrossRef] [PubMed]
- Casey, M.J.; Stewart, R.A. Pediatric Cancer Models in Zebrafish. Trends Cancer 2020, 6, 407–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwiatkowska, I.; Hermanowicz, J.M.; Iwinska, Z.; Kowalczuk, K.; Iwanowska, J.; Pawlak, D. Zebrafish—An Optimal Model in Experimental Oncology. Molecules 2022, 27, 4223. https://doi.org/10.3390/molecules27134223
Kwiatkowska I, Hermanowicz JM, Iwinska Z, Kowalczuk K, Iwanowska J, Pawlak D. Zebrafish—An Optimal Model in Experimental Oncology. Molecules. 2022; 27(13):4223. https://doi.org/10.3390/molecules27134223
Chicago/Turabian StyleKwiatkowska, Iwona, Justyna Magdalena Hermanowicz, Zaneta Iwinska, Krystyna Kowalczuk, Jolanta Iwanowska, and Dariusz Pawlak. 2022. "Zebrafish—An Optimal Model in Experimental Oncology" Molecules 27, no. 13: 4223. https://doi.org/10.3390/molecules27134223
APA StyleKwiatkowska, I., Hermanowicz, J. M., Iwinska, Z., Kowalczuk, K., Iwanowska, J., & Pawlak, D. (2022). Zebrafish—An Optimal Model in Experimental Oncology. Molecules, 27(13), 4223. https://doi.org/10.3390/molecules27134223






