The Development and Application of Opto-Chemical Tools in the Zebrafish
Abstract
1. Introduction
2. An Overview of Established Opto-Chemical Tools
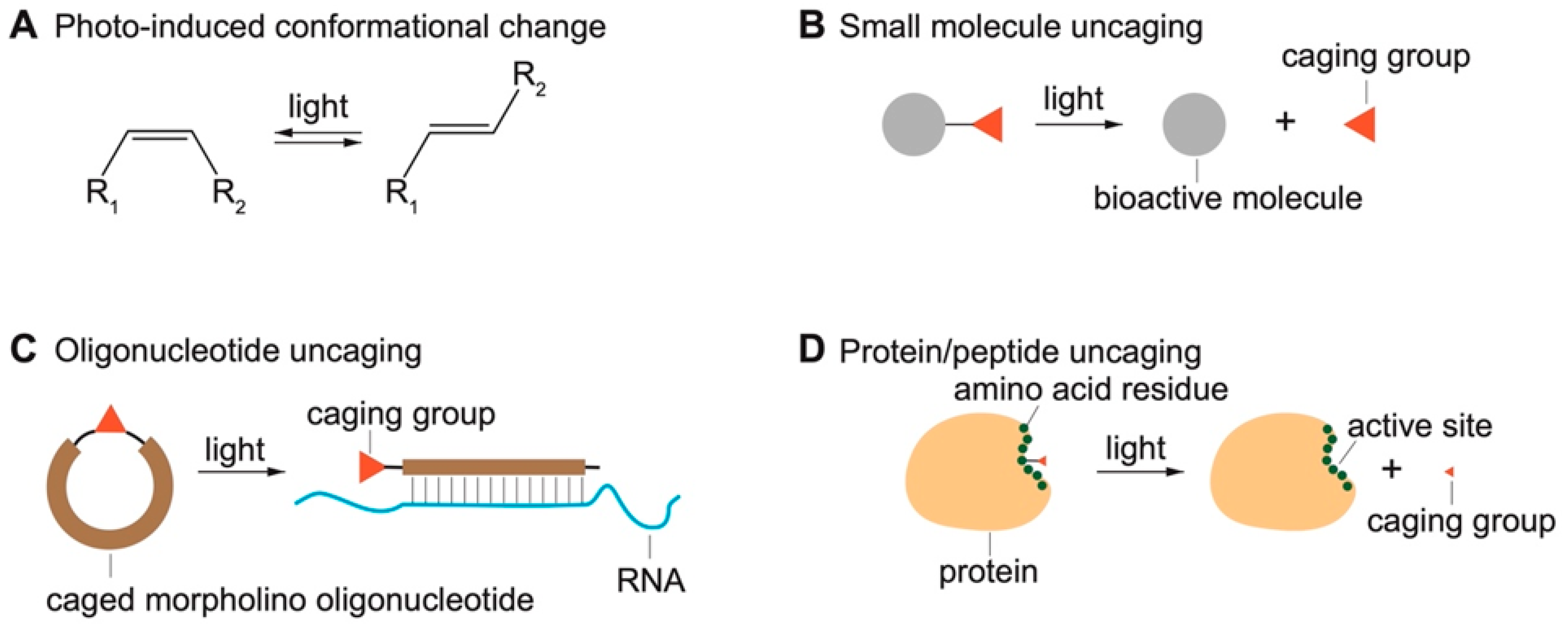
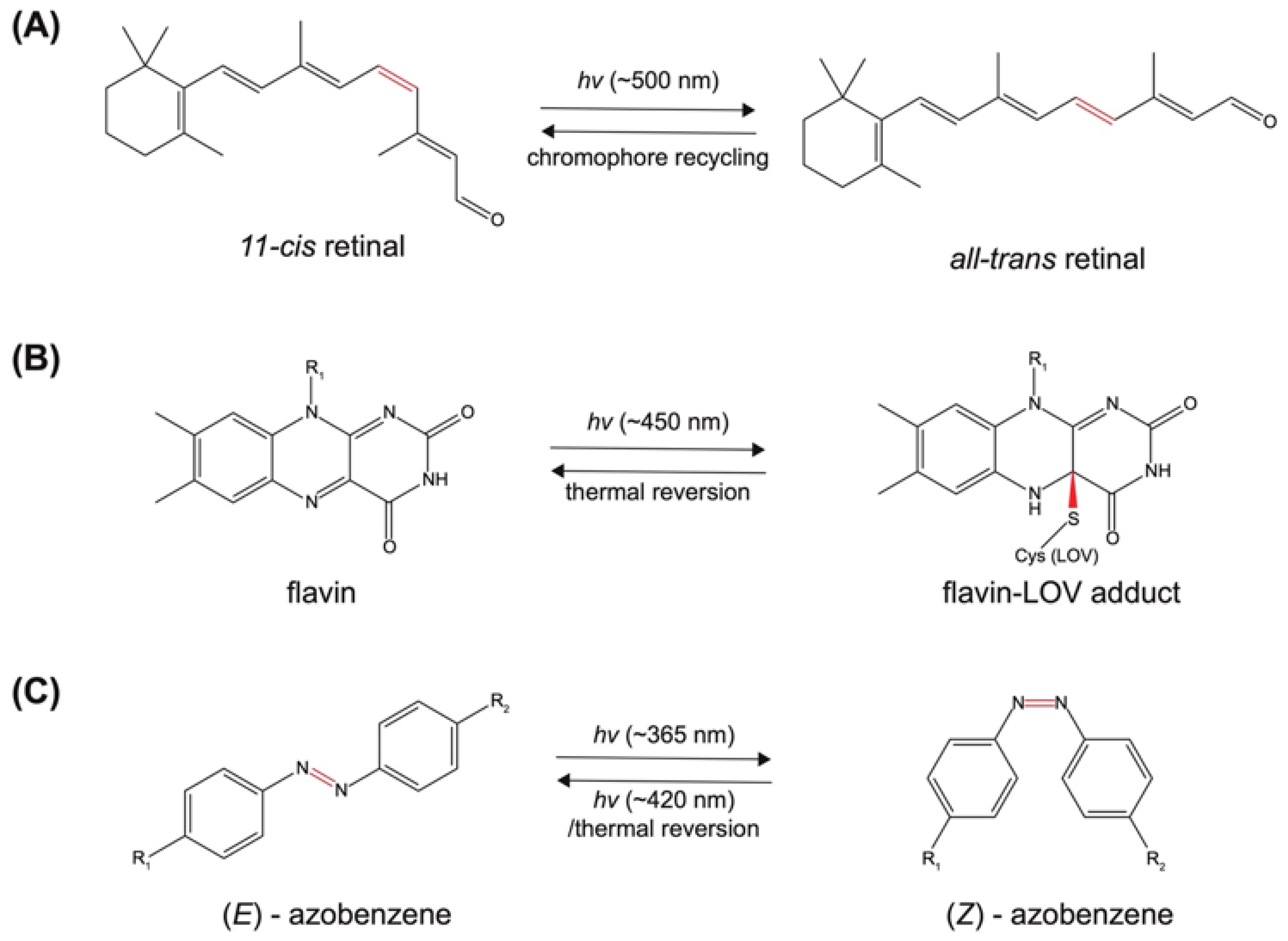
2.1. Photo-Induced Conformational Change
2.1.1. Photoreceptor-Derived Photoswitches
2.1.2. Non-Photoreceptor Based Photoswitches
2.2. Light-Induced Uncaging
2.2.1. Photocaged Small-Molecule Actuators and Probes
2.2.2. Caged Oligonucleotides
2.2.3. Caged Peptides and Proteins
| Caging Group | Chemical Structure | Typical Activation Wavelength (nm) | Examples and References |
|---|---|---|---|
| 2-Nitrobenzyl derivatives | 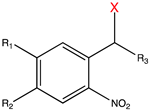 | 250–450 | ATP [65], glutamate [72], retinoic acid [81], rapamycin [84,168], doxycycline [86], Cyc [89], Ca2+ [78], auxin [96], PROTAC [97], siRNA [107], MO [114,118], gRNA [122], Lys [135], Ser [135], Cys [151], Tyr [161], Gln [169], Glu [170], Ans [171], small-molecule inhibitor [172]. |
| Coumarin derivatives | 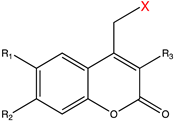 | 340–470 | Glutamate [73], GABA [77], retinoic acid [81], tamoxifen [88], Cyc [89], PROTAC [98], mRNA [104], MO [118], Cys [173], Lys [174], Gly [175], small-molecule inhibitor [176]. |
| 7-Nitroindolinyl derivatives | 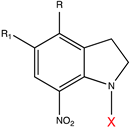 | 300–450 | GABA [76], Asp [133], Glu [133], acetic acid [177], Ca2+ [178], auxin [179]. |
| BODIPY derivatives | 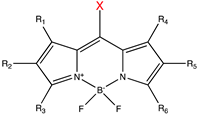 | 500–650 | Histamine [180], dopamine [180], acetic acid [181], alcohols [182], small-molecule inhibitor [183]. |
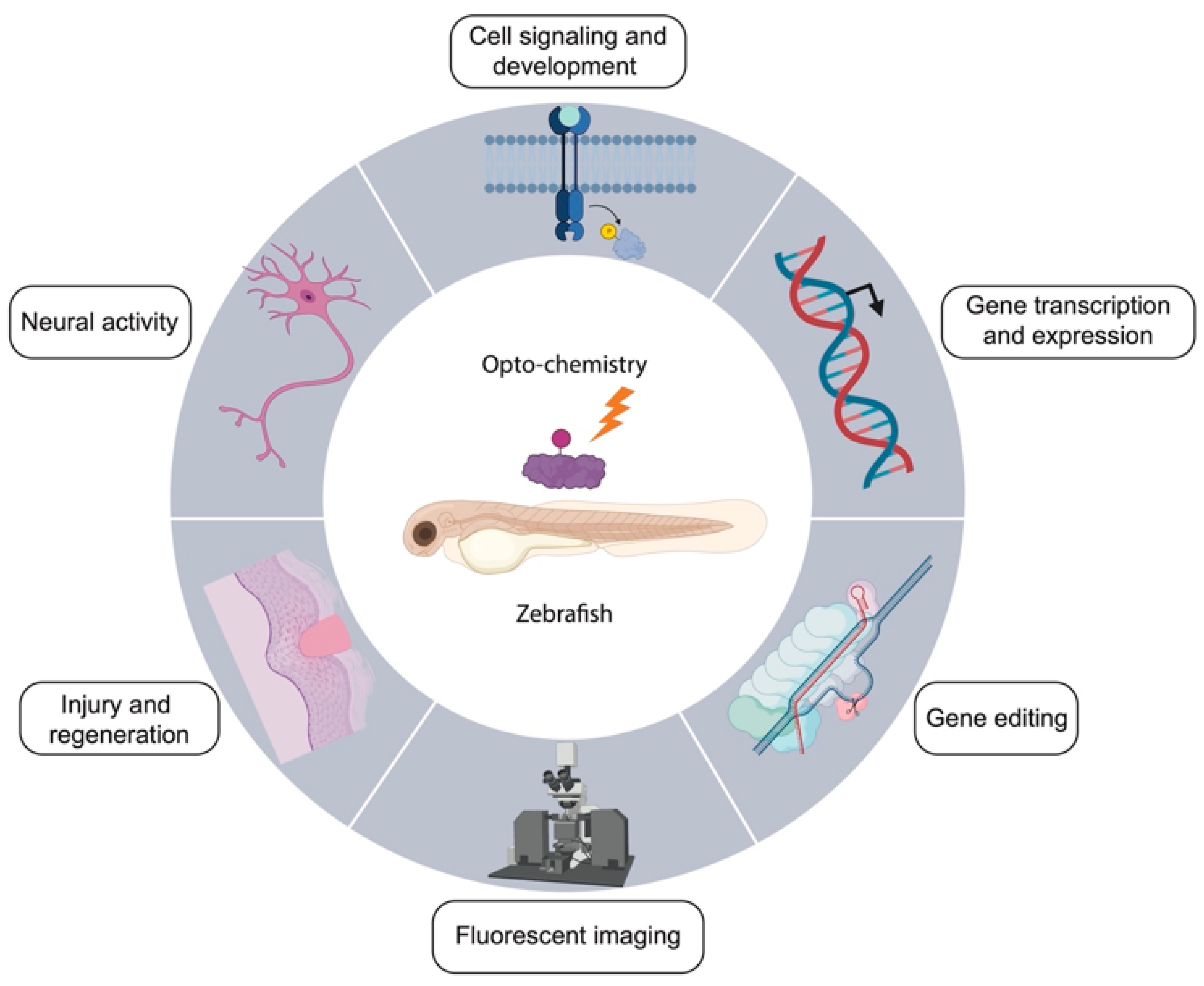
3. Applications of Opto-Chemical Tools in Zebrafish
3.1. Modulation of Neural Activity
3.2. Cell Signaling and Development
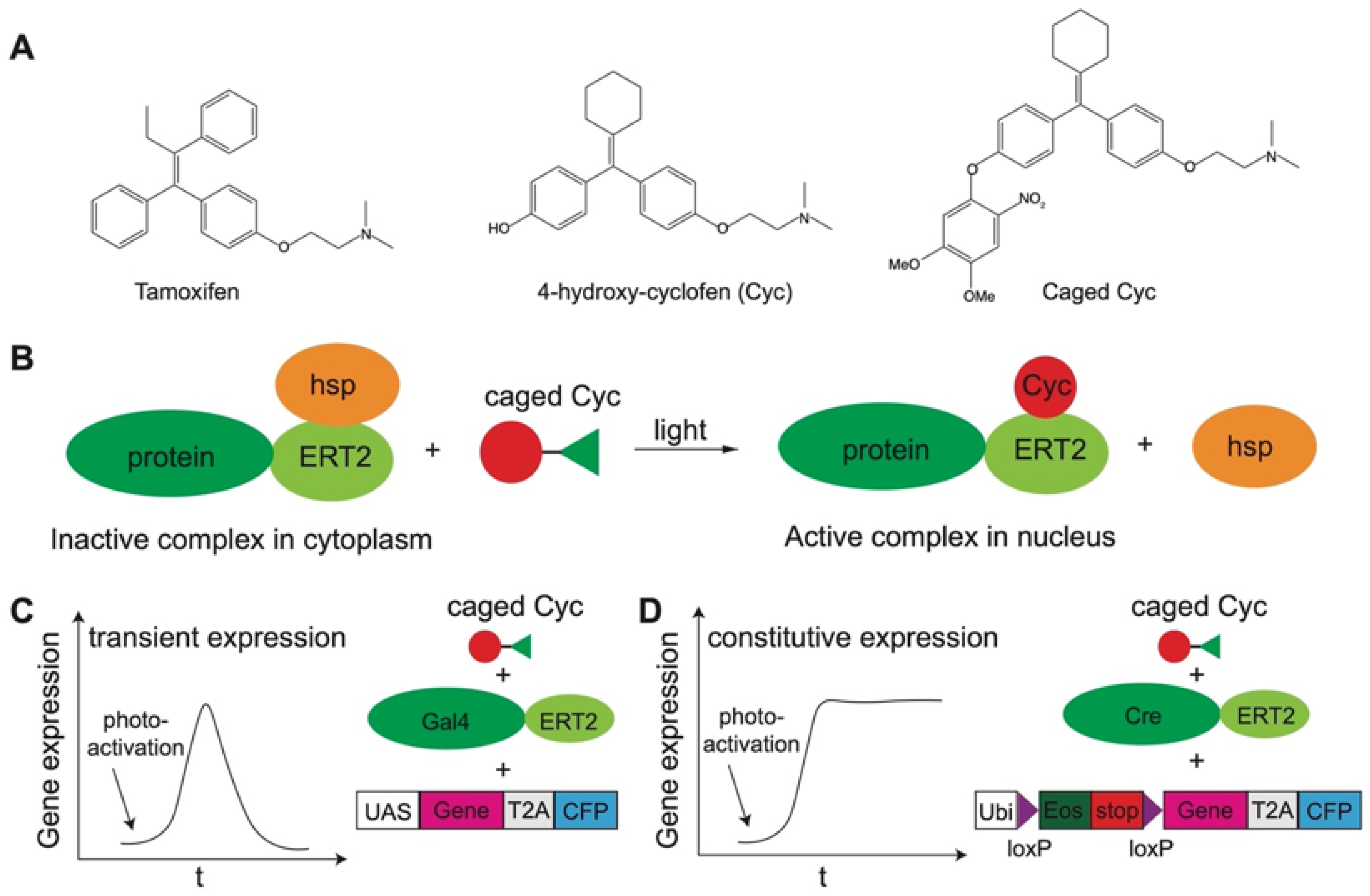
3.3. Gene Expression
3.4. Gene Editing
3.5. Fluorescent Imaging
3.6. Injury and Regeneration
| Study | Photocontrol Modules | Biological Targets |
|---|---|---|
| Modulation of neural activity | Rhodamine-based photoswitch [189], azobenzene-derived photoswitches [190,191,192,194,258] | TRPA1 ligand [189], glutamate receptor modulators [190,191,194,258], GIRK ligand [192] |
| Cell signaling and development | 2-Nitrobenzyl-based caging [81,168,172,208], coumarin-based caging [81,210], cis-trans isomerization [205]. | Caged Ca2+ [204], caged retinoic acid [81], 13-cis retinoic acid [205], caged rapamycin [168], caged RET kinase inhibitor [172], cMOs [114,115,207,208,209], caged amino acids [210] |
| Gene expression | 2-Nitrobenzyl-based caging [89,90,114,115,214], coumarin-based caging [89,90,215] | Caged Cyc [89,90,214], caged amino acids [215], cMOs [114,116,118,207,208,209]. |
| Gene editing | 6-Nitropiperonyloxymethylene-based caging [122,124]. | Caged gRNAs [122,124,126]. |
| Fluorescent imaging | 2-Nitrobenzy-based caging [208,232,236,259], 2,3-dimethyl 2,3-dinitrobutane-based caging [234], O6-benzylguanine-based caging [237]. | Caged fluorescein [208,232,234,236,237,259], Kaede [205,230,231,260], mEos [214,261,262], Dronpa [230,238], Dendra2 [239,240,241], rhodamine [233,235]. |
| Injury and regeneration | Azobenzene-derived photoswitches [257]. | β1-adrenoceptor ligands [257]. |
4. Challenges and Future Directions of Opto-Chemical Tools in Zebrafish
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Glossary
| Asn | asparagine |
| Asp | aspartate |
| ATP | adenosine triphosphate |
| BLUF | sensors of blue light using FAD |
| BODIPY | boron-dipyrromethene |
| Ca2+ | calcium |
| CALI | chromophore-assisted light inactivation |
| cAMP | cyclic adenosine monophosphate |
| cMOs | caged morpholino oligonucleotides |
| CFP | cyan fluorescent protein |
| CRISPR | clustered regularly interspaced short palindromic repeats |
| CRISPRi | CRISPR interference |
| crRNA | CRISPR RNA |
| CRY | cryptochrome |
| Cyc | 4-hydroxy-cyclofen |
| Cys | cysteine |
| DNA | deoxyribonucleic acid |
| eIF4E | the eukaryotic translation initiation factor 4E |
| ERT2 | modified estrogen receptor |
| FAD | flavin adenine dinucleotide |
| FGF | fibroblast growth factor |
| FKBP | FK506 binding protein |
| FMN | flavin mononucleotide |
| FRB | FKBP-rapamycin binding protein |
| GABA | gamma-aminobutyric acid |
| GFP | green fluorescent protein |
| GIRK | G-protein coupled inwardly rectifying potassium channel |
| Gln | glutamine |
| Glu | glutamate |
| Gly | glycine |
| gRNA | guide RNA |
| hsp | heat shock proteins |
| LiGluR | light-gated ionotropic glutamate receptor |
| LOV | light-oxygen-voltage sensing domain |
| Lys | lysine |
| MEK/ERK | mitogen-activated protein kinase kinase/extracellular signal-regulated kinase |
| MOs | morpholino oligonucleotides |
| mRNA | messenger ribonucleic acid |
| Ntl | no tail |
| PALM | photoactivated localization microscopy |
| PROTACs | proteolysis-targeting chimeras |
| PSM | presomitic mesoderm |
| RESOLFT | reversible saturable optical linear fluorescence transitions |
| RET | Rearranged during transfection |
| RISC | RNA-induced silencing complex |
| RNA | ribonucleic acid |
| ROS | reactive oxygen species |
| Ser | serine |
| siRNA | small interfering ribonucleic acid |
| STED | stimulated emission depletion |
| STORM | stochastic optical reconstruction microscopy |
| TDP-43 | trans-activation response element DNA-binding protein 43 |
| TGF-β | transforming growth factor β |
| TRPA1 | transient receptor potential A1 channel |
| Tyr | tyrosine |
| UAAs | unnatural amino acids |
| UAS | upstream activating sequence |
| Ubi | ubiquitin promoter |
| UV | ultraviolet |
References
- Heisenberg, C.P.; Bellaiche, Y. Forces in tissue morphogenesis and patterning. Cell 2013, 153, 948–962. [Google Scholar] [CrossRef]
- Housden, B.E.; Perrimon, N. Spatial and temporal organization of signaling pathways. Trends Biochem. Sci. 2014, 39, 457–464. [Google Scholar] [CrossRef]
- Garcia, E.; Ismail, S. Spatiotemporal Regulation of Signaling: Focus on T Cell Activation and the Immunological Synapse. Int. J. Mol. Sci. 2020, 21, 3283. [Google Scholar] [CrossRef]
- Lehner, B. Genotype to phenotype: Lessons from model organisms for human genetics. Nat. Rev. Genet. 2013, 14, 168–178. [Google Scholar] [CrossRef]
- Goldstein, B.; King, N. The Future of Cell Biology: Emerging Model Organisms. Trends Cell Biol. 2016, 26, 818–824. [Google Scholar] [CrossRef][Green Version]
- Ankeny, R.; Leonelli, S. Model Organisms (Elements in the Philosophy of Biology); Cambridge University Press: Cambridge, UK, 2021. [Google Scholar]
- Miklosi, A.; Andrew, R.J. The Zebrafish as a Model for Behavioral Studies. Zebrafish 2006, 3, 227–234. [Google Scholar] [CrossRef]
- Lieschke, G.J.; Currie, P.D. Animal models of human disease: Zebrafish swim into view. Nat. Rev. Genet. 2007, 8, 353–367. [Google Scholar] [CrossRef]
- Hirata, H. Zebrafish muscular disease models towards drug discovery. Expert Opin. Drug Discov. 2009, 4, 507–513. [Google Scholar] [CrossRef]
- White, R.; Rose, K.; Zon, L. Zebrafish cancer: The state of the art and the path forward. Nat. Rev. Cancer 2013, 13, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Choi, T.-Y.; Choi, T.-I.; Lee, Y.-R.; Choe, S.-K.; Kim, C.-H. Zebrafish as an animal model for biomedical research. Exp. Mol. Med. 2021, 53, 310–317. [Google Scholar] [CrossRef]
- Marrs, J.A.; Sarmah, S. The Genius of the Zebrafish Model: Insights on Development and Disease. Biomedicines 2021, 9, 577. [Google Scholar] [CrossRef] [PubMed]
- Patton, E.E.; Zon, L.I.; Langenau, D.M. Zebrafish disease models in drug discovery: From preclinical modelling to clinical trials. Nat. Rev. Drug Discov. 2021, 20, 611–628. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, J.B.; He, K.J.; Wang, F.; Liu, C.F. Advances of Zebrafish in Neurodegenerative Disease: From Models to Drug Discovery. Front. Pharmacol. 2021, 12, 713963. [Google Scholar] [CrossRef] [PubMed]
- Fenno, L.; Yizhar, O.; Deisseroth, K. The development and application of optogenetics. Annu. Rev. Neurosci. 2011, 34, 389–412. [Google Scholar] [CrossRef] [PubMed]
- Packer, A.M.; Roska, B.; Hausser, M. Targeting neurons and photons for optogenetics. Nat. Neurosci. 2013, 16, 805–815. [Google Scholar] [CrossRef]
- Hausser, M. Optogenetics: The age of light. Nat. Methods 2014, 11, 1012–1014. [Google Scholar] [CrossRef]
- Duebel, J.; Marazova, K.; Sahel, J.A. Optogenetics. Curr. Opin. Ophthalmol. 2015, 26, 226–232. [Google Scholar] [CrossRef]
- Tan, P.; He, L.; Huang, Y.; Zhou, Y. Optophysiology: Illuminating cell physiology with optogenetics. Physiol. Rev. 2022, 102, 1263–1325. [Google Scholar] [CrossRef]
- Bansal, A.; Shikha, S.; Zhang, Y. Towards translational optogenetics. Nat. Biomed. Eng. 2022. [Google Scholar] [CrossRef]
- Szymanski, W.; Beierle, J.M.; Kistemaker, H.A.; Velema, W.A.; Feringa, B.L. Reversible photocontrol of biological systems by the incorporation of molecular photoswitches. Chem. Rev. 2013, 113, 6114–6178. [Google Scholar] [CrossRef]
- Terakita, A. The opsins. Genome Biol. 2005, 6, 213. [Google Scholar] [CrossRef] [PubMed]
- Sexton, T.; Buhr, E.; Van Gelder, R.N. Melanopsin and Mechanisms of Non-visual Ocular Photoreception. J. Biol. Chem. 2012, 287, 1649–1656. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Shalitin, D. Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol. 2003, 54, 469–496. [Google Scholar] [CrossRef]
- Moglich, A.; Yang, X.; Ayers, R.A.; Moffat, K. Structure and function of plant photoreceptors. Annu. Rev. Plant Biol. 2010, 61, 21–47. [Google Scholar] [CrossRef] [PubMed]
- Christie, J.M.; Gawthorne, J.; Young, G.; Fraser, N.J.; Roe, A.J. LOV to BLUF: Flavoprotein contributions to the optogenetic toolkit. Mol. Plant 2012, 5, 533–544. [Google Scholar] [CrossRef]
- Rizzini, L.; Favory, J.J.; Cloix, C.; Faggionato, D.; O’Hara, A.; Kaiserli, E.; Baumeister, R.; Schafer, E.; Nagy, F.; Jenkins, G.I.; et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science 2011, 332, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Hu, Q.; Yan, Z.; Chen, W.; Yan, C.; Huang, X.; Zhang, J.; Yang, P.; Deng, H.; Wang, J.; et al. Structural basis of ultraviolet-B perception by UVR8. Nature 2012, 484, 214–219. [Google Scholar] [PubMed]
- Jenkins, G.I. The UV-B photoreceptor UVR8: From structure to physiology. Plant Cell 2014, 26, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Su, Y.S.; Lagarias, J.C. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 2006, 57, 837–858. [Google Scholar] [CrossRef] [PubMed]
- Nagatani, A. Phytochrome: Structural basis for its functions. Curr. Opin. Plant Biol. 2010, 13, 565–570. [Google Scholar] [CrossRef]
- Blaner, W.S. Cellular metabolism and actions of 13-cis-retinoic acid. J. Am. Acad. Dermatol. 2001, 45, S129–S135. [Google Scholar] [CrossRef] [PubMed]
- Niederreither, K.; Dolle, P. Retinoic acid in development: Towards an integrated view. Nat. Rev. Genet. 2008, 9, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Kane, M.A. Analysis, occurrence, and function of 9-cis-retinoic acid. Biochim. Biophys. Acta 2012, 1821, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Pronkin, P.; Tatikolov, A. Isomerization and Properties of Isomers of Carbocyanine Dyes. Sci 2019, 1, 19. [Google Scholar] [CrossRef]
- Beija, M.; Afonso, C.A.M.; Martinho, J.M.G. Synthesis and applications of Rhodamine derivatives as fluorescent probes. Chem. Soc. Rev. 2009, 38, 2410–2433. [Google Scholar] [CrossRef] [PubMed]
- Bandara, H.M.; Burdette, S.C. Photoisomerization in different classes of azobenzene. Chem. Soc. Rev. 2012, 41, 1809–1825. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.X.; Lin, M.Z. Photoswitchable fluorescent proteins: Ten years of colorful chemistry and exciting applications. Curr. Opin. Chem. Biol. 2013, 17, 682–690. [Google Scholar] [CrossRef]
- Adam, V.; Berardozzi, R.; Byrdin, M.; Bourgeois, D. Phototransformable fluorescent proteins: Future challenges. Curr. Opin. Chem. Biol. 2014, 20, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Sadovski, O.; Beharry, A.A.; Zhang, F.; Woolley, G.A. Spectral tuning of azobenzene photoswitches for biological applications. Angew. Chem. Int. Ed. 2009, 48, 1484–1486. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.; Babalhavaeji, A.; Collins, C.V.; Jarrah, K.; Sadovski, O.; Dai, Q.; Woolley, G.A. Near-Infrared Photoswitching of Azobenzenes under Physiological Conditions. J. Am. Chem. Soc. 2017, 139, 13483–13486. [Google Scholar] [CrossRef] [PubMed]
- Bozovic, O.; Jankovic, B.; Hamm, P. Using azobenzene photocontrol to set proteins in motion. Nat. Rev. Chem. 2022, 6, 112–124. [Google Scholar] [CrossRef]
- Yamada, M.D.; Nakajima, Y.; Maeda, H.; Maruta, S. Photocontrol of kinesin ATPase activity using an azobenzene derivative. J. Biochem. 2007, 142, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Timm, K.A.; Arndt, K.M.; Woolley, G.A. Photocontrol of coiled-coil proteins in living cells. Angew. Chem. Int. Ed. 2010, 49, 3943–3946. [Google Scholar] [CrossRef] [PubMed]
- Wachtveitl, J.; Zumbusch, A. Azobenzene: An optical switch for in vivo experiments. ChemBioChem 2011, 12, 1169–1170. [Google Scholar] [CrossRef] [PubMed]
- Beharry, A.A.; Wong, L.; Tropepe, V.; Woolley, G.A. Fluorescence imaging of azobenzene photoswitching in vivo. Angew. Chem. Int. Ed. 2011, 50, 1325–1327. [Google Scholar] [CrossRef] [PubMed]
- Mourot, A.; Fehrentz, T.; Le Feuvre, Y.; Smith, C.M.; Herold, C.; Dalkara, D.; Nagy, F.; Trauner, D.; Kramer, R.H. Rapid optical control of nociception with an ion-channel photoswitch. Nat. Methods 2012, 9, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Nevola, L.; Martin-Quiros, A.; Eckelt, K.; Camarero, N.; Tosi, S.; Llobet, A.; Giralt, E.; Gorostiza, P. Light-regulated stapled peptides to inhibit protein-protein interactions involved in clathrin-mediated endocytosis. Angew. Chem. Int. Ed. 2013, 52, 7704–7708. [Google Scholar] [CrossRef]
- Tochitsky, I.; Helft, Z.; Meseguer, V.; Fletcher, R.B.; Vessey, K.A.; Telias, M.; Denlinger, B.; Malis, J.; Fletcher, E.L.; Kramer, R.H. How Azobenzene Photoswitches Restore Visual Responses to the Blind Retina. Neuron 2016, 92, 100–113. [Google Scholar] [CrossRef]
- Blacklock, K.M.; Yachnin, B.J.; Woolley, G.A.; Khare, S.D. Computational Design of a Photocontrolled Cytosine Deaminase. J. Am. Chem. Soc. 2018, 140, 14–17. [Google Scholar] [CrossRef] [PubMed]
- Peddie, V.; Abell, A.D. Photocontrol of peptide secondary structure through non-azobenzene photoswitches. J. Photochem. Photobiol. C Photochem. Rev. 2019, 40, 1–20. [Google Scholar] [CrossRef]
- Han, W.G.; Lovell, T.; Liu, T.; Noodleman, L. Density functional studies of the ground- and excited-state potential-energy curves of stilbene cis-trans isomerization. ChemPhysChem 2002, 3, 167–178. [Google Scholar] [CrossRef]
- Otolski, C.J.; Mohan Raj, A.; Sharma, G.; Prabhakar, R.; Ramamurthy, V.; Elles, C.G. Ultrafast trans --> cis Photoisomerization Dynamics of Alkyl-Substituted Stilbenes in a Supramolecular Capsule. J. Phys. Chem. A 2019, 123, 5061–5071. [Google Scholar] [CrossRef]
- Regner, N.; Herzog, T.T.; Haiser, K.; Hoppmann, C.; Beyermann, M.; Sauermann, J.; Engelhard, M.; Cordes, T.; Ruck-Braun, K.; Zinth, W. Light-switchable hemithioindigo-hemistilbene-containing peptides: Ultrafast spectroscopy of the Z --> E isomerization of the chromophore and the structural dynamics of the peptide moiety. J. Phys. Chem. B 2012, 116, 4181–4191. [Google Scholar] [CrossRef] [PubMed]
- Maerz, B.; Wiedbrauk, S.; Oesterling, S.; Samoylova, E.; Nenov, A.; Mayer, P.; de Vivie-Riedle, R.; Zinth, W.; Dube, H. Making fast photoswitches faster--using Hammett analysis to understand the limit of donor-acceptor approaches for faster hemithioindigo photoswitches. Chemistry 2014, 20, 13984–13992. [Google Scholar] [CrossRef]
- Klajn, R. Spiropyran-based dynamic materials. Chem. Soc. Rev. 2014, 43, 148–184. [Google Scholar] [CrossRef]
- Zhang, X.; Heng, S.; Abell, A.D. Photoregulation of alpha-Chymotrypsin Activity by Spiropyran-Based Inhibitors in Solution and Attached to an Optical Fiber. Chemistry 2015, 21, 10703–10713. [Google Scholar] [CrossRef] [PubMed]
- Irie, M.; Fukaminato, T.; Matsuda, K.; Kobatake, S. Photochromism of diarylethene molecules and crystals: Memories, switches, and actuators. Chem. Rev. 2014, 114, 12174–12277. [Google Scholar] [CrossRef] [PubMed]
- Kowalik, L.; Chen, J.K. Illuminating developmental biology through photochemistry. Nat. Chem. Biol. 2017, 13, 587–598. [Google Scholar] [CrossRef]
- Ankenbruck, N.; Courtney, T.; Naro, Y.; Deiters, A. Optochemical Control of Biological Processes in Cells and Animals. Angew. Chem. Int. Ed. 2018, 57, 2768–2798. [Google Scholar] [CrossRef]
- Bardhan, A.; Deiters, A. Development of photolabile protecting groups and their application to the optochemical control of cell signaling. Curr. Opin. Struct. Biol. 2019, 57, 164–175. [Google Scholar] [CrossRef]
- Mangubat-Medina, A.E.; Ball, Z.T. Triggering biological processes: Methods and applications of photocaged peptides and proteins. Chem. Soc. Rev. 2021, 50, 10403–10421. [Google Scholar] [CrossRef] [PubMed]
- Weinstain, R.; Slanina, T.; Kand, D.; Klan, P. Visible-to-NIR-Light Activated Release: From Small Molecules to Nanomaterials. Chem. Rev. 2020, 120, 13135–13272. [Google Scholar] [CrossRef] [PubMed]
- Engels, J.; Schlaeger, E.J. Synthesis, structure, and reactivity of adenosine cyclic 3′,5′-phosphate benzyl triesters. J. Med. Chem. 1977, 20, 907–911. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, J.H.; Forbush, B., 3rd; Hoffman, J.F. Rapid photolytic release of adenosine 5′-triphosphate from a protected analogue: Utilization by the Na:K pump of human red blood cell ghosts. Biochemistry 1978, 17, 1929–1935. [Google Scholar] [CrossRef] [PubMed]
- Ellis-Davies, G.C. Caged compounds: Photorelease technology for control of cellular chemistry and physiology. Nat. Methods 2007, 4, 619–628. [Google Scholar] [CrossRef]
- Young, D.D.; Deiters, A. Photochemical control of biological processes. Org. Biomol. Chem. 2007, 5, 999–1005. [Google Scholar] [CrossRef]
- Meng, X.M.; Chen, X.Y.; Fu, Y.; Guo, Q.X. Photolysis of Caged Compounds and Its Applications to Chemical Biology. Prog. Chem. 2008, 20, 2034–2044. [Google Scholar]
- Puliti, D.; Warther, D.; Orange, C.; Specht, A.; Goeldner, M. Small photoactivatable molecules for controlled fluorescence activation in living cells. Bioorg. Chem. 2011, 19, 1023–1029. [Google Scholar] [CrossRef]
- Ellis-Davies, G.C.R. Useful Caged Compounds for Cell Physiology. Acc. Chem. Res. 2020, 53, 1593–1604. [Google Scholar] [CrossRef]
- Laczi, D.; Johnstone, M.D.; Fleming, C.L. Photoresponsive Small Molecule Inhibitors for the Remote Control of Enzyme Activity. Chem.-Asian J. 2022, 17, e202200200. [Google Scholar] [CrossRef]
- Callaway, E.M.; Katz, L.C. Photostimulation Using Caged Glutamate Reveals Functional Circuitry in Living Brain-Slices. Proc. Natl. Acad. Sci. USA 1993, 90, 7661–7665. [Google Scholar] [CrossRef] [PubMed]
- Furuta, T.; Wang, S.S.; Dantzker, J.L.; Dore, T.M.; Bybee, W.J.; Callaway, E.M.; Denk, W.; Tsien, R.Y. Brominated 7-hydroxycoumarin-4-ylmethyls: Photolabile protecting groups with biologically useful cross-sections for two photon photolysis. Proc. Natl. Acad. Sci. USA 1999, 96, 1193–1200. [Google Scholar] [CrossRef] [PubMed]
- Ellis-Davies, G.C.R.; Matsuzaki, M.; Paukert, M.; Kasai, H.; Bergles, D.E. 4-carboxymethoxy-5,7-dinitroindolinyl-glu: An improved caged glutamate for expeditious ultraviolet and two-photon photolysis in brain slices. J. Neurosci. 2007, 27, 6601–6604. [Google Scholar] [CrossRef] [PubMed]
- Guruge, C.; Ouedraogo, Y.P.; Comitz, R.L.; Ma, J.X.; Losonczy, A.; Nesnas, N. Improved Synthesis of Caged Glutamate and Caging Each Functional Group. ACS Chem. Neurosci. 2018, 9, 2713–2721. [Google Scholar] [CrossRef]
- Matsuzaki, M.; Hayama, T.; Kasai, H.; Ellis-Davies, G.C.R. Two-photon uncaging of gamma-aminobutyric acid in intact brain tissue. Nat. Chem. Biol. 2010, 6, 255–257. [Google Scholar] [CrossRef]
- Amatrudo, J.M.; Olson, J.P.; Lur, G.; Chiu, C.Q.; Higley, M.J.; Ellis-Davies, G.C.R. Wavelength-Selective One- and Two-Photon Uncaging of GABA. ACS Chem. Neurosci. 2014, 5, 64–70. [Google Scholar] [CrossRef]
- Ellis-Davies, G.C.R. Neurobiology with caged calcium. Chem. Rev. 2008, 108, 1603–1613. [Google Scholar] [CrossRef]
- Kong, D.; Lv, Z.; Häring, M.; Lin, B.; Wolf, F.; Großhans, J. In vivo optochemical control of cell contractility at single-cell resolution. EMBO Rep. 2019, 20, e47755. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.W.; Somlyo, A.V.; Goldman, Y.E.; Somlyo, A.P.; Trentham, D.R. Kinetics of Smooth and Skeletal-Muscle Activation by Laser-Pulse Photolysis of Caged Inositol 1,4,5-Trisphosphate. Nature 1987, 327, 249–252. [Google Scholar] [CrossRef] [PubMed]
- Neveu, P.; Aujard, I.; Benbrahim, C.; Le Saux, T.; Allemand, J.F.; Vriz, S.; Bensimon, D.; Jullien, L. A caged retinoic acid for one- and two-photon excitation in zebrafish embryos. Angew. Chem. Int. Ed. 2008, 47, 3744–3746. [Google Scholar] [CrossRef]
- Li, W.H.; Zheng, G.H. Photoactivatable fluorophores and techniques for biological imaging applications. Photochem. Photobiol. Sci. 2012, 11, 460–471. [Google Scholar] [CrossRef]
- Mayer, S.V.; Murnauer, A.; von Wrisberg, M.K.; Jokisch, M.L.; Lang, K. Photo-induced and Rapid Labeling of Tetrazine-Bearing Proteins via Cyclopropenone-Caged Bicyclononynes. Angew. Chem. Int. Ed. 2019, 58, 15876–15882. [Google Scholar] [CrossRef]
- Karginov, A.V.; Zou, Y.; Shirvanyants, D.; Kota, P.; Dokholyan, N.V.; Young, D.D.; Hahn, K.M.; Deiters, A. Light Regulation of Protein Dimerization and Kinase Activity in Living Cells Using Photocaged Rapamycin and Engineered FKBP. J. Am. Chem. Soc. 2011, 133, 420–423. [Google Scholar] [CrossRef]
- Cambridge, S.B.; Curten, B.; Bonhoeffer, T. A caged doxycycline analog for photoactivated gene expression with high spatiotemporal resolution. Mol. Biol. Cell 2002, 13, 407a. [Google Scholar]
- Cambridge, S.B.; Geissler, D.; Keller, S.; Curten, B. A caged doxycycline analogue for photoactivated gene expression. Angew. Chem. Int. Ed. 2006, 45, 2229–2231. [Google Scholar] [CrossRef] [PubMed]
- Link, K.H.; Shi, Y.H.; Koh, J.T. Light activated recombination. J. Am. Chem. Soc. 2005, 127, 13088–13089. [Google Scholar] [CrossRef] [PubMed]
- Wong, P.T.; Roberts, E.W.; Tang, S.Z.; Mukherjee, J.; Cannon, J.; Nip, A.J.; Corbin, K.; Krummel, M.F.; Choi, S.K. Control of an Unusual Photo-Claisen Rearrangement in Coumarin Caged Tamoxifen through an Extended Spacer. ACS Chem. Biol. 2017, 12, 1001–1010. [Google Scholar] [CrossRef]
- Sinha, D.K.; Neveu, P.; Gagey, N.; Aujard, I.; Benbrahim-Bouzidi, C.; Le Saux, T.; Rampon, C.; Gauron, C.; Goetz, B.; Dubruille, S.; et al. Photocontrol of Protein Activity in Cultured Cells and Zebrafish with One- and Two-Photon Illumination. ChemBioChem 2010, 11, 653–663. [Google Scholar] [CrossRef]
- Sinha, D.K.; Neveu, P.; Gagey, N.; Aujard, I.; Le Saux, T.; Rampon, C.; Gauron, C.; Kawakami, K.; Leucht, C.; Bally-Cuif, L.; et al. Photoactivation of the CreER(T2) Recombinase for Conditional Site-Specific Recombination with High Spatiotemporal Resolution. Zebrafish 2010, 7, 199–204. [Google Scholar] [CrossRef]
- Fournier, L.; Aujard, I.; Le Saux, T.; Maurin, S.; Beaupierre, S.; Baudin, J.B.; Jullien, L. Coumarinylmethyl Caging Groups with Redshifted Absorption. Chem.-Eur. J. 2013, 19, 17494–17507. [Google Scholar] [CrossRef]
- Zhang, W.T.; Hamouri, F.; Feng, Z.P.; Aujard, I.; Ducos, B.; Ye, S.X.; Weiss, S.; Volovitch, M.; Vriz, S.; Jullien, L.; et al. Control of Protein Activity and Gene Expression by Cyclofen-OH Uncaging. ChemBioChem 2018, 19, 1232–1238. [Google Scholar] [CrossRef]
- Klocker, N.; Weissenboeck, F.P.; van Dulmen, M.; Spacek, P.; Huwel, S.; Rentmeister, A. Photocaged 5′ cap analogues for optical control of mRNA translation in cells. Nat. Chem. 2022, 14, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Hoffman-Kim, D.; Diefenbach, T.J.; Eustace, B.K.; Jay, D.G. Chromophore-assisted laser inactivation. Methods Cell Biol. 2007, 82, 335–354. [Google Scholar]
- Jacobson, K.; Rajfur, Z.; Vitriol, E.; Hahn, K. Chromophore-assisted laser inactivation in cell biology. Trends Cell Biol. 2008, 18, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Delacour, Q.; Li, C.; Plamont, M.A.; Billon-Denis, E.; Aujard, I.; Le Saux, T.; Jullien, L.; Gautier, A. Light-Activated Proteolysis for the Spatiotemporal Control of Proteins. ACS Chem. Biol. 2015, 10, 1643–1647. [Google Scholar] [CrossRef]
- Xue, G.; Wang, K.; Zhou, D.; Zhong, H.; Pan, Z. Light-Induced Protein Degradation with Photocaged PROTACs. J. Am. Chem. Soc. 2019, 141, 18370–18374. [Google Scholar] [CrossRef]
- Naro, Y.; Darrah, K.; Deiters, A. Optical Control of Small Molecule-Induced Protein Degradation. J. Am. Chem. Soc. 2020, 142, 2193–2197. [Google Scholar] [CrossRef]
- Reynders, M.; Matsuura, B.S.; Berouti, M.; Simoneschi, D.; Marzio, A.; Pagano, M.; Trauner, D. PHOTACs enable optical control of protein degradation. Sci. Adv. 2020, 6, eaay5064. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Manna, D. Optochemical Control of Protein Degradation. ChemBioChem 2020, 21, 2250–2252. [Google Scholar] [CrossRef] [PubMed]
- Reynders, M.; Trauner, D. Optical control of targeted protein degradation. Cell Chem. Biol. 2021, 28, 969–986. [Google Scholar] [CrossRef]
- Ruble, B.K.; Yeldell, S.B.; Dmochowski, I.J. Caged oligonucleotides for studying biological systems. J. Inorg. Biochem. 2015, 150, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Monroe, W.T.; McQuain, M.M.; Chang, M.S.; Alexander, J.S.; Haselton, F.R. Targeting expression with light using caged DNA. J. Biol. Chem. 1999, 274, 20895–20900. [Google Scholar] [CrossRef]
- Ando, H.; Furuta, T.; Tsien, R.Y.; Okamoto, H. Photo-mediated gene activation using caged RNA/DNA in zebrafish embryos. Nat. Genet. 2001, 28, 317–325. [Google Scholar] [CrossRef]
- Shah, S.; Rangarajan, S.; Friedman, S.H. Light activated RNA interference. Angew. Chem. Int. Ed. 2005, 44, 1328–1332. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.N.; Chavli, R.V.; Marques, J.T.; Conrad, P.G.; Wang, D.; He, W.H.; Belisle, B.E.; Zhang, A.G.; Pastor, L.M.; Witney, F.R.; et al. Light controllable siRNAs regulate gene suppression and phenotypes in cells. Biochim. Biophys. Acta BBA-Biomembr. 2006, 1758, 394–403. [Google Scholar] [CrossRef]
- Mikat, V.; Heckel, A. Light-dependent RNA interference with nucleobase-caged siRNAs. RNA 2007, 13, 2341–2347. [Google Scholar] [CrossRef]
- Jain, P.K.; Shah, S.; Friedman, S.H. Patterning of Gene Expression Using New Photolabile Groups Applied to Light Activated RNAi. J. Am. Chem. Soc. 2011, 133, 440–446. [Google Scholar] [CrossRef]
- Govan, J.M.; Young, D.D.; Lusic, H.; Liu, Q.; Lively, M.O.; Deiters, A. Optochemical control of RNA interference in mammalian cells. Nucleic Acids Res. 2013, 41, 10518–10528. [Google Scholar] [CrossRef]
- Kala, A.; Jain, P.K.; Karunakaran, D.; Shah, S.; Friedman, S.H. The synthesis of tetra-modified RNA for the multidimensional control of gene expression via light-activated RNA interference. Nat. Protoc. 2014, 9, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Bill, B.R.; Petzold, A.M.; Clark, K.J.; Schimmenti, L.A.; Ekker, S.C. A Primer for Morpholino Use in Zebrafish. Zebrafish 2009, 6, 69–77. [Google Scholar] [CrossRef]
- Shestopalov, I.A.; Chen, J.K. Oligonucleotide-Based Tools for Studying Zebrafish Development. Zebrafish 2010, 7, 31–40. [Google Scholar] [CrossRef]
- Corey, D.R.; Abrams, J.M. Morpholino antisense oligonucleotides: Tools for investigating vertebrate development. Genome Biol. 2001, 2, reviews1015.1. [Google Scholar] [CrossRef]
- Shestopalov, I.A.; Sinha, S.; Chen, J.K. Light-controlled gene silencing in zebrafish embryos. Nat. Chem. Biol. 2007, 3, 650–651. [Google Scholar] [CrossRef]
- Tang, X.; Maegawa, S.; Weinberg, E.S.; Dmochowski, I.J. Regulating gene expression in zebrafish embryos using light-activated, negatively charged peptide nucleic acids. J. Am. Chem. Soc. 2007, 129, 11000–11001. [Google Scholar] [CrossRef]
- Yamazoe, S.; Shestopalov, I.A.; Provost, E.; Leach, S.D.; Chen, J.K. Cyclic caged morpholinos: Conformationally gated probes of embryonic gene function. Angew. Chem. Int. Ed. 2012, 51, 6908–6911. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, L.; Wang, P.; Lv, C.; Yang, Z.; Tang, X. Manipulation of gene expression in zebrafish using caged circular morpholino oligomers. Nucleic Acids Res. 2012, 40, 11155–11162. [Google Scholar] [CrossRef] [PubMed]
- Yamazoe, S.; Liu, Q.; McQuade, L.E.; Deiters, A.; Chen, J.K. Sequential Gene Silencing Using Wavelength-Selective Caged Morpholino Oligonucleotides. Angew. Chem. Int. Ed. 2014, 53, 10114–10118. [Google Scholar] [CrossRef] [PubMed]
- Nasevicius, A.; Ekker, S.C. Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 2000, 26, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Bedell, V.M.; Westcot, S.E.; Ekker, S.C. Lessons from morpholino-based screening in zebrafish. Brief. Funct. Genom. 2011, 10, 181–188. [Google Scholar] [CrossRef]
- Darrah, K.E.; Deiters, A. Translational control of gene function through optically regulated nucleic acids. Chem. Soc. Rev. 2021, 50, 13253–13267. [Google Scholar] [CrossRef]
- Zhou, W.; Brown, W.; Bardhan, A.; Delaney, M.; Ilk, A.S.; Rauen, R.R.; Kahn, S.I.; Tsang, M.; Deiters, A. Spatiotemporal Control of CRISPR/Cas9 Function in Cells and Zebrafish using Light-Activated Guide RNA. Angew. Chem. Int. Ed. 2020, 59, 8998–9003. [Google Scholar] [CrossRef]
- Liu, Y.; Zou, R.S.; He, S.; Nihongaki, Y.; Li, X.; Razavi, S.; Wu, B.; Ha, T. Very fast CRISPR on demand. Science 2020, 368, 1265–1269. [Google Scholar] [CrossRef] [PubMed]
- Moroz-Omori, E.V.; Satyapertiwi, D.; Ramel, M.C.; Hogset, H.; Sunyovszki, I.K.; Liu, Z.; Wojciechowski, J.P.; Zhang, Y.; Grigsby, C.L.; Brito, L.; et al. Photoswitchable gRNAs for Spatiotemporally Controlled CRISPR-Cas-Based Genomic Regulation. ACS Cent. Sci. 2020, 6, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Modell, A.E.; Siriwardena, S.U.; Shoba, V.M.; Li, X.; Choudhary, A. Chemical and optical control of CRISPR-associated nucleases. Curr. Opin. Chem. Biol. 2021, 60, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.Y.; Brown, W.; Bardhan, A.; Tsang, M.; Deiters, A. Optical Control of Base Editing and Transcription through Light-Activated Guide RNA. ChemPhotoChem 2021, 5, 984–988. [Google Scholar] [CrossRef]
- Carlson-Stevermer, J.; Kelso, R.; Kadina, A.; Joshi, S.; Rossi, N.; Walker, J.; Stoner, R.; Maures, T. CRISPRoff enables spatio-temporal control of CRISPR editing. Nat. Commun. 2020, 11, 5041. [Google Scholar] [CrossRef]
- Zou, R.S.; Liu, Y.; Wu, B.; Ha, T. Cas9 deactivation with photocleavable guide RNAs. Mol. Cell. 2021, 81, 1553–1565.e8. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, J.; Hua, X.; Meng, X.; Cai, H.; Wang, R.; Shi, J.; Deng, H.; Liu, L.; Li, Y.-M. Photocaging of Activity-Based Ubiquitin Probes via a C-Terminal Backbone Modification Strategy. Angew. Chem. Int. Ed. 2022, 134, e202203792. [Google Scholar]
- Odaka, M.; Furuta, T.; Kobayashi, Y.; Iwamura, M. Synthesis of caged compounds of L-leucyl-L-leucine methyl ester, an apoptosis inducer, and their cytotoxic activity. Biochem. Biophys. Res. Commun. 1995, 213, 652–656. [Google Scholar] [CrossRef]
- Tatsu, Y.; Shigeri, Y.; Sogabe, S.; Yumoto, N.; Yoshikawa, S. Solid-Phase Synthesis of Caged Peptides Using Tyrosine Modified with a Photocleavable Protecting Group: Application to the Synthesis of Caged Neuropeptide Y. Biochem. Biophys. Res. Commun. 1996, 227, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Veldhuyzen, W.F.; Nguyen, Q.; McMaster, G.; Lawrence, D.S. A light-activated probe of intracellular protein kinase activity. J. Am. Chem. Soc. 2003, 125, 13358–13359. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Cheng, J.-Y.; Zheng, J.-S. An Fmoc-compatible method for synthesis of peptides containing photocaged aspartic acid or glutamic acid. Tetrahedron Lett. 2015, 56, 4582–4585. [Google Scholar] [CrossRef]
- Mahmoodi, M.M.; Abate-Pella, D.; Pundsack, T.J.; Palsuledesai, C.C.; Goff, P.C.; Blank, D.A.; Distefano, M.D. Nitrodibenzofuran: A One- and Two-Photon Sensitive Protecting Group That Is Superior to Brominated Hydroxycoumarin for Thiol Caging in Peptides. J. Am. Chem. Soc. 2016, 138, 5848–5859. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Wan, Z.; Gao, Y.; Zheng, J.S.; Wang, J.; Si, Y.Y.; Chen, X.; Qi, H.; Liu, L.; Liu, W. Total chemical synthesis of photoactivatable proteins for light-controlled manipulation of antigen-antibody interactions. Chem. Sci. 2016, 7, 1891–1895. [Google Scholar] [CrossRef] [PubMed]
- Zou, K.; Cheley, S.; Givens, R.S.; Bayley, H. Catalytic subunit of protein kinase A caged at the activating phosphothreonine. J. Am. Chem. Soc. 2002, 124, 8220–8229. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, M.; Ichetovkin, I.; Song, X.; Condeelis, J.S.; Lawrence, D.S. A new strategy for caging proteins regulated by kinases. J. Am. Chem. Soc. 2002, 124, 2440–2441. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.M.; Xu, W.; Lawrence, D.S. Construction of a photoactivatable profluorescent enzyme via propinquity labeling. J. Am. Chem. Soc. 2011, 133, 2331–2333. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mangubat-Medina, A.E.; Martin, S.C.; Hanaya, K.; Ball, Z.T. A Vinylogous Photocleavage Strategy Allows Direct Photocaging of Backbone Amide Structure. J. Am. Chem. Soc. 2018, 140, 8401–8404. [Google Scholar] [CrossRef]
- Liu, C.C.; Schultz, P.G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010, 79, 413–444. [Google Scholar] [CrossRef]
- Chin, J.W. Expanding and reprogramming the genetic code of cells and animals. Annu. Rev. Biochem. 2014, 83, 379–408. [Google Scholar] [CrossRef]
- Lang, K.; Chin, J.W. Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins. Chem. Rev. 2014, 114, 4764–4806. [Google Scholar] [CrossRef] [PubMed]
- Chin, J.W. Expanding and reprogramming the genetic code. Nature 2017, 550, 53–60. [Google Scholar] [CrossRef]
- Drienovská, I.; Roelfes, G. Expanding the enzyme universe with genetically encoded unnatural amino acids. Nat. Catal. 2020, 3, 193–202. [Google Scholar] [CrossRef]
- Smolskaya, S.; Andreev, Y.A. Site-Specific Incorporation of Unnatural Amino Acids into Escherichia coli Recombinant Protein: Methodology Development and Recent Achievement. Biomolecules 2019, 9, 255. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.C.; Silverman, S.K.; England, P.M.; Dougherty, D.A.; Lester, H.A. Flash decaging of tyrosine sidechains in an ion channel. Neuron 1998, 20, 619–624. [Google Scholar] [CrossRef]
- Hoppmann, C.; Wang, L. Chapter Twelve—Genetically encoding photoswitchable click amino acids for general optical control of conformation and function of proteins. In Methods in Enzymology; Deiters, A., Ed.; Academic Press: Cambridge, MA, USA, 2019; Volume 624, pp. 249–264. [Google Scholar]
- Feng, Z.; Zhang, W.; Xu, J.; Gauron, C.; Ducos, B.; Vriz, S.; Volovitch, M.; Jullien, L.; Weiss, S.; Bensimon, D. Optical control and study of biological processes at the single-cell level in a live organism. Rep. Prog. Phys. 2013, 76, 072601. [Google Scholar] [CrossRef]
- Baker, A.S.; Deiters, A. Optical Control of Protein Function through Unnatural Amino Acid Mutagenesis and Other Optogenetic Approaches. ACS Chem. Biol. 2014, 9, 1398–1407. [Google Scholar] [CrossRef]
- Courtney, T.; Deiters, A. Recent advances in the optical control of protein function through genetic code expansion. Curr. Opin. Chem. Biol. 2018, 46, 99–107. [Google Scholar] [CrossRef]
- Kang, J.Y.; Kawaguchi, D.; Coin, I.; Xiang, Z.; O’Leary, D.D.; Slesinger, P.A.; Wang, L. In vivo expression of a light-activatable potassium channel using unnatural amino acids. Neuron 2013, 80, 358–370. [Google Scholar] [CrossRef]
- Zhu, S.; Riou, M.; Yao, C.A.; Carvalho, S.; Rodriguez, P.C.; Bensaude, O.; Paoletti, P.; Ye, S. Genetically encoding a light switch in an ionotropic glutamate receptor reveals subunit-specific interfaces. Proc. Natl. Acad. Sci. USA 2014, 111, 6081–6086. [Google Scholar] [CrossRef]
- Klippenstein, V.; Mony, L.; Paoletti, P. Probing Ion Channel Structure and Function Using Light-Sensitive Amino Acids. Trends Biochem. Sci. 2018, 43, 436–451. [Google Scholar] [CrossRef] [PubMed]
- Lemke, E.A.; Summerer, D.; Geierstanger, B.H.; Brittain, S.M.; Schultz, P.G. Control of protein phosphorylation with a genetically encoded photocaged amino acid. Nat. Chem. Biol. 2007, 3, 769–772. [Google Scholar] [CrossRef] [PubMed]
- Courtney, T.M.; Deiters, A. Optical control of protein phosphatase function. Nat. Commun. 2019, 10, 4384. [Google Scholar] [CrossRef]
- Ryan, A.; Zhou, W.; Shoger, K.E.; Courtney, T.M.; Gottschalk, R.A.; Deiters, A. Optogenetic Control of Phosphatase and Kinase Function. FASEB J. 2022, 36. [Google Scholar] [CrossRef]
- Hemphill, J.; Chou, C.; Chin, J.W.; Deiters, A. Genetically encoded light-activated transcription for spatiotemporal control of gene expression and gene silencing in mammalian cells. J. Am. Chem. Soc. 2013, 135, 13433–13439. [Google Scholar] [CrossRef]
- Davis, L.; Radman, I.; Goutou, A.; Tynan, A.; Baxter, K.; Xi, Z.Y.; O’Shea, J.M.; Chin, J.W.; Greiss, S. Precise optical control of gene expression in C elegans using improved genetic code expansion and Cre recombinase. eLife 2021, 10, e67075. [Google Scholar] [CrossRef] [PubMed]
- Walker, O.S.; Elsässer, S.J.; Mahesh, M.; Bachman, M.; Balasubramanian, S.; Chin, J.W. Photoactivation of Mutant Isocitrate Dehydrogenase 2 Reveals Rapid Cancer-Associated Metabolic and Epigenetic Changes. J. Am. Chem. Soc. 2016, 138, 718–721. [Google Scholar] [CrossRef] [PubMed]
- Wolffgramm, J.; Buchmuller, B.; Palei, S.; Muñoz-López, Á.; Kanne, J.; Janning, P.; Schweiger, M.R.; Summerer, D. Light-Activation of DNA-Methyltransferases. Angew. Chem. Int. Ed. 2021, 60, 13507–13512. [Google Scholar] [CrossRef]
- Luo, J.; Arbely, E.; Zhang, J.; Chou, C.; Uprety, R.; Chin, J.W.; Deiters, A. Genetically encoded optical activation of DNA recombination in human cells. Chem. Commun. 2016, 52, 8529–8532. [Google Scholar] [CrossRef]
- Gautier, A.; Nguyen, D.P.; Lusic, H.; An, W.; Deiters, A.; Chin, J.W. Genetically encoded photocontrol of protein localization in mammalian cells. J. Am. Chem. Soc. 2010, 132, 4086–4088. [Google Scholar] [CrossRef]
- Ryan, A.; Hammond, G.R.V.; Deiters, A. Optical Control of Phosphoinositide Binding: Rapid Activation of Subcellular Protein Translocation and Cell Signaling. ACS Synth. Biol. 2021, 10, 2886–2895. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Jia, H.Y.; Ramirez-Diaz, D.A.; Budisa, N.; Schwille, P. Fine-Tuning Protein Self-Organization by Orthogonal Chemo-Optogenetic Tools. Angew. Chem. Int. Ed. 2021, 60, 4501–4506. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Ji, A.; Ai, H.W. Light activation of protein splicing with a photocaged fast intein. J. Am. Chem. Soc. 2015, 137, 2155–2158. [Google Scholar] [CrossRef]
- Cruz-Samperio, R.; Mart, R.J.; Luk, L.Y.P.; Tsai, Y.H.; Jones, A.T.; Allemann, R.K. Spatio-Temporal Photoactivation of Cytotoxic Proteins. ChemBioChem 2022, 23, e202200115. [Google Scholar] [CrossRef]
- He, J.; Fan, Z.; Tian, Y.; Yang, W.; Zhou, Y.; Zhu, Q.; Zhang, W.; Qin, W.; Yi, W. Spatiotemporal Activation of Protein O-GlcNAcylation in Living Cells. J. Am. Chem. Soc. 2022, 144, 4289–4293. [Google Scholar] [CrossRef] [PubMed]
- Courtney, T.M.; Darrah, K.E.; Horst, T.J.; Tsang, M.; Deiters, A. Blue Light Activated Rapamycin for Optical Control of Protein Dimerization in Cells and Zebrafish Embryos. ACS Chem. Biol. 2021, 16, 2434–2443. [Google Scholar] [CrossRef]
- Awad, L.; Jejelava, N.; Burai, R.; Lashuel, H.A. A New Caged-Glutamine Derivative as a Tool To Control the Assembly of Glutamine-Containing Amyloidogenic Peptides. ChemBioChem 2016, 17, 2353–2360. [Google Scholar] [CrossRef]
- Olson, J.P.; Kwon, H.B.; Takasaki, K.T.; Chiu, C.Q.; Higley, M.J.; Sabatini, B.L.; Ellis-Davies, G.C. Optically selective two-photon uncaging of glutamate at 900 nm. J. Am. Chem. Soc. 2013, 135, 5954–5957. [Google Scholar] [CrossRef]
- Ramesh, D.; Wieboldt, R.; Billington, A.P.; Carpenter, B.K.; Hess, G.P. Photolabile precursors of biological amides: Synthesis and characterization of caged o-nitrobenzyl derivatives of glutamine, asparagine, glycinamide, and.gamma.-aminobutyramide. J. Org. Chem. 1993, 58, 4599–4605. [Google Scholar] [CrossRef]
- Bliman, D.; Nilsson, J.R.; Kettunen, P.; Andréasson, J.; Grøtli, M. A Caged Ret Kinase Inhibitor and its Effect on Motoneuron Development in Zebrafish Embryos. Sci. Rep. 2015, 5, 13109. [Google Scholar] [CrossRef]
- Kahlstatt, J.; Reiss, P.; Halbritter, T.; Essen, L.O.; Koert, U.; Heckel, A. A light-triggered transmembrane porin. Chem. Commun. 2018, 54, 9623–9626. [Google Scholar] [CrossRef]
- Luo, J.; Uprety, R.; Naro, Y.; Chou, C.; Nguyen, D.P.; Chin, J.W.; Deiters, A. Genetically encoded optochemical probes for simultaneous fluorescence reporting and light activation of protein function with two-photon excitation. J. Am. Chem. Soc. 2014, 136, 15551–15558. [Google Scholar] [CrossRef] [PubMed]
- Shembekar, V.R.; Chen, Y.; Carpenter, B.K.; Hess, G.P. Coumarin-caged glycine that can be photolyzed within 3 microseconds by visible light. Biochemistry 2007, 46, 5479–5484. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, H.J.; Perdicakis, B.; Fishlock, D.; Lajoie, G.A.; Jervis, E.; Guy Guillemette, J. Photo-Control of nitric oxide synthase activity using a caged isoform specific inhibitor. Bioorg. Med. Chem. 2002, 10, 1919–1927. [Google Scholar] [CrossRef]
- Cueto Diaz, E.; Picard, S.; Klausen, M.; Hugues, V.; Pagano, P.; Genin, E.; Blanchard-Desce, M. Cooperative Veratryle and Nitroindoline Cages for Two-Photon Uncaging in the NIR. Chem. Eur. J. 2016, 22, 10848–10859. [Google Scholar] [CrossRef]
- Papageorgiou, G.; Corrie, J.E.T. Synthesis and Photolytic Assessment of Nitroindolinyl-Caged Calcium Ion Chelators. Molecules 2022, 27, 2645. [Google Scholar] [CrossRef]
- Hayashi, K.-i.; Kusaka, N.; Yamasaki, S.; Zhao, Y.; Nozaki, H. Development of 4-methoxy-7-nitroindolinyl (MNI)-caged auxins which are extremely stable in planta. Bioorg. Med. Chem. Lett. 2015, 25, 4464–4471. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, N.; Liu, P.; Miller, E.W.; Weinstain, R. meso-Methylhydroxy BODIPY: A scaffold for photo-labile protecting groups. Chem. Commun. 2015, 51, 6369–6372. [Google Scholar] [CrossRef]
- Goswami, P.P.; Syed, A.; Beck, C.L.; Albright, T.R.; Mahoney, K.M.; Unash, R.; Smith, E.A.; Winter, A.H. BODIPY-Derived Photoremovable Protecting Groups Unmasked with Green Light. J. Am. Chem. Soc. 2015, 137, 3783–3786. [Google Scholar] [CrossRef]
- Peterson, J.A.; Fischer, L.J.; Gehrmann, E.J.; Shrestha, P.; Yuan, D.; Wijesooriya, C.S.; Smith, E.A.; Winter, A.H. Direct Photorelease of Alcohols from Boron-Alkylated BODIPY Photocages. J. Org. Chem. 2020, 85, 5712–5717. [Google Scholar] [CrossRef]
- Toupin, N.P.; Arora, K.; Shrestha, P.; Peterson, J.A.; Fischer, L.J.; Rajagurubandara, E.; Podgorski, I.; Winter, A.H.; Kodanko, J.J. BODIPY-Caged Photoactivated Inhibitors of Cathepsin B Flip the Light Switch on Cancer Cell Apoptosis. ACS Chem. Biol. 2019, 14, 2833–2840. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Madisen, L. Mouse transgenic approaches in optogenetics. Prog. Brain Res. 2012, 196, 193–213. [Google Scholar] [PubMed]
- White, R.M.; Sessa, A.; Burke, C.; Bowman, T.; LeBlanc, J.; Ceol, C.; Bourque, C.; Dovey, M.; Goessling, W.; Burns, C.E.; et al. Transparent Adult Zebrafish as a Tool for In Vivo Transplantation Analysis. Cell Stem Cell 2008, 2, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Andrasfalvy, B.K.; Zemelman, B.V.; Tang, J.; Vaziri, A. Two-photon single-cell optogenetic control of neuronal activity by sculpted light. Proc. Natl. Acad. Sci. USA 2010, 107, 11981–11986. [Google Scholar] [CrossRef]
- Hoover, E.E.; Squier, J.A. Advances in multiphoton microscopy technology. Nat. Photonics 2013, 7, 93–101. [Google Scholar] [CrossRef]
- Hontani, Y.; Xia, F.; Xu, C. Multicolor three-photon fluorescence imaging with single-wavelength excitation deep in mouse brain. Sci. Adv. 2021, 7, eabf3531. [Google Scholar] [CrossRef]
- Kokel, D.; Cheung, C.Y.J.; Mills, R.; Coutinho-Budd, J.; Huang, L.; Setola, V.; Sprague, J.; Jin, S.; Jin, Y.N.; Huang, X.-P.; et al. Photochemical activation of TRPA1 channels in neurons and animals. Nat. Chem. Biol. 2013, 9, 257–263. [Google Scholar] [CrossRef]
- Rovira, X.; Trapero, A.; Pittolo, S.; Zussy, C.; Faucherre, A.; Jopling, C.; Giraldo, J.; Pin, J.-P.; Gorostiza, P.; Goudet, C.; et al. OptoGluNAM4.1, a Photoswitchable Allosteric Antagonist for Real-Time Control of mGlu4 Receptor Activity. Cell Chem. Biol. 2016, 23, 929–934. [Google Scholar] [CrossRef]
- Gómez-Santacana, X.; Pittolo, S.; Rovira, X.; Lopez, M.; Zussy, C.; Dalton, J.A.; Faucherre, A.; Jopling, C.; Pin, J.P.; Ciruela, F.; et al. Illuminating Phenylazopyridines To Photoswitch Metabotropic Glutamate Receptors: From the Flask to the Animals. ACS Cent. Sci. 2017, 3, 81–91. [Google Scholar] [CrossRef]
- Barber, D.M.; Schönberger, M.; Burgstaller, J.; Levitz, J.; Weaver, C.D.; Isacoff, E.Y.; Baier, H.; Trauner, D. Optical control of neuronal activity using a light-operated GIRK channel opener (LOGO). Chem. Sci. 2016, 7, 2347–2352. [Google Scholar] [CrossRef]
- Volgraf, M.; Gorostiza, P.; Numano, R.; Kramer, R.H.; Isacoff, E.Y.; Trauner, D. Allosteric control of an ionotropic glutamate receptor with an optical switch. Nat. Chem. Biol. 2006, 2, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Szobota, S.; Gorostiza, P.; Del Bene, F.; Wyart, C.; Fortin, D.L.; Kolstad, K.D.; Tulyathan, O.; Volgraf, M.; Numano, R.; Aaron, H.L.; et al. Remote Control of Neuronal Activity with a Light-Gated Glutamate Receptor. Neuron 2007, 54, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Wyart, C.; Del Bene, F.; Warp, E.; Scott, E.K.; Trauner, D.; Baier, H.; Isacoff, E.Y. Optogenetic dissection of a behavioural module in the vertebrate spinal cord. Nature 2009, 461, 407–410. [Google Scholar] [CrossRef]
- Janovjak, H.; Szobota, S.; Wyart, C.; Trauner, D.; Isacoff, E.Y. A light-gated, potassium-selective glutamate receptor for the optical inhibition of neuronal firing. Nat. Neurosci. 2010, 13, 1027–1032. [Google Scholar] [CrossRef]
- Levitz, J.; Pantoja, C.; Gaub, B.; Janovjak, H.; Reiner, A.; Hoagland, A.; Schoppik, D.; Kane, B.; Stawski, P.; Schier, A.F.; et al. Optical control of metabotropic glutamate receptors. Nat. Neurosci. 2013, 16, 507–516. [Google Scholar] [CrossRef]
- Berlin, S.; Szobota, S.; Reiner, A.; Carroll, E.C.; Kienzler, M.A.; Guyon, A.; Xiao, T.; Trauner, D.; Isacoff, E.Y. A family of photoswitchable NMDA receptors. eLife 2016, 5, e12040. [Google Scholar] [CrossRef]
- Patton, E.E.; Zon, L.I. The art and design of genetic screens: Zebrafish. Nat. Rev. Genet. 2001, 2, 956–966. [Google Scholar] [CrossRef]
- Fuentes, R.; Letelier, J.; Tajer, B.; Valdivia, L.E.; Mullins, M.C. Fishing forward and reverse: Advances in zebrafish phenomics. Mech. Dev. 2018, 154, 296–308. [Google Scholar] [CrossRef] [PubMed]
- Schroter, C.; Herrgen, L.; Cardona, A.; Brouhard, G.J.; Feldman, B.; Oates, A.C. Dynamics of zebrafish somitogenesis. Dev. Dyn. 2008, 237, 545–553. [Google Scholar] [CrossRef]
- Naganathan, S.R.; Oates, A.C. Patterning and mechanics of somite boundaries in zebrafish embryos. Semin. Cell Dev. Biol. 2020, 107, 170–178. [Google Scholar] [CrossRef]
- Ducos, B.; Bensimon, D.; Scerbo, P. Vertebrate Cell Differentiation, Evolution, and Diseases: The Vertebrate-Specific Developmental Potential Guardians VENTX/NANOG and POU5/OCT4 Enter the Stage. Cells 2022, 11, 2299. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.F.; Miller, A.L.; Korzh, V.; Chong, S.-W.; Sleptsova-Freidrich, I.; Webb, S.E. Visualization of stochastic Ca2+ signals in the formed somites during the early segmentation period in intact, normally developing zebrafish embryos. Dev. Growth Differ. 2009, 51, 617–637. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Feng, Z.; Sinha, D.; Ducos, B.; Ebenstein, Y.; Tadmor, A.D.; Gauron, C.; Le Saux, T.; Lin, S.; Weiss, S.; et al. Spatiotemporal manipulation of retinoic acid activity in zebrafish hindbrain development via photo-isomerization. Development 2012, 139, 3355–3362. [Google Scholar] [CrossRef] [PubMed]
- Valdivia, L.E.; Lamb, D.B.; Horner, W.; Wierzbicki, C.; Tafessu, A.; Williams, A.M.; Gestri, G.; Krasnow, A.M.; Vleeshouwer-Neumann, T.S.; Givens, M.; et al. Antagonism between Gdf6a and retinoic acid pathways controls timing of retinal neurogenesis and growth of the eye in zebrafish. Development 2016, 143, 1087–1098. [Google Scholar] [CrossRef] [PubMed]
- Payumo, A.Y.; Walker, W.J.; McQuade, L.E.; Yamazoe, S.; Chen, J.K. Optochemical dissection of T-box gene-dependent medial floor plate development. ACS Chem. Biol. 2015, 10, 1466–1475. [Google Scholar] [CrossRef]
- Shestopalov, I.A.; Pitt, C.L.; Chen, J.K. Spatiotemporal resolution of the Ntla transcriptome in axial mesoderm development. Nat. Chem. Biol. 2012, 8, 270–276. [Google Scholar] [CrossRef]
- Payumo, A.Y.; McQuade, L.E.; Walker, W.J.; Yamazoe, S.; Chen, J.K. Tbx16 regulates hox gene activation in mesodermal progenitor cells. Nat. Chem. Biol. 2016, 12, 694–701. [Google Scholar] [CrossRef]
- Liu, J.; Hemphill, J.; Samanta, S.; Tsang, M.; Deiters, A. Genetic Code Expansion in Zebrafish Embryos and Its Application to Optical Control of Cell Signaling. J. Am. Chem. Soc. 2017, 139, 9100–9103. [Google Scholar] [CrossRef] [PubMed]
- Cruz, F.G.; Koh, J.T.; Link, K.H. Light-Activated Gene Expression. J. Am. Chem. Soc. 2000, 122, 8777–8778. [Google Scholar] [CrossRef]
- Lin, W.; Albanese, C.; Pestell, R.G.; Lawrence, D.S. Spatially discrete, light-driven protein expression. Chem. Biol. 2002, 9, 1347–1353. [Google Scholar] [CrossRef]
- Young, D.D.; Deiters, A. Photochemical activation of protein expression in bacterial cells. Angew. Chem. Int. Ed. 2007, 46, 4290–4292. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Nam, S.; Hamouri, F.; Aujard, I.; Ducos, B.; Vriz, S.; Volovitch, M.; Jullien, L.; Lin, S.; Weiss, S.; et al. Optical Control of Tumor Induction in the Zebrafish. Sci. Rep. 2017, 7, 9195. [Google Scholar] [CrossRef]
- Brown, W.; Liu, J.; Tsang, M.; Deiters, A. Cell-Lineage Tracing in Zebrafish Embryos with an Expanded Genetic Code. ChemBioChem 2018, 19, 1244–1249. [Google Scholar] [CrossRef] [PubMed]
- Deiters, A.; Garner, R.A.; Lusic, H.; Govan, J.M.; Dush, M.; Nascone-Yoder, N.M.; Yoder, J.A. Photocaged morpholino oligomers for the light-regulation of gene function in zebrafish and Xenopus embryos. J. Am. Chem. Soc. 2010, 132, 15644–15650. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Wang, H.; La Russa, M.; Qi, L.S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016, 85, 227–264. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Doudna, J.A. Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 2016, 34, 933–941. [Google Scholar] [CrossRef]
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018, 9, 1911. [Google Scholar] [CrossRef]
- Dow, L.E.; Fisher, J.; O’Rourke, K.P.; Muley, A.; Kastenhuber, E.R.; Livshits, G.; Tschaharganeh, D.F.; Socci, N.D.; Lowe, S.W. Inducible in vivo genome editing with CRISPR-Cas9. Nat. Biotechnol. 2015, 33, 390–394. [Google Scholar] [CrossRef]
- Polstein, L.R.; Gersbach, C.A. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol. 2015, 11, 198–200. [Google Scholar] [CrossRef]
- Davis, K.M.; Pattanayak, V.; Thompson, D.B.; Zuris, J.A.; Liu, D.R. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat. Chem. Biol. 2015, 11, 316–318. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.K.; Ramanan, V.; Schepers, A.G.; Dalvie, N.S.; Panda, A.; Fleming, H.E.; Bhatia, S.N. Development of Light-Activated CRISPR Using Guide RNAs with Photocleavable Protectors. Angew. Chem. Int. Ed. 2016, 55, 12440–12444. [Google Scholar] [CrossRef] [PubMed]
- Maji, B.; Moore, C.L.; Zetsche, B.; Volz, S.E.; Zhang, F.; Shoulders, M.D.; Choudhary, A. Multidimensional chemical control of CRISPR-Cas9. Nat. Chem. Biol. 2017, 13, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ling, X.Y.; Su, X.X.; Zhang, S.L.; Wang, J.; Zhang, P.J.; Feng, W.J.; Zhu, Y.Y.; Liu, T.; Tang, X.J. Optical Control of a CRISPR/Cas9 System for Gene Editing by Using Photolabile crRNA. Angew. Chem. Int. Ed. 2020, 59, 20895–20899. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wei, L.; Wang, J.Q.; Ji, H.; Xiong, W.; Liu, J.; Yin, P.; Tian, T.; Zhou, X. Light-Driven Activation of RNA-Guided Nucleic Acid Cleavage. ACS Chem. Biol. 2020, 15, 1455–1463. [Google Scholar] [CrossRef]
- Hemphill, J.; Borchardt, E.K.; Brown, K.; Asokan, A.; Deiters, A. Optical Control of CRISPR/Cas9 Gene Editing. J. Am. Chem. Soc. 2015, 137, 5642–5645. [Google Scholar] [CrossRef]
- Ko, S.K.; Chen, X.; Yoon, J.; Shin, I. Zebrafish as a good vertebrate model for molecular imaging using fluorescent probes. Chem. Soc. Rev. 2011, 40, 2120–2130. [Google Scholar] [CrossRef]
- Hatta, K.; Tsujii, H.; Omura, T. Cell tracking using a photoconvertible fluorescent protein. Nat. Protoc. 2006, 1, 960–967. [Google Scholar] [CrossRef]
- Kwan, K.M.; Otsuna, H.; Kidokoro, H.; Carney, K.R.; Saijoh, Y.; Chien, C.-B. A complex choreography of cell movements shapes the vertebrate eye. Development 2012, 139, 359–372. [Google Scholar] [CrossRef]
- Zhong, T.P.; Childs, S.; Leu, J.P.; Fishman, M.C. Gridlock signalling pathway fashions the first embryonic artery. Nature 2001, 414, 216–220. [Google Scholar] [CrossRef]
- Childs, S.; Chen, J.-N.; Garrity, D.M.; Fishman, M.C. Patterning of angiogenesis in the zebrafish embryo. Development 2002, 129, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Vogeli, K.M.; Jin, S.W.; Martin, G.R.; Stainier, D.Y. A common progenitor for haematopoietic and endothelial lineages in the zebrafish gastrula. Nature 2006, 443, 337–339. [Google Scholar] [CrossRef]
- Warga, R.M.; Kane, D.A.; Ho, R.K. Fate mapping embryonic blood in zebrafish: Multi- and unipotential lineages are segregated at gastrulation. Dev. Cell 2009, 16, 744–755. [Google Scholar] [CrossRef] [PubMed]
- Lam, E.Y.N.; Hall, C.J.; Crosier, P.S.; Crosier, K.E.; Flores, M.V. Live imaging of Runx1 expression in the dorsal aorta tracks the emergence of blood progenitors from endothelial cells. Blood 2010, 116, 909–914. [Google Scholar] [CrossRef]
- Campos, C.; Kamiya, M.; Banala, S.; Johnsson, K.; González-Gaitán, M. Labelling cell structures and tracking cell lineage in zebrafish using SNAP-tag. Dev. Dyn. 2011, 240, 820–827. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.L.; Yeung, E.; McGuire, S.E.; Wu, A.Y.; Toettcher, J.E.; Burdine, R.D.; Shvartsman, S.Y. Optimizing photoswitchable MEK. Proc. Natl. Acad. Sci. USA 2019, 116, 25756–25763. [Google Scholar] [CrossRef] [PubMed]
- Morales, R.A.; Allende, M.L. Peripheral macrophages promote tissue regeneration in zebrafish by fine-tuning the inflammatory response. Front. Immunol. 2019, 10, 253. [Google Scholar] [CrossRef]
- Tauzin, S.; Starnes, T.W.; Becker, F.B.; Lam, P.-y.; Huttenlocher, A. Redox and Src family kinase signaling control leukocyte wound attraction and neutrophil reverse migration. J. Cell Biol. 2014, 207, 589–598. [Google Scholar] [CrossRef]
- Bischel, L.L.; Mader, B.R.; Green, J.M.; Huttenlocher, A.; Beebe, D.J. Zebrafish Entrapment By Restriction Array (ZEBRA) device: A low-cost, agarose-free zebrafish mounting technique for automated imaging. Lab Chip 2013, 13, 1732–1736. [Google Scholar] [CrossRef]
- Schermelleh, L.; Ferrand, A.; Huser, T.; Eggeling, C.; Sauer, M.; Biehlmaier, O.; Drummen, G.P.C. Super-resolution microscopy demystified. Nat. Cell Biol. 2019, 21, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Bates, M.; Zhuang, X.W. Super-Resolution Fluorescence Microscopy. Annu. Rev. Biochem. 2009, 78, 993–1016. [Google Scholar] [CrossRef] [PubMed]
- Sigal, Y.M.; Zhou, R.B.; Zhuang, X.W. Visualizing and discovering cellular structures with super-resolution microscopy. Science 2018, 361, 880–887. [Google Scholar] [CrossRef]
- Jing, Y.Y.; Zhang, C.S.; Yu, B.; Lin, D.Y.; Qu, J.L. Super-Resolution Microscopy: Shedding New Light on In Vivo Imaging. Front. Chem. 2021, 9, 740. [Google Scholar]
- Wang, S.Y.; Moffitt, J.R.; Dempsey, G.T.; Xie, X.S.; Zhuang, X.W. Characterization and development of photoactivatable fluorescent proteins for single-molecule-based superresolution imaging. Proc. Natl. Acad. Sci. USA 2014, 111, 8452–8457. [Google Scholar] [CrossRef] [PubMed]
- Minoshima, M.; Kikuchi, K. Photostable and photoswitching fluorescent dyes for super-resolution imaging. J. Biol. Inorg. Chem. 2017, 22, 639–652. [Google Scholar] [CrossRef]
- Raut, P.; Weller, S.R.; Obeng, B.; Soos, B.L.; West, B.E.; Potts, C.M.; Sangroula, S.; Kinney, M.S.; Burnell, J.E.; King, B.L.; et al. Cetylpyridinium chloride (CPC) reduces zebrafish mortality from influenza infection: Super-resolution microscopy reveals CPC interference with multiple protein interactions with phosphatidylinositol 4,5-bisphosphate in immune function. Toxicol. Appl. Pharmacol. 2022, 440, 115913. [Google Scholar] [CrossRef]
- Gabor, K.A.; Kim, D.; Kim, C.H.; Hess, S.T. Nanoscale imaging of caveolin-1 membrane domains in vivo. PLoS ONE 2015, 10, e0117225. [Google Scholar] [CrossRef]
- Gabor, K.A.; Stevens, C.R.; Pietraszewski, M.J.; Gould, T.J.; Shim, J.; Yoder, J.A.; Lam, S.H.; Gong, Z.; Hess, S.T.; Kim, C.H. Super resolution microscopy reveals that caveolin-1 is required for spatial organization of CRFB1 and subsequent antiviral signaling in zebrafish. PLoS ONE 2013, 8, e68759. [Google Scholar] [CrossRef]
- Middel, V.; Zhou, L.; Takamiya, M.; Beil, T.; Shahid, M.; Roostalu, U.; Grabher, C.; Rastegar, S.; Reischl, M.; Nienhaus, G.U. Dysferlin-mediated phosphatidylserine sorting engages macrophages in sarcolemma repair. Nat. Commun. 2016, 7, 12875. [Google Scholar] [CrossRef]
- Gemberling, M.; Bailey, T.J.; Hyde, D.R.; Poss, K.D. The zebrafish as a model for complex tissue regeneration. Trends Genet. 2013, 29, 611–620. [Google Scholar] [CrossRef]
- Marques, I.J.; Lupi, E.; Mercader, N. Model systems for regeneration: Zebrafish. Development 2019, 146, dev167692. [Google Scholar] [CrossRef]
- Mruk, K.; Ciepla, P.; Piza, P.A.; Alnaqib, M.A.; Chen, J.K. Targeted cell ablation in zebrafish using optogenetic transcriptional control. Development 2020, 147, dev183640. [Google Scholar] [CrossRef]
- Xiao, Y.; Tian, W.L.; Lopez-Schier, H. Optogenetic stimulation of neuronal repair. Curr. Biol. 2015, 25, R1068–R1069. [Google Scholar] [CrossRef]
- Asakawa, K.; Handa, H.; Kawakami, K. Optogenetic modulation of TDP-43 oligomerization accelerates ALS-related pathologies in the spinal motor neurons. Nat. Commun. 2020, 11, 1004. [Google Scholar] [CrossRef]
- Duran-Corbera, A.; Faria, M.; Ma, Y.; Prats, E.; Dias, A.; Catena, J.; Martinez, K.L.; Raldua, D.; Llebaria, A.; Rovira, X. A Photoswitchable Ligand Targeting the β1-Adrenoceptor Enables Light-Control of the Cardiac Rhythm. Angew. Chem. Int. Ed. 2022, 61, e202203449. [Google Scholar] [CrossRef]
- Pittolo, S.; Gómez-Santacana, X.; Eckelt, K.; Rovira, X.; Dalton, J.; Goudet, C.; Pin, J.-P.; Llobet, A.; Giraldo, J.; Llebaria, A.; et al. An allosteric modulator to control endogenous G protein-coupled receptors with light. Nat. Chem. Biol. 2014, 10, 813–815. [Google Scholar] [CrossRef]
- Clanton, J.A.; Shestopalov, I.; Chen, J.K.; Gamse, J.T. Lineage labeling of zebrafish cells with laser uncagable fluorescein dextran. J. Vis. Exp. 2011, 50, e2672. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, S.-i.; Hatta, K. Ca2+-imaging and photo-manipulation of the simple gut of zebrafish larvae in vivo. Sci. Rep. 2022, 12, 2018. [Google Scholar] [CrossRef] [PubMed]
- Mandal, A.; Wong, H.C.; Pinter, K.; Mosqueda, N.; Beirl, A.; Lomash, R.M.; Won, S.; Kindt, K.S.; Drerup, C.M. Retrograde Mitochondrial Transport Is Essential for Organelle Distribution and Health in Zebrafish Neurons. J. Neurosci. 2021, 41, 1371–1392. [Google Scholar] [CrossRef]
- De Simone, A.; Evanitsky, M.N.; Hayden, L.; Cox, B.D.; Wang, J.; Tornini, V.A.; Ou, J.; Chao, A.; Poss, K.D.; Di Talia, S. Control of osteoblast regeneration by a train of Erk activity waves. Nature 2021, 590, 129–133. [Google Scholar] [CrossRef]
- Godley, B.F.; Shamsi, F.A.; Liang, F.Q.; Jarrett, S.G.; Davies, S.; Boulton, M. Blue light induces mitochondrial DNA damage and free radical production in epithelial cells. J. Biol. Chem. 2005, 280, 21061–21066. [Google Scholar] [CrossRef]
- Owen, S.F.; Liu, M.H.; Kreitzer, A.C. Thermal constraints on in vivo optogenetic manipulations. Nat. Neurosci. 2019, 22, 1061–1065. [Google Scholar] [CrossRef] [PubMed]
- Villamizar, N.; Vera, L.M.; Foulkes, N.S.; Sánchez-Vázquez, F.J. Effect of lighting conditions on zebrafish growth and development. Zebrafish 2014, 11, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Sigurgeirsson, B.; Þorsteinsson, H.; Sigmundsdóttir, S.; Lieder, R.; Sveinsdóttir, H.S.; Sigurjónsson, Ó.E.; Halldórsson, B.; Karlsson, K. Sleep–wake dynamics under extended light and extended dark conditions in adult zebrafish. Behav. Brain Res. 2013, 256, 377–390. [Google Scholar] [CrossRef]
- Jia, S.; Sletten, E.M. Spatiotemporal Control of Biology: Synthetic Photochemistry Toolbox with Far-Red and Near-Infrared Light. ACS Chem. Biol. 2021. [Google Scholar] [CrossRef]
- Vorobev, A.Y.; Moskalensky, A.E. Long-wavelength photoremovable protecting groups: On the way to in vivo application. Comput. Struct. Biotechnol. J. 2020, 18, 27–34. [Google Scholar] [CrossRef]
- Lecoq, J.; Orlova, N.; Grewe, B.F. Wide. fast. deep: Recent advances in multiphoton microscopy of in vivo neuronal activity. J. Neurosci. 2019, 39, 9042–9052. [Google Scholar] [CrossRef]
- Williams, R.M.; Zipfel, W.R.; Webb, W.W. Multiphoton microscopy in biological research. Curr. Opin. Chem. Biol. 2001, 5, 603–608. [Google Scholar] [CrossRef]
- Pap, E.H.W.; Drummen, G.P.C.; Winter, V.J.; Kooij, T.W.A.; Rijken, P.; Wirtz, K.W.A.; Op den Kamp, J.A.F.; Hage, W.J.; Post, J.A. Ratio-fluorescence microscopy of lipid oxidation in living cells using C11-BODIPY581/591. FEBS Lett. 1999, 453, 278–282. [Google Scholar] [CrossRef]
- Smith, R.A.J.; Hartley, R.C.; Murphy, M.P. Mitochondria-Targeted Small Molecule Therapeutics and Probes. Antioxid. Redox Signal. 2011, 15, 3021–3038. [Google Scholar] [CrossRef]
- Neto, B.A.; Corrêa, J.R.; Silva, R.G. Selective mitochondrial staining with small fluorescent probes: Importance, design, synthesis, challenges and trends for new markers. RSC Adv. 2013, 3, 5291–5301. [Google Scholar] [CrossRef]
- Tian, L.; Yang, Y.; Wysocki, L.M.; Arnold, A.C.; Hu, A.; Ravichandran, B.; Sternson, S.M.; Looger, L.L.; Lavis, L.D. Selective esterase-ester pair for targeting small molecules with cellular specificity. Proc. Natl. Acad. Sci. USA 2012, 109, 4756–4761. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Tiwari, K.; Tiwari, R.; Pramanik, S.K.; Das, A. Small Molecule as Fluorescent Probes for Monitoring Intracellular Enzymatic Transformations. Chem. Rev. 2019, 119, 11718–11760. [Google Scholar] [CrossRef] [PubMed]
- Kok, F.O.; Shin, M.; Ni, C.W.; Gupta, A.; Grosse, A.S.; van Impel, A.; Kirchmaier, B.C.; Peterson-Maduro, J.; Kourkoulis, G.; Male, I.; et al. Reverse Genetic Screening Reveals Poor Correlation between Morpholino-Induced and Mutant Phenotypes in Zebrafish. Dev. Cell 2015, 32, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Stainier, D.Y.R.; Raz, E.; Lawson, N.D.; Ekker, S.C.; Burdine, R.D.; Eisen, J.S.; Ingham, P.W.; Schulte-Merker, S.; Yelon, D.; Weinstein, B.M.; et al. Guidelines for morpholino use in zebrafish. PLoS Genet. 2017, 13, e1007000. [Google Scholar] [CrossRef]
- Watson, E.E.; Russo, F.; Moreau, D.; Winssinger, N. Optochemical Control of Therapeutic Agents through Photocatalyzed Isomerization. Angew. Chem. 2022, 134, e202203390. [Google Scholar]
- Liu, Y.; Long, K.; Kang, W.; Wang, T.; Wang, W. Optochemical Control of Immune Checkpoint Blockade via Light-Triggered PD-L1 Dimerization. Adv. NanoBiomed Res. 2022, 2, 2200017. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, Z.; Ducos, B.; Scerbo, P.; Aujard, I.; Jullien, L.; Bensimon, D. The Development and Application of Opto-Chemical Tools in the Zebrafish. Molecules 2022, 27, 6231. https://doi.org/10.3390/molecules27196231
Feng Z, Ducos B, Scerbo P, Aujard I, Jullien L, Bensimon D. The Development and Application of Opto-Chemical Tools in the Zebrafish. Molecules. 2022; 27(19):6231. https://doi.org/10.3390/molecules27196231
Chicago/Turabian StyleFeng, Zhiping, Bertrand Ducos, Pierluigi Scerbo, Isabelle Aujard, Ludovic Jullien, and David Bensimon. 2022. "The Development and Application of Opto-Chemical Tools in the Zebrafish" Molecules 27, no. 19: 6231. https://doi.org/10.3390/molecules27196231
APA StyleFeng, Z., Ducos, B., Scerbo, P., Aujard, I., Jullien, L., & Bensimon, D. (2022). The Development and Application of Opto-Chemical Tools in the Zebrafish. Molecules, 27(19), 6231. https://doi.org/10.3390/molecules27196231






