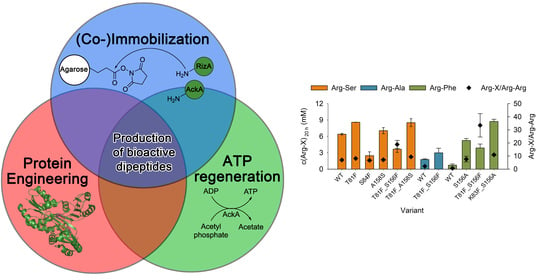
Co-Immobilization of RizA Variants with Acetate Kinase for the Production of Bioactive Arginyl Dipeptides
Abstract
:1. Introduction
2. Results and Discussion
2.1. (Co-)Immobilization Conditions
2.2. Co-Immobilization of RizA Variants
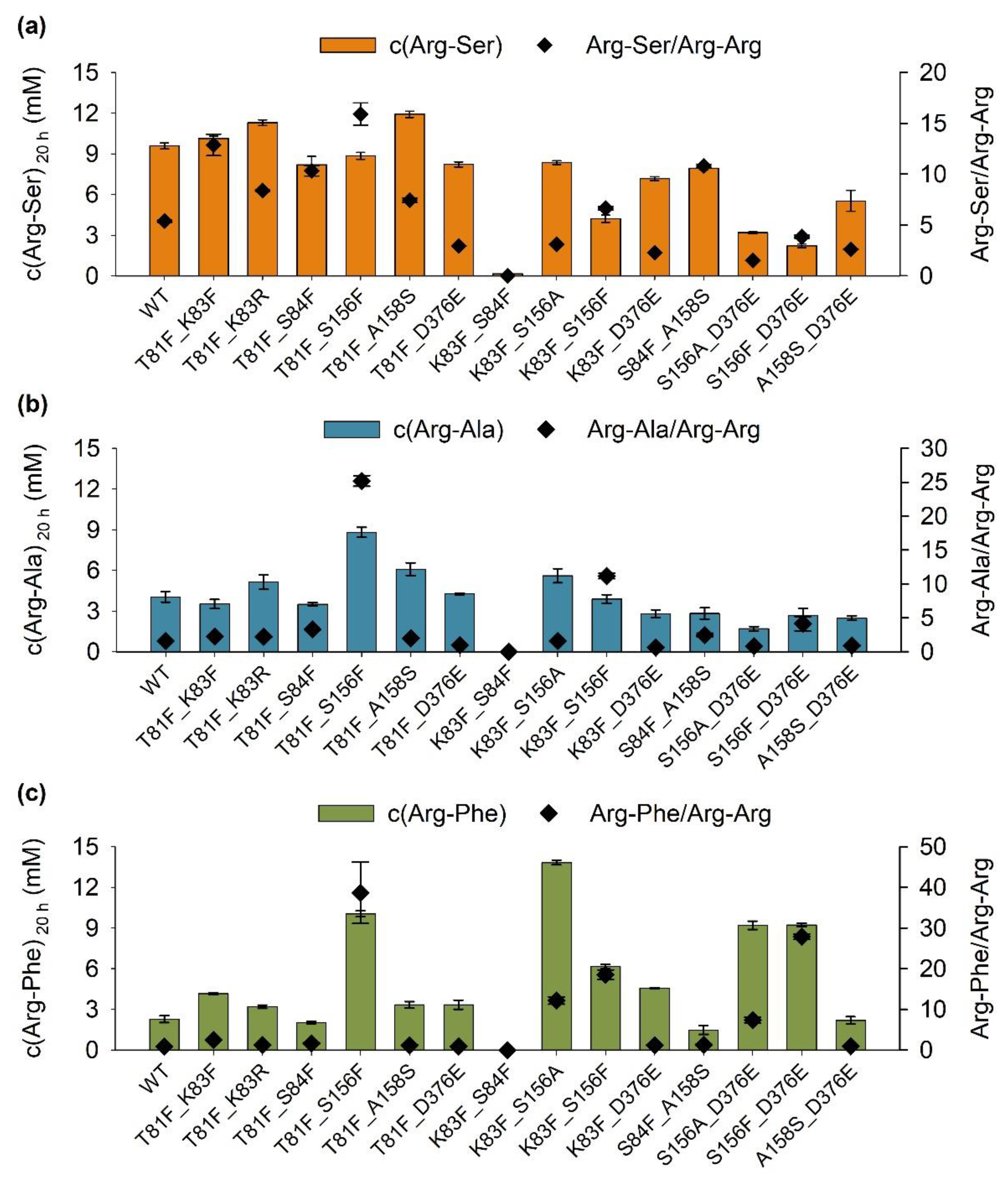
2.2.1. Combination of Mutations to Yield Improved Variants
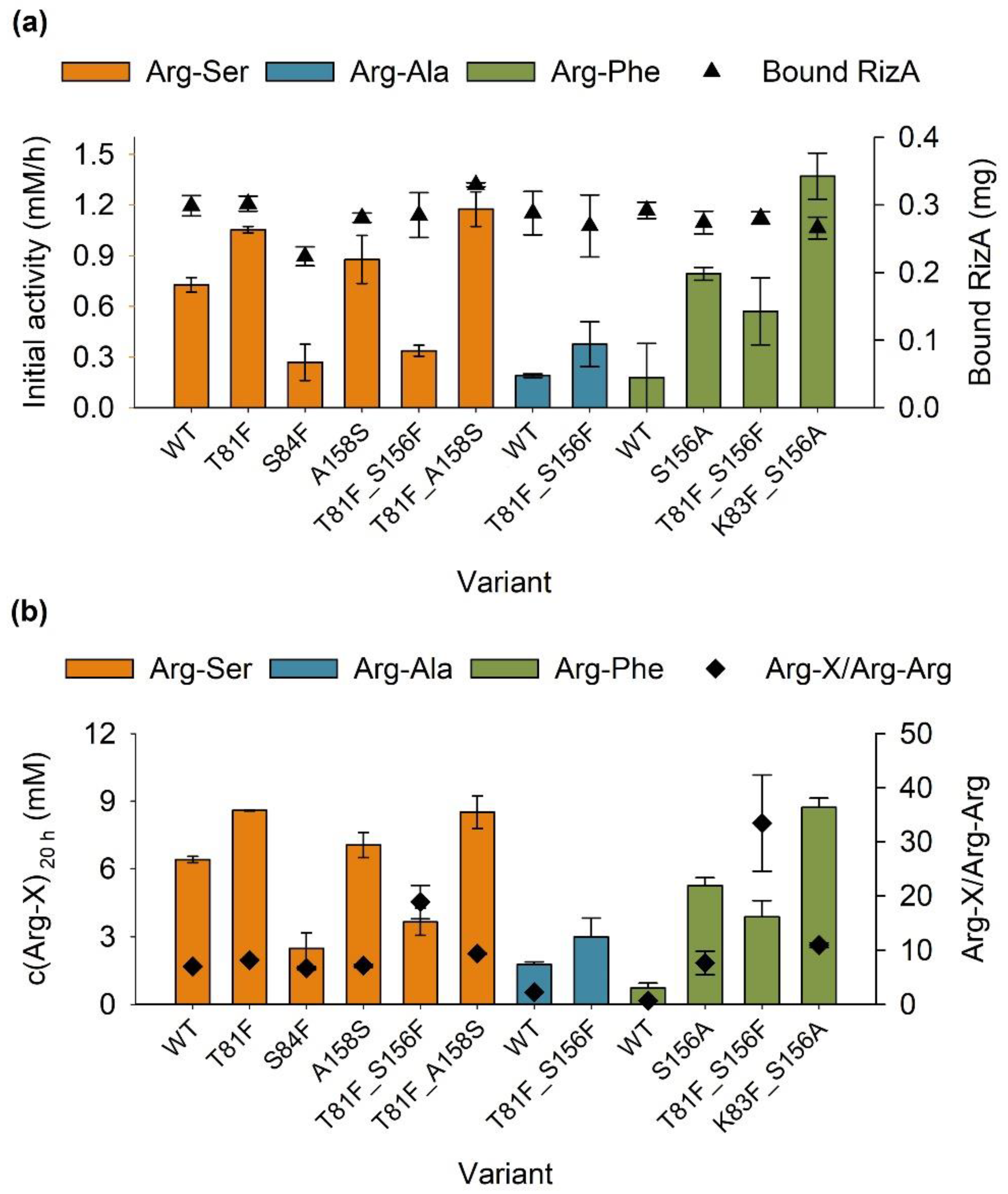
2.2.2. Immobilization of RizA Variants
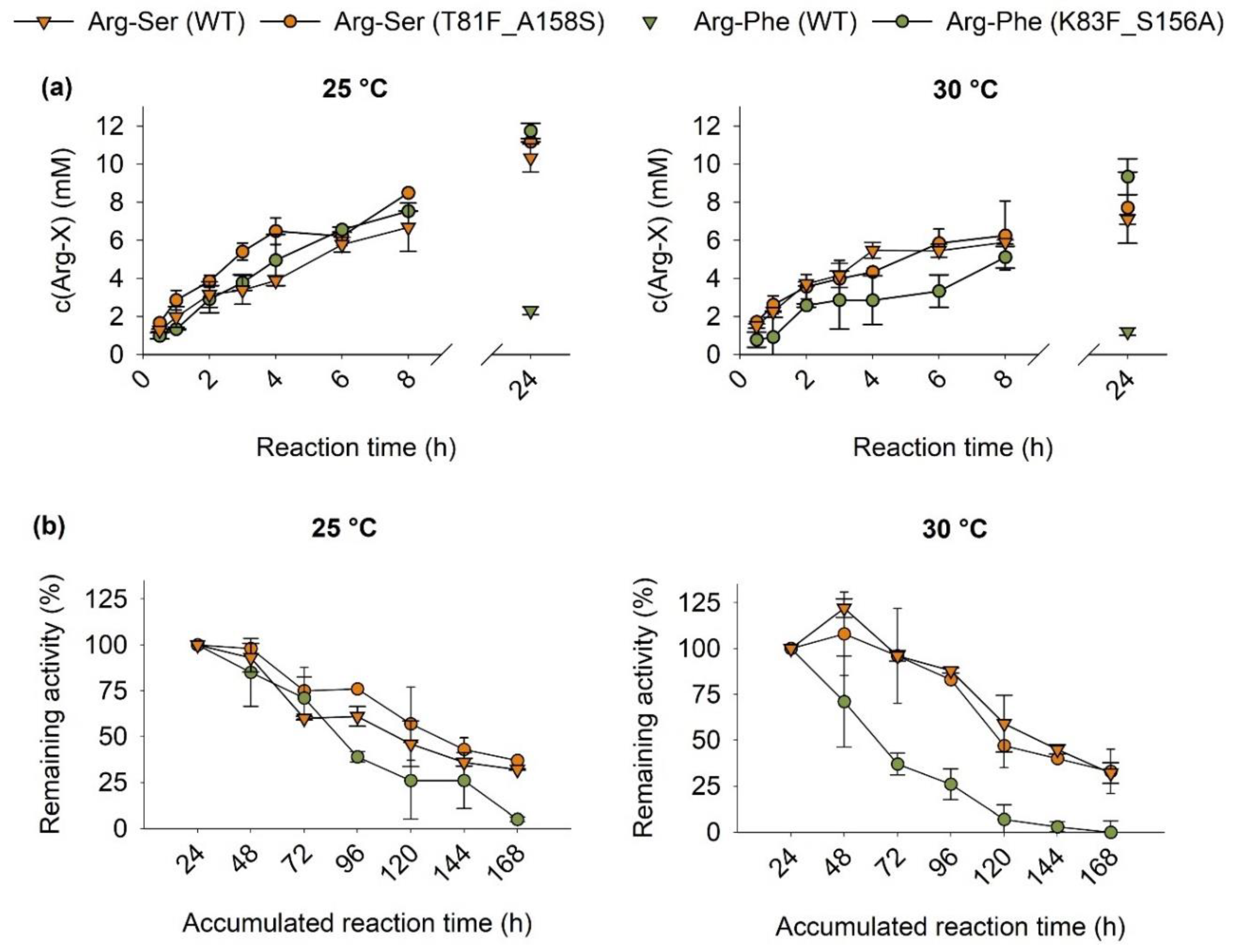
2.3. Characterization of the Immobilisates
3. Materials and Methods
3.1. Chemicals, Reagents and Strains
3.2. Mutagenesis
3.3. Production of Soluble Enzymes
3.4. Immobilization
3.5. Biocatalysis
3.6. Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Wang, T.; Zhang, Y.-F.; Ning, L.-X.; Wang, Y.-F.; Liu, X.-H.; Li, R.; Chen, X.-E. l-amino acid ligase: A promising alternative for the biosynthesis of L-dipeptides. Enzyme Microb. Technol. 2020, 136, 109537. [Google Scholar] [CrossRef] [PubMed]
- Tabata, K.; Ikeda, H.; Hashimoto, S.-I. ywfE in Bacillus subtilis Codes for a Novel Enzyme L-Amino Acid Ligase. J. Bacteriol. 2005, 187, 5195–5202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kino, K.; Nakazawa, Y.; Yagasaki, M. Dipeptide synthesis by l-amino acid ligase from Ralstonia solanacearum. Biochem. Biophys. Res. Commun. 2008, 371, 536–540. [Google Scholar] [CrossRef] [PubMed]
- Kino, K.; Noguchi, A.; Nakazawa, Y.; Yagasaki, M. A novel l-amino acid ligase from Bacillus licheniformis. J. Biosci. Bioeng. 2008, 106, 313–315. [Google Scholar] [CrossRef]
- Arai, T.; Arimura, Y.; Ishikura, S.; Kino, K. L-Amino Acid Ligase from Pseudomonas syringae Producing Tabtoxin Can Be Used for Enzymatic Synthesis of Various Functional Peptides. Appl. Environ. Microbiol. 2013, 79, 5023–5029. [Google Scholar] [CrossRef] [Green Version]
- Kino, K.; Kotanaka, Y.; Arai, T.; Yagasaki, M. A Novel L-Amino Acid Ligase from Bacillus subtilis NBRC3134, a Microorganism Producing Peptide-Antibiotic Rhizocticin. Biosci. Biotechnol. Biochem. 2009, 73, 901–907. [Google Scholar] [CrossRef] [Green Version]
- Schindler, A.; Dunkel, A.; Stähler, F.; Backes, M.; Ley, J.; Meyerhof, W.; Hofmann, T. Discovery of Salt Taste Enhancing Arginyl Dipeptides in Protein Digests and Fermented Fish Sauces by Means of a Sensomics Approach. J. Agric. Food Chem. 2011, 59, 12578–12588. [Google Scholar] [CrossRef]
- Xu, J.-J.; Elkaddi, N.; Garcia-Blanco, A.; Spielman, A.I.; Bachmanov, A.A.; Chung, H.Y.; Ozdener, M.H. Arginyl dipeptides increase the frequency of NaCl-elicited responses via epithelial sodium channel alpha and delta subunits in cultured human fungiform taste papillae cells. Sci. Rep. 2017, 7, 7483. [Google Scholar] [CrossRef]
- Harth, L.; Krah, U.; Linke, D.; Dunkel, A.; Hofmann, T.; Berger, R.G. Salt Taste Enhancing L-Arginyl Dipeptides from Casein and Lysozyme Released by Peptidases of Basidiomycota. J. Agric. Food Chem. 2018, 66, 2344–2353. [Google Scholar] [CrossRef]
- Kagebayashi, T.; Kontani, N.; Yamada, Y.; Mizushige, T.; Arai, T.; Kino, K.; Ohinata, K. Novel CCK-dependent vasorelaxing dipeptide, Arg-Phe, decreases blood pressure and food intake in rodents. Mol. Nutr. Food Res. 2012, 56, 1456–1463. [Google Scholar] [CrossRef]
- Crans, D.C.; Whitesides, G.M. A convenient synthesis of disodium acetyl phosphate for use in in situ ATP cofactor regeneration. J. Org. Chem. 1983, 48, 3130–3132. [Google Scholar] [CrossRef]
- Alissandratos, A.; Caron, K.; Loan, T.D.; Hennessy, J.E.; Easton, C.J. ATP Recycling with Cell Lysate for Enzyme-Catalyzed Chemical Synthesis, Protein Expression and PCR. ACS Chem. Biol. 2016, 11, 3289–3293. [Google Scholar] [CrossRef] [PubMed]
- Bordewick, S.; Mast, T.A.; Berger, R.G.; Ersoy, F. Recombinant Production of Arginyl Dipeptides by l-Amino Acid Ligase RizA Coupled with ATP Regeneration. Catalysts 2021, 11, 1290. [Google Scholar] [CrossRef]
- Sheldon, R.A. Enzyme Immobilization: The Quest for Optimum Performance. Adv. Syn. Catal. 2007, 349, 1289–1307. [Google Scholar] [CrossRef]
- Datta, S.; Christena, L.R.; Rajaram, Y.R.S. Enzyme immobilization: An overview on techniques and support materials. 3 Biotech 2013, 3, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohamad, N.R.; Marzuki, N.H.C.; Buang, N.A.; Huyop, F.; Wahab, R.A. An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotechnol. Biotechnol. Equip. 2015, 29, 205–220. [Google Scholar] [CrossRef]
- Garcia-Galan, C.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R.; Rodrigues, R.C. Potential of Different Enzyme Immobilization Strategies to Improve Enzyme Performance. Adv. Syn. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Yushkova, E.D.; Nazarova, E.A.; Matyuhina, A.V.; Noskova, A.O.; Shavronskaya, D.O.; Vinogradov, V.V.; Skvortsova, N.N.; Krivoshapkina, E.F. Application of Immobilized Enzymes in Food Industry. J. Agric. Food Chem. 2019, 67, 11553–11567. [Google Scholar] [CrossRef]
- Rodrigues, D.S.; Mendes, A.A.; Adriano, W.S.; Gonçalves, L.R.B.; Giordano, R.L.C. Multipoint covalent immobilization of microbial lipase on chitosan and agarose activated by different methods. J. Mol. Catal. B Enzym. 2008, 51, 100–109. [Google Scholar] [CrossRef]
- Bilal, M.; Asgher, M.; Cheng, H.; Yan, Y.; Iqbal, H.M.N. Multi-point enzyme immobilization, surface chemistry, and novel platforms: A paradigm shift in biocatalyst design. Crit. Rev. Biotechnol. 2019, 39, 202–219. [Google Scholar] [CrossRef]
- Ruzicka, J.; Carroll, A.D.; Lähdesmäki, I. Immobilization of proteins on agarose beads, monitored in real time by bead injection spectroscopy. Analyst 2006, 131, 799–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ThermoScientific. Pierce NHS-Activated Agarose Dry Resin Manual. Available online: https://assets.thermofisher.com/TFS-Assets%2FLSG%2Fmanuals%2FMAN0011707_Pierce_NHSActiv_Agarose_Dry_Resin_UG.pdf (accessed on 24 May 2022).
- Kalkhof, S.; Sinz, A. Chances and pitfalls of chemical cross-linking with amine-reactive N-hydroxysuccinimide esters. Anal. Bioanal. Chem. Res. 2008, 392, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhang, Z.; Pei, X.; Xia, X. Covalent Immobilization of L-Asparaginase and Optimization of Its Enzyme Reactor for Reducing Acrylamide Formation in a Heated Food Model System. Front. Bioeng. Biotechnol. 2020, 8, 584758. [Google Scholar] [CrossRef] [PubMed]
- Bié, J.; Sepodes, B.; Fernandes, P.C.B.; Ribeiro, M.H.L. Enzyme Immobilization and Co-Immobilization: Main Framework, Advances and Some Applications. Processes 2022, 10, 494. [Google Scholar] [CrossRef]
- Zucca, P.; Fernandez-Lafuente, R.; Sanjust, E. Agarose and Its Derivatives as Supports for Enzyme Immobilization. Molecules 2016, 21, 1577. [Google Scholar] [CrossRef] [Green Version]
- Kazenwadel, F.; Franzreb, M.; Rapp, B.E. Synthetic enzyme supercomplexes: Co-immobilization of enzyme cascades. Anal. Methods 2015, 7, 4030–4037. [Google Scholar] [CrossRef] [Green Version]
- Basso, A.; Brown, M.S.; Cruz-Izquierdo, A.; Martinez, C.A.; Serban, S. Optimization of Metal Affinity Ketoreductase Immobilization for Application in Batch and Flow Processes. Org. Process Res. Dev. 2022. [Google Scholar] [CrossRef]
- Petrovičová, T.; Markošová, K.; Hegyi, Z.; Smonou, I.; Rosenberg, M.; Rebroš, M. Co-Immobilization of Ketoreductase and Glucose Dehydrogenase. Catalysts 2018, 8, 168. [Google Scholar] [CrossRef] [Green Version]
- Bachosz, K.; Piasecki, A.; Zdarta, A.; Kaczorek, E.; Pinelo, M.; Zdarta, J.; Jesionowski, T. Enzymatic membrane reactor in xylose bioconversion with simultaneous cofactor regeneration. Bioorg. Chem. 2022, 123, 105781. [Google Scholar] [CrossRef]
- Plž, M.; Petrovičová, T.; Rebroš, M. Semi-Continuous Flow Biocatalysis with Affinity Co-Immobilized Ketoreductase and Glucose Dehydrogenase. Molecules 2020, 25, 4278. [Google Scholar] [CrossRef]
- Cui, C.; Ming, H.; Li, L.; Li, M.; Gao, J.; Han, T.; Wang, Y. Fabrication of an in-situ co-immobilized enzyme in mesoporous silica for synthesizing GSH with ATP regeneration. Mol. Catal. 2020, 486, 110870. [Google Scholar] [CrossRef]
- Huffman, M.A.; Fryszkowska, A.; Alvizo, O.; Borra-Garske, M.; Campos, K.R.; Canada, K.A.; Devine, P.N.; Duan, D.; Forstater, J.H.; Grosser, S.T.; et al. Design of an in vitro biocatalytic cascade for the manufacture of islatravir. Science 2019, 366, 1255–1259. [Google Scholar] [CrossRef] [PubMed]
- Bernal, C.; Rodríguez, K.; Martínez, R. Integrating enzyme immobilization and protein engineering: An alternative path for the development of novel and improved industrial biocatalysts. Biotechnol. Adv. 2018, 36, 1470–1480. [Google Scholar] [CrossRef] [PubMed]
- Bordewick, S.; Berger, R.G.; Ersoy, F. Mutagenesis of the L-Amino Acid Ligase RizA Increased the Production of Bioactive Dipeptides. Catalysts 2021, 11, 1385. [Google Scholar] [CrossRef]
- Frost, R.G.; Monthony, J.F.; Engelhorn, S.C.; Siebert, C.J. Covalent immobilization of proteins to N-hydroxysuccinimide ester derivatives of agarose: Effect of protein charge on immobilization. Biochim. Biophys. Acta Proteins Proteom. 1981, 670, 163–169. [Google Scholar] [CrossRef]
- Cline, G.W.; Hanna, S.B. The aminolysis of N-hydroxysuccinimide esters. A structure-reactivity study. J. Am. Chem. Soc. 1987, 109, 3087–3091. [Google Scholar] [CrossRef]
- Lim, C.Y.; Owens, N.A.; Wampler, R.D.; Ying, Y.; Granger, J.H.; Porter, M.D.; Takahashi, M.; Shimazu, K. Succinimidyl Ester Surface Chemistry: Implications of the Competition between Aminolysis and Hydrolysis on Covalent Protein Immobilization. Langmuir 2014, 30, 12868–12878. [Google Scholar] [CrossRef]
- Reetz, M.T. The Importance of Additive and Non-Additive Mutational Effects in Protein Engineering. Angew. Chem. Int. Ed. 2013, 52, 2658–2666. [Google Scholar] [CrossRef]
- Behrens, G.A.; Hummel, A.; Padhi, S.K.; Schätzle, S.; Bornscheuer, U.T. Discovery and Protein Engineering of Biocatalysts for Organic Synthesis. Adv. Syn. Catal. 2011, 353, 2191–2215. [Google Scholar] [CrossRef]
- Nakajima, H.; Suzuki, K.; Imahori, K. Purification and Properties of Acetate Kinase from Bacillus stearothermophilus. J. Biochem. 1978, 84, 193–203. [Google Scholar] [CrossRef]
- Leibrock, E.; Bayer, P.; Lüdemann, H.D. Nonenzymatic hydrolysis of adenosinetriphosphate (ATP) at high temperatures and high pressures. Biophys. Chem. 1995, 54, 175–180. [Google Scholar] [CrossRef]
- Whicher, A.; Camprubi, E.; Pinna, S.; Herschy, B.; Lane, N. Acetyl Phosphate as a Primordial Energy Currency at the Origin of Life. Orig. Life Evol. Biosph. 2018, 48, 159–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dean, R.L. Kinetic studies with alkaline phosphatase in the presence and absence of inhibitors and divalent cations. Biochem. Mol. Biol. Educ. 2002, 30, 401–407. [Google Scholar] [CrossRef]
- Tasnádi, G.; Jud, W.; Hall, M.; Baldenius, K.; Ditrich, K.; Faber, K. Evaluation of Natural and Synthetic Phosphate Donors for the Improved Enzymatic Synthesis of Phosphate Monoesters. Adv. Syn. Catal. 2018, 360, 2394–2401. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, Y.H.P. Cell-free protein synthesis energized by slowly-metabolized maltodextrin. BMC Biotechnol. 2009, 9, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Zhang, Y.-H.P.J. Enzymatic regeneration and conservation of ATP: Challenges and opportunities. Crit. Rev. Biotechnol. 2021, 41, 16–33. [Google Scholar] [CrossRef] [PubMed]
- Naramittanakul, A.; Buttranon, S.; Petchsuk, A.; Chaiyen, P.; Weeranoppanant, N. Development of a continuous-flow system with immobilized biocatalysts towards sustainable bioprocessing. React. Chem. Eng. 2021, 6, 1771–1790. [Google Scholar] [CrossRef]
- Porta, R.; Benaglia, M.; Puglisi, A. Flow Chemistry: Recent Developments in the Synthesis of Pharmaceutical Products. Org. Process Res. Dev. 2016, 20, 2–25. [Google Scholar] [CrossRef] [Green Version]
- Britton, J.; Majumdar, S.; Weiss, G.A. Continuous flow biocatalysis. Chem. Soc. Rev. 2018, 47, 5891–5918. [Google Scholar] [CrossRef]
- Thompson, M.P.; Peñafiel, I.; Cosgrove, S.C.; Turner, N.J. Biocatalysis Using Immobilized Enzymes in Continuous Flow for the Synthesis of Fine Chemicals. Org. Process Res. Dev. 2019, 23, 9–18. [Google Scholar] [CrossRef]
- Rottmann, E.; Hauke, K.F.; Krings, U.; Berger, R.G. Enzymatic acrylamide mitigation in French fries–An industrial-scale case study. Food Control 2021, 123, 107739. [Google Scholar] [CrossRef]



| Dipeptide | Variant | Temperature (°C) | c(Arg-X)24 h (mM) | c(Arg-Arg)24 h (mM) | Arg-X/ Arg-Arg | Yield (Arg-X)24 h (%) | >50% Activity * |
|---|---|---|---|---|---|---|---|
| Arg-Ser | Wild type | 25 | 10.3 ± 0.8 | 1.9 ± 0.3 | 5.4 ± 1.1 | 21 | >96 h |
| 30 | 7.1 ± 0.2 | 1.1 ± 0.0 | 6.7 ± 0.3 | 14 | >120 h | ||
| T81F_A158S | 25 | 11.2 ± 0.0 | 1.6 ± 0.1 | 7.2 ± 0.6 | 22 | >120 h | |
| 30 | 7.7 ± 1.9 | 1.0 ± 0.0 | 8.1 ± 0.4 | 15 | >96 h | ||
| Arg-Phe | Wild type | 25 | 2.3 ± 0.2 | 1.5 ± 0.3 | 1.5 ± 0.2 | 5 | n.d.† |
| 30 | 1.2 ± 0.3 | 1.1 ± 0.0 | 1.1 ± 0.3 | 2 | n.d.† | ||
| K83F_S156A | 25 | 11.8 ± 0.4 | 0.7 ± 0.0 | 16 ± 0.2 | 24 | >72 h | |
| 30 | 9.3 ± 0.9 | 0.4 ± 0.2 | 24 ± 15 | 19 | >48 h |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bordewick, S.; Berger, R.G.; Ersoy, F. Co-Immobilization of RizA Variants with Acetate Kinase for the Production of Bioactive Arginyl Dipeptides. Molecules 2022, 27, 4352. https://doi.org/10.3390/molecules27144352
Bordewick S, Berger RG, Ersoy F. Co-Immobilization of RizA Variants with Acetate Kinase for the Production of Bioactive Arginyl Dipeptides. Molecules. 2022; 27(14):4352. https://doi.org/10.3390/molecules27144352
Chicago/Turabian StyleBordewick, Sven, Ralf G. Berger, and Franziska Ersoy. 2022. "Co-Immobilization of RizA Variants with Acetate Kinase for the Production of Bioactive Arginyl Dipeptides" Molecules 27, no. 14: 4352. https://doi.org/10.3390/molecules27144352
APA StyleBordewick, S., Berger, R. G., & Ersoy, F. (2022). Co-Immobilization of RizA Variants with Acetate Kinase for the Production of Bioactive Arginyl Dipeptides. Molecules, 27(14), 4352. https://doi.org/10.3390/molecules27144352