The Expression Profiles of the Salvia miltiorrhiza 3-Hydroxy-3-methylglutaryl-coenzyme A Reductase 4 Gene and Its Influence on the Biosynthesis of Tanshinones
Abstract
:1. Introduction
2. Results
2.1. Organ-Specific Expression of S. miltiorrhiza HMGR4 Gene
2.2. Potential Regulators of S. miltiorrhiza HMGR4 Gene Expression
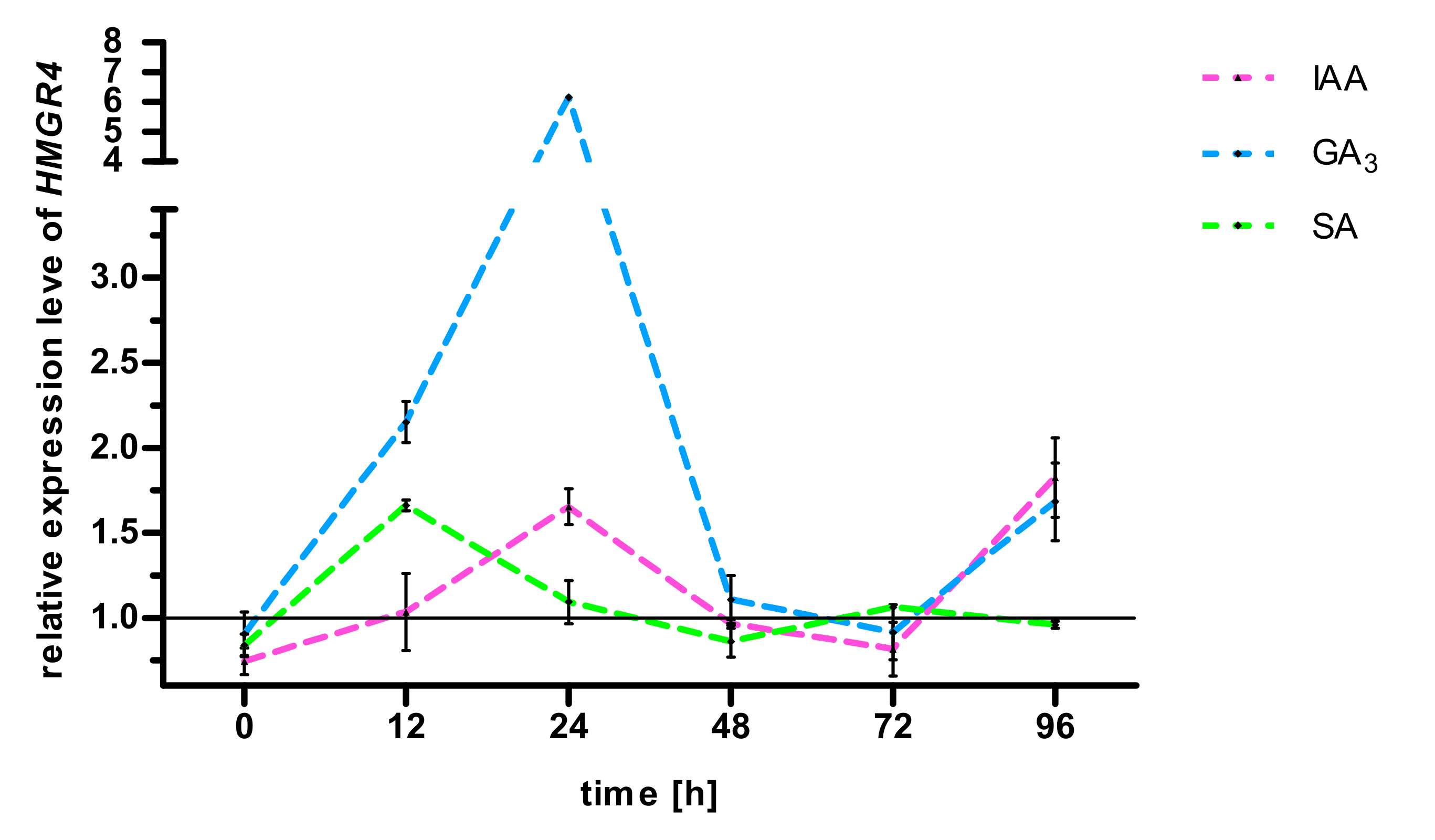
2.3. Effect of GA3, IAA, SA on S. miltiorrhiza HMGR4 Gene Expression
2.4. Impact of pRI201-AN-HMGR4 Transformation on S. miltiorrhiza HMGR4 Gene Expression
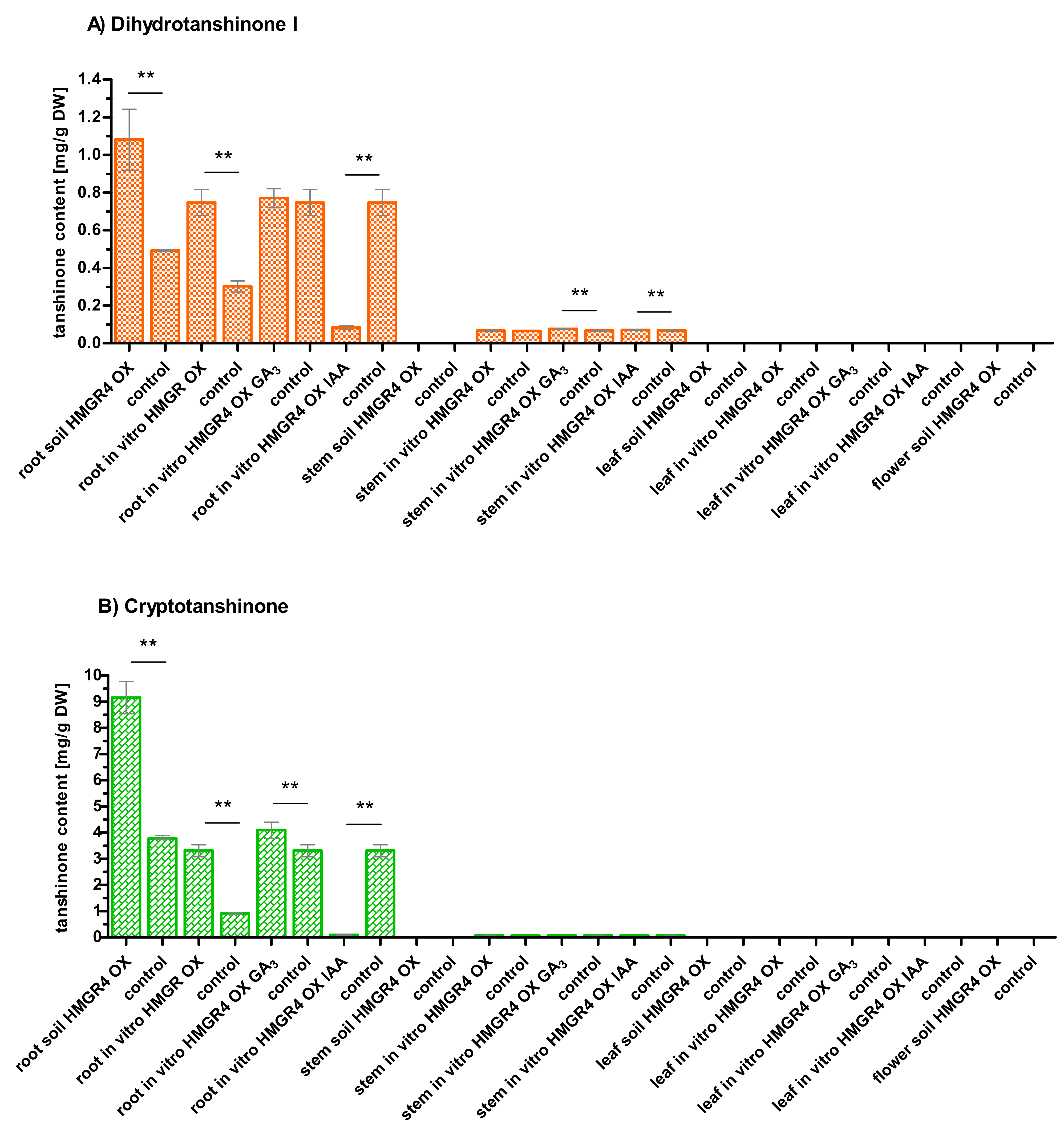
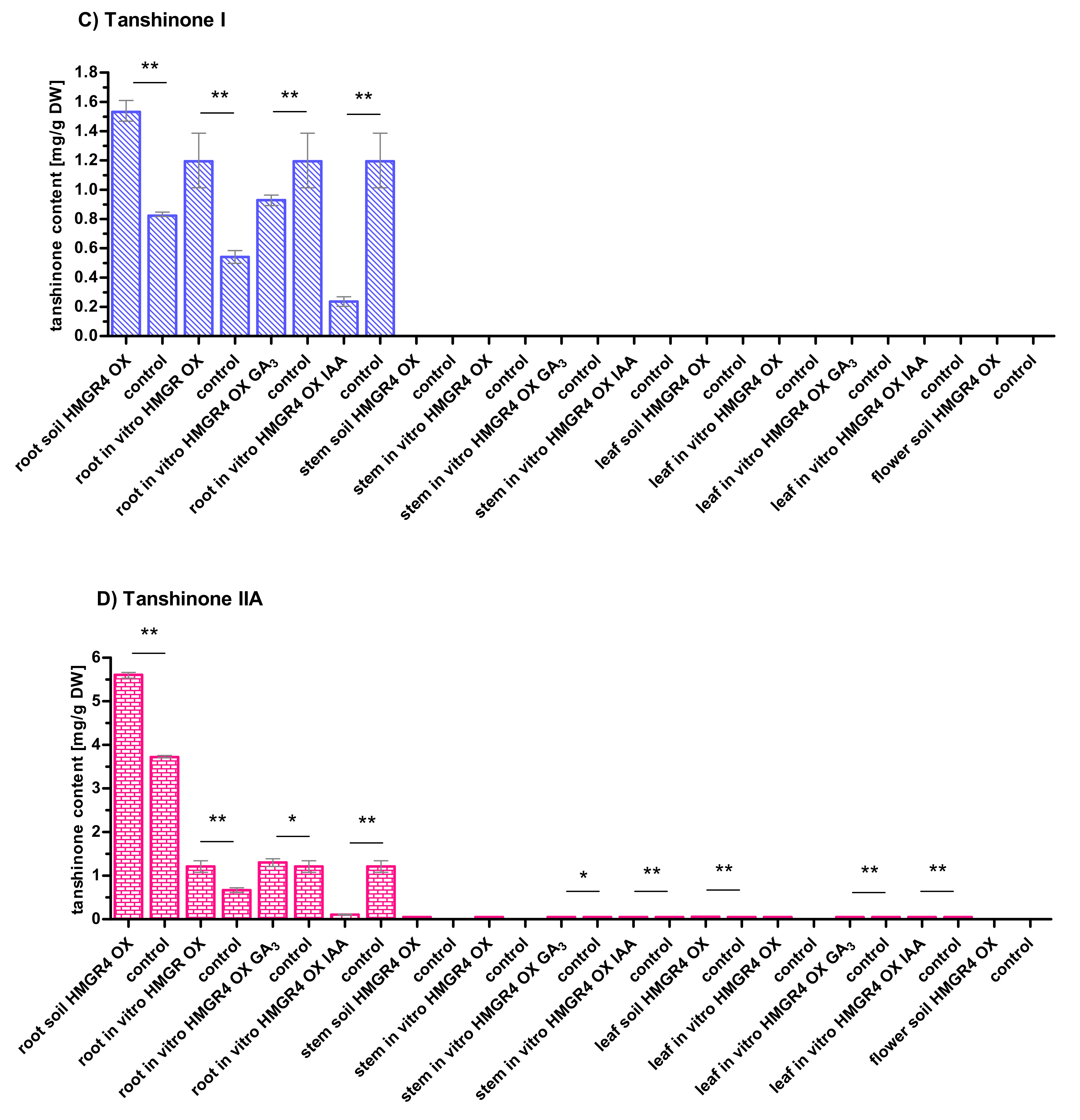
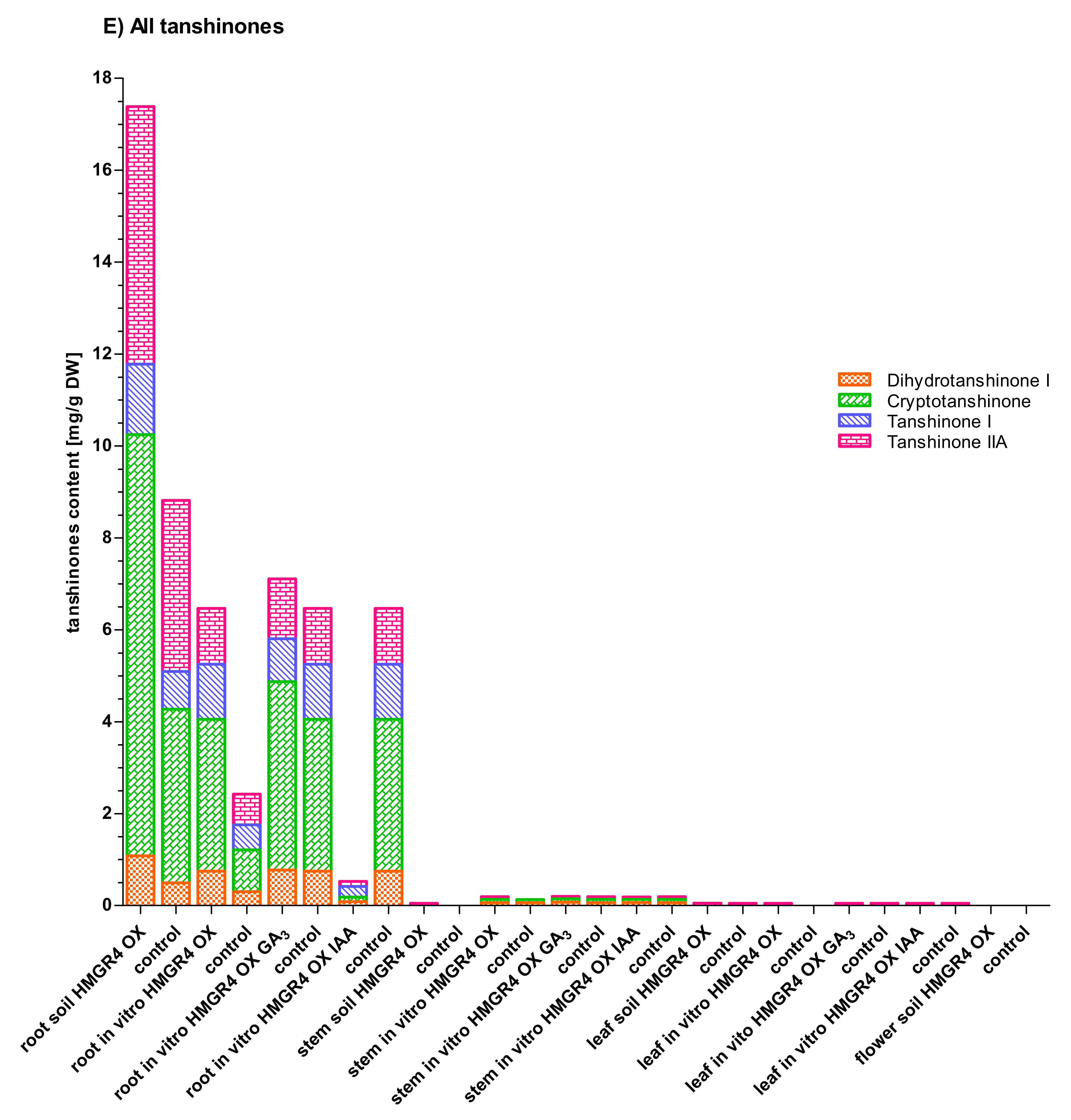
2.5. Influence of HMGR4 Overexpression on the Biosynthesis of Tanshinones in S. miltiorrhiza
2.6. Organ-Dependent Accumulation of Tanshinones in S. miltiorrhiza
2.7. Impact of Growth Environment on the Biosynthesis of Tanshinones in S. miltiorrhiza
2.8. Effect of GA3 and IAA on the Biosynthesis of Tanshinones in S. miltiorrhiza
3. Discussion
4. Materials and Methods
4.1. Establishment of S. miltiorrhiza Culture and Treatments
4.2. Selection of Potential Regulators of S. miltiorrhiza HMGR4 Gene Expression
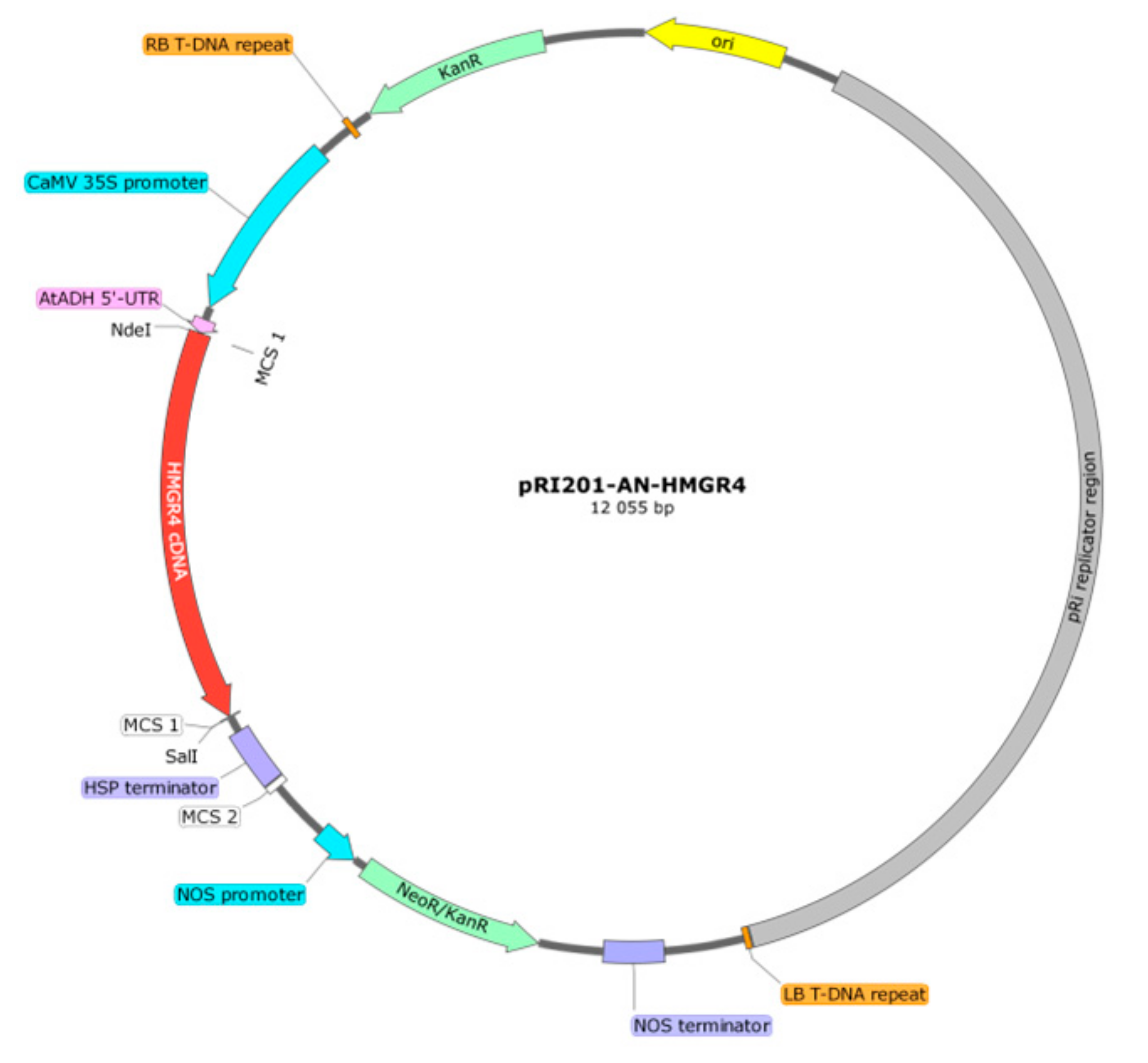
4.3. Preparation of pRI201-AN-HMGR4 Overexpression Construct
4.4. Transformation, Selection, Regeneration and Treatments of S. miltiorrhiza Culture
4.5. RNA Isolation, Reverse Transcription and Quantitative Real-Time PCR
4.6. Quantitative Analysis of Tanshinones
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Cheng, T.O. Cardiovascular effects of Danshen. Int. J. Cardiol. 2007, 121, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wan, R.; Lin, Y.; Yang, L.; Chen, Y.; Liu, C. Recent advance on research and application of Salvia miltiorrhiza. Asian J. Pharmacodyn. Pharmacokinet. 2007, 7, 99–130. [Google Scholar]
- Zhang, H.; Long, M.; Wu, Z.; Han, X.; Yu, Y. Sodium tanshinone IIA silate as an add-on therapy in patients with unstable angina pectoris. J. Thorac. Dis. 2014, 6, 1794–1799. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.Q.; Zhang, N.; Xu, Y.; Chen, W.N.; Zhu, M.L.; Song, N.; Ren, L.; Cao, H.M.; Wang, J.Y.; Yang, G.L. Tanshinone IIA affects the HDL subfractions distribution not serum lipid levels: Involving in intake and efflux of cholesterol. Arch. Biochem. Biophys. 2016, 592, 50–59. [Google Scholar] [CrossRef]
- Chiu, S.C.; Huang, S.Y.; Chen, S.P.; Su, C.C.; Chiu, T.L.; Pang, C.Y. Tanshinone IIA inhibits human prostate cancer cells growth by induction of endoplasmic reticulum stress in vitro and in vivo. Prostate Cancer Prostatic Dis. 2013, 16, 315–322. [Google Scholar] [CrossRef]
- Wang, S.; Jing, H.; Yang, H.; Liu, Z.; Guo, H.; Chai, L.; Hu, L. Tanshinone I selectively suppresses pro-inflammatory genes expression in activated microglia and prevents nigrostriatal dopaminergic neurodegeneration in a mouse model of Parkinson’s disease. J. Ethnopharmacol. 2015, 164, 247–255. [Google Scholar] [CrossRef]
- Kim, S.Y.; Moon, T.C.; Chang, H.W.; Son, K.H.; Kang, S.S.; Kim, H.P. Effects of tanshinone I isolated from Salvia miltiorrhiza bunge on arachidonic acid metabolism and in vivo inflammatory responses. Phytother. Res. 2002, 16, 616–620. [Google Scholar] [CrossRef]
- Tang, F.; Wu, X.; Wang, T.; Wang, P.; Li, R.; Zhang, H.; Gao, J.; Chen, S.; Bao, L.; Huang, H.; et al. Tanshinone II A attenuates atherosclerotic calcification in rat model by inhibition of oxidative stress. Vascul. Pharmacol. 2007, 46, 427–438. [Google Scholar] [CrossRef]
- Wang, C.; Du, X.; Yang, R.; Liu, J.; Xu, D.; Shi, J.; Chen, L.; Shao, R.; Fan, G.; Gao, X.; et al. The prevention and treatment effects of tanshinone IIA on oestrogen/androgen-induced benign prostatic hyperplasia in rats. J. Steroid Biochem. Mol. Biol. 2015, 145, 28–37. [Google Scholar] [CrossRef]
- Wang, L.; Cheng, L.; Zhang, B.; Wang, N.; Wang, F. Tanshinone prevents alveolar bone loss in ovariectomized osteoporosis rats by up-regulating phosphoglycerate dehydrogenase. Toxicol. Appl. Pharmacol. 2019, 376, 9–16. [Google Scholar] [CrossRef]
- Honda, G.; Koezuka, Y.; Tabata, M. Isolation of an antidermatophytic substance from the root of Salvia miltiorrhiza. Chem. Pharm. Bull. 1988, 36, 408–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, A.R.; Wu, W.L.; Chang, W.L.; Lin, H.C.; King, M.L. Isolation and bioactivity of new tanshinones. J. Nat. Prod. 1987, 50, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.D.; Cao, Y.F.; Che, Y.Y.; Li, J.; Shang, Z.P.; Zhao, W.J.; Qiao, Y.J.; Zhang, J.Y. Danshen: A phytochemical and pharmacological overview. Chin. J. Nat. Med. 2019, 17, 59–80. [Google Scholar] [CrossRef]
- Lu, S. The Salvia Miltiorrhiza Genome; Springer: Cham, Switzerland, 2019; pp. 129–139. [Google Scholar]
- Dai, Z.; Cui, G.; Zhou, S.F.; Zhang, X.; Huang, L. Cloning and characterization of a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Salvia miltiorrhiza involved in diterpenoid tanshinone accumulation. J. Plant Physiol. 2011, 168, 148–157. [Google Scholar] [CrossRef]
- Ma, Y.; Yuan, L.; Wu, B.; Li, X.; Chen, S.; Lu, S. Genome-wide identification and characterization of novel genes involved in terpenoid biosynthesis in Salvia miltiorrhiza. J. Exp. Bot. 2012, 63, 2809–2823. [Google Scholar] [CrossRef] [Green Version]
- Liao, P.; Zhou, W.; Zhang, L.; Wang, J.; Yan, X.; Zhang, Y.; Zhang, R.; Li, L.; Zhou, G.; Kai, G. Molecular cloning, characterization and expression analysis of a new gene encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase from Salvia miltiorrhiza. Acta Physiol. Plant. 2009, 31, 565–572. [Google Scholar] [CrossRef]
- Wang, J.W.; Wu, J.Y. Tanshinone biosynthesis in Salvia miltiorrhiza and production in plant tissue cultures. Appl. Microbiol. Biotechnol. 2010, 88, 437–449. [Google Scholar] [CrossRef]
- Cheng, Q.; He, Y.; Li, G.; Liu, Y.; Gao, W.; Huang, L. Effects of combined elicitors on tanshinone metabolic profiling and SmCPS expression in Salvia miltiorrhiza hairy root cultures. Molecules 2013, 18, 7473–7485. [Google Scholar] [CrossRef]
- Hao, X.; Shi, M.; Cui, L.; Xu, C.; Zhang, Y.; Kai, G. Effects of methyl jasmonate and salicylic acid on tanshinone production and biosynthetic gene expression in transgenic Salvia miltiorrhiza hairy roots. Biotechnol. Appl. Biochem. 2015, 62, 24–31. [Google Scholar] [CrossRef]
- Wu, C.T.; Mulabagal, V.; Nalawade, S.M.; Chen, C.L.; Yang, T.F.; Tsay, H.S. Isolation and quantitative analysis of cryptotanshinone, an active quinoid diterpene formed in callus of Salvia miltiorrhiza BUNGE. Biol. Pharm. Bull. 2003, 26, 845–848. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Liu, Q.; Yao, T.; Fu, X. Shedding light on integrative GA signaling. Curr. Opin. Plant Biol. 2014, 21, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Chakrabarty, S.K. Gibberellic acid in plant: Still a mystery unresolved. Plant Signal. Behav. 2013, 8, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Wu, K.; Xu, H.; Gao, X.; Fu, X. New insights into gibberellin signaling in regulating plant growth-metabolic coordination. Curr. Opin. Plant Biol. 2021, 63, 102074. [Google Scholar] [CrossRef]
- Li, W.; Liu, C.; Liu, J.; Bai, Z.; Liang, Z. Transcriptomic analysis reveals the GRAS family genes respond to gibberellin in Salvia miltiorrhiza hairy roots. BMC Genom. 2020, 21, 727. [Google Scholar] [CrossRef]
- Li, W.; Bai, Z.; Pei, T.; Yang, D.; Mao, R.; Zhang, B.; Liu, C.; Liang, Z. SmGRAS1 and SmGRAS2 Regulate the Biosynthesis of Tanshinones and Phenolic Acids in Salvia miltiorrhiza. Front. Plant Sci. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Grones, P.; Robert, S. Auxin signaling: A big question to be addressed by small molecules. J. Exp. Bot. 2018, 69, 313–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leyser, O. Auxin signaling. Plant Physiol. 2018, 176, 465–479. [Google Scholar] [CrossRef] [Green Version]
- Freire-Rios, A.; Tanaka, K.; Crespo, I.; van der Wijk, E.; Sizentsova, Y.; Levitsky, V.; Lindhoud, S.; Fontana, M.; Hohlbein, J.; Boer, D.R.; et al. Architecture of DNA elements mediating ARF transcription factor binding and auxin-responsive gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2020, 117, 24557–24566. [Google Scholar] [CrossRef]
- Xu, Z.; Ji, A.; Song, J.; Chen, S. Genome-wide analysis of auxin response factor gene family members in medicinal model plant Salvia miltiorrhiza. Biol. Open 2016, 5, 848–857. [Google Scholar] [CrossRef] [Green Version]
- Kuai, X.; MacLeod, B.J.; Després, C. Integrating data on the Arabidopsis NPR1/NPR3/NPR4 salicylic acid receptors; a differentiating argument. Front. Plant Sci. 2015, 6, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Zhang, D.; Chu, J.Y.; Boyle, P.; Wang, Y.; Brindle, I.D.; De Luca, V.; Després, C. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012, 1, 639–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.M.; Trifa, Y.; Silva, H.; Pontier, D.; Lam, E.; Shah, J.; Klessig, D.F. NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant Microbe Interact. 2000, 13, 191–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fonseca, J.P.; Menossi, M.; Thibaud-Nissen, F.; Town, C.D. Functional analysis of a TGA factor-binding site located in the promoter region controlling salicylic acid-induced NIMIN-1 expression in Arabidopsis. Genet. Mol. Res. 2010, 9, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Busta, L.; Zhang, Q.; Ding, P.; Jetter, R.; Zhang, Y. TGACG-BINDING FACTOR 1 (TGA1) and TGA4 regulate salicylic acid and pipecolic acid biosynthesis by modulating the expression of SYSTEMIC ACQUIRED RESISTANCE DEFICIENT 1 (SARD1) and CALMODULIN-BINDING PROTEIN 60g (CBP60g). New Phytol. 2018, 217, 344–354. [Google Scholar] [CrossRef] [Green Version]
- Blanco, F.; Garretón, V.; Frey, N.; Dominguez, C.; Pérez-Acle, T.; Van der Straeten, D.; Jordana, X.; Holuigue, L. Identification of NPR1-dependent and independent genes early induced by salicylic acid treatment in Arabidopsis. Plant Mol. Biol. 2005, 59, 927–944. [Google Scholar] [CrossRef]
- Cao, W.; Wang, Y.; Shi, M.; Hao, X.; Zhao, W.; Wang, Y.; Ren, J.; Kai, G. Transcription factor SmWRKY1 positively promotes the biosynthesis of tanshinones in Salvia miltiorrhiza. Front. Plant Sci. 2018, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.P.; Lin, J.J.; Li, W.H. Positional distribution of transcription factor binding sites in Arabidopsis thaliana. Sci. Rep. 2016, 6, 25164. [Google Scholar] [CrossRef]
- Xu, Z.; Peters, R.J.; Weirather, J.; Luo, H.; Liao, B.; Zhang, X.; Zhu, Y.; Ji, A.; Zhang, B.; Hu, S.; et al. Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant J. 2015, 82, 951–961. [Google Scholar] [CrossRef]
- Szymczyk, P.; Grąbkowska, R.; Skała, E.; Żebrowska, M.; Balcerczak, E.; Jeleń, A. Isolation and characterization of a 3-hydroxy-3-methylglutaryl coenzyme A reductase 2 promoter from Salvia miltiorrhiza. J. Plant Biochem. Biotechnol. 2018, 27, 223–236. [Google Scholar] [CrossRef]
- Lv, D.; Zhang, Y. Isolation and functional analysis of apple MdHMGR1 and MdHMGR4 gene promoters in transgenic Arabidopsis thaliana. Plant Cell Tissue Organ Cult. 2017, 129, 133–143. [Google Scholar] [CrossRef]
- Kasahara, H.; Hanada, A.; Kuzuyama, T.; Takagi, M.; Kamiya, Y.; Yamaguchi, S. Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of gibberellins in Arabidopsis. J. Biol. Chem. 2002, 277, 45188–45194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, S.; Meng, X.; Liao, Y.; Yu, T.; Cao, J.; Tan, J.; Xu, F.; Cheng, S. Characterization and functional analysis of two novel 3-hydroxy-3-methylglutaryl-coenzyme A reductase genes (GbHMGR2 and GbHMGR3) from Ginkgo biloba. Sci. Rep. 2019, 9, 14109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Li, B.; Luo, L.; Cao, F.; Yang, B.; Gao, J.; Yan, Y.; Zhang, G.; Peng, L.; Hu, B. Increased phenolic acid and tanshinone production and transcriptional responses of biosynthetic genes in hairy root cultures of Salvia przewalskii Maxim. treated with methyl jasmonate and salicylic acid. Mol. Biol. Rep. 2020, 47, 8565–8578. [Google Scholar] [CrossRef]
- Kai, G.; Xu, H.; Zhou, C.; Liao, P.; Xiao, J.; Luo, X.; You, L.; Zhang, L. Metabolic engineering tanshinone biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. Metab. Eng. 2011, 13, 319–327. [Google Scholar] [CrossRef]
- Li, S.; Zhu, N.; Tang, C.; Duan, H.; Wang, Y.; Zhao, G.; Liu, J.; Ye, Y. Differential distribution of characteristic constituents in root, stem and leaf tissues of Salvia miltiorrhiza using MALDI mass spectrometry imaging. Fitoterapia 2020, 146. [Google Scholar] [CrossRef]
- Hang, L.; Wang, J.; Yang, D.; Shu, Z.; Liang, Z. Distribution traits of bioactives in different parts of Salvia miltiorrhiza Bunge. and Salvia miltiorrhiza Bunge. f. alba. J. Northwest A F Univ. 2008, 36, 217–222. [Google Scholar]
- Leivar, P.; Antolín-Llovera, M.; Ferrero, S.; Closa, M.; Arró, M.; Ferrer, A.; Boronat, A.; Campos, N. Multilevel control of Arabidopsis 3-hydroxy-3-methylglutaryl coenzyme A reductase by protein phosphatase 2A. Plant Cell 2011, 23, 1494–1511. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Long, C.; Lam, E. Roles of plant-associated microbiota in traditional herbal medicine. Trends Plant Sci. 2018, 23, 559–562. [Google Scholar] [CrossRef]
- Jeliazkov (Zheljazkov), V.D.; Cantrell, C.L. Medicinal and Aromatic Crops: Production, Phytochemistry, and Utilization; American Chemical Society: Washington, DC, USA, 2016; pp. 75–90. [Google Scholar]
- Yan, Y.; Zhang, S.; Zhang, J.; Ma, P.; Duan, J.; Liang, Z. Effect and mechanism of endophytic bacteria on growth and secondary metabolite synthesis in Salvia miltiorrhiza hairy roots. Acta Physiol. Plant. 2014, 36, 1095–1105. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, L.; Wu, J. Promotion of Salvia miltiorrhiza hairy root growth and tanshinone production by polysaccharide–protein fractions of plant growth-promoting rhizobacterium Bacillus cereus. Process Biochem. 2010, 45, 1517–1522. [Google Scholar] [CrossRef]
- George, E.F.; Hall, M.A.; De Klerk, G. Plant Propagation by Tissue Culture, 3rd ed.; Springer: Dordrecht, The Netherlands, 2008; pp. 465–477. [Google Scholar]
- Debergh, P.C.; Zimmerman, R.H. Micropropagation. Technology and Application, 1st ed.; Kluwer Academic Publishers: London, UK, 1991; pp. 71–93. [Google Scholar]
- Li, W.; Xing, B.; Mao, R.; Bai, Z.; Yang, D.; Xu, J.; Liang, Z. SmGRAS3 negatively responds to GA signaling while promotes tanshinones biosynthesis in Salvia miltiorrhiza. Ind. Crops Prod. 2020, 144. [Google Scholar] [CrossRef]
- Gupta, S.K.; Liu, R.; Liaw, S.; Chan, H.; Tsay, H. Enhanced tanshinone production in hairy roots of ‘Salvia miltiorrhiza Bunge’ under the influence of plant growth regulators in liquid culture. Bot. Stud. 2011, 52, 435–443. [Google Scholar]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Gamborg, O.L.; Miller, R.A.; Ojima, K. Nutrient requirements of suspension cultures of Soybean root cells. Exp. Cell Res. 1968, 50, 151–158. [Google Scholar] [CrossRef]
- Chow, C.N.; Zheng, H.Q.; Wu, N.Y.; Chien, C.H.; Huang, H.D.; Lee, T.Y.; Chiang-Hsieh, Y.F.; Hou, P.F.; Yang, T.Y.; Chang, W.C. PlantPAN 2.0: An update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 2016, 44, D1154–D1160. [Google Scholar] [CrossRef] [Green Version]
- UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Wang, K. Agrobacterium Protocols, 2nd ed.; Humana Press: Totowa, NJ, USA, 2006; pp. 43–53. [Google Scholar]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.G.; Smith, J.A.; Struhl, K. Current Protocols in Molecular Biology; John Wiley & Sons: New York, NY, USA, 2003; pp. 1.6.1–1.6.10, 12.11.11–12.11.10. [Google Scholar]
- Peña, L. Transgenic Plants. Methods and Protocols; Humana Press: Totowa, NJ, USA, 2005. [Google Scholar]
- Yang, Y.; Hou, S.; Cui, G.; Chen, S.; Wei, J.; Huang, L. Characterization of reference genes for quantitative real-time PCR analysis in various tissues of Salvia miltiorrhiza. Mol. Biol. Rep. 2010, 37, 507–513. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 2002–2007. [Google Scholar] [CrossRef]
- Wan, X.; Wang, Y.; Row, K.H. Separation of tanshinone I, tanshinone IIA, and cryptotanshinone from Salvia miltiorrhiza Bunge by normal phase HPLC. J. Liq. Chromatogr. Relat. Technol. 2009, 32, 544–552. [Google Scholar] [CrossRef]
- Yuan, L.; Perry, S.E. Plant Transcription Factors, 1st ed.; Humana Press: Totowa, NJ, USA, 2011; pp. 45–66. [Google Scholar]
- Luján, M.A.; Soria-García, Á.; Claver, A.; Lorente, P.; Rubio, M.C.; Picorel, R.; Alfonso, M. Different Cis-Regulatory Elements Control the Tissue-Specific Contribution of Plastid ω-3 Desaturases to Wounding and Hormone Responses. Front. Plant Sci. 2021, 12, 727292. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yang, Q.; Fu, H.; Chen, K.; Zhao, S.; Zhang, C.; Cai, T.; Wang, L.; Lu, W.; Dang, H.; et al. Identification of Key Gene Networks and Deciphering Transcriptional Regulators Associated With Peanut Embryo Abortion Mediated by Calcium Deficiency. Front. Plant Sci. 2022, 13, 814015. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Liu, Z.; Xie, Q.; Fang, J.; Wang, C.; Li, J.; Wang, C.; Gao, C. Construction of two regulatory networks related to salt stress and lignocellulosic synthesis under salt stress based on a Populus davidiana × P. bolleana transcriptome analysis. Plant Mol. Biol. 2022. [Google Scholar] [CrossRef] [PubMed]

| TF Family Name | TF Gene Name and Locus | TFBSs Number |
|---|---|---|
| GA3 | ||
| AT-Hook | AHL25; At4g35390 | 6 |
| MYB-related | CCA1; At2g46830 | 1 |
| RVE8; At3g09600 | 2 | |
| RVE4; At5g02840 | ||
| Homeodomain; HD-ZIP | ATHB-23; At1g26960 | 1 |
| bHLH | PIF3; At1g09530 | 2 |
| GATA | GATA22; At4g26150 | 62 |
| GATA21; At5g56860 | ||
| MADS box; MIKC | SOC1; At2g45660 | 8 |
| AGL24; At4g24540 | 15 | |
| MADS box | AGL71; At5g51870 | 13 |
| AGL72; At5g51860 | 14 | |
| AGL42; At5g62165 | ||
| NF-YC | NFYC3; At1g54830 | 48 |
| IAA | ||
| MYB-related | CCA1; At2g46830 | 1 |
| RVE 8; At3g09600 | 2 | |
| Homeodomain; HD-ZIP | HAT2; At5g47370 | 2 |
| ATHB-20; At3g01220 | 3 | |
| CAMTA | CAMTA1; At5g09410 | 2 |
| MADS box; MIKC | AGL14; At4g11880 | 17 |
| SA | ||
| NAC; NAM | NAC081; At5g08790 | 1 |
| NAC062; At3g49530 | 2 | |
| MYB | MYB46; At5g12870 | 4 |
| Myb/SANT; MYB | MYB3; At1g22640 | 1 |
| MYB-related | CCA1; At2g46830 | 1 |
| RVE8; At3g09600 | 2 | |
| RVE4; At5g02840 | ||
| Dof | DOF1.1; At1g07640 | 34 |
| DOF3.4; At3g50410 | ||
| bZIP | TGA2; At5g06950 | 32 |
| bHLH | LRL1; At2g24260 | 2 |
| CG-1; CAMTA | CAMTA2; At5g64220 | 3 |
| CAMTA | CAMTA4; At1g67310 | 2 |
| CAMTA6; At3g16940 | ||
| CSD | CSP2; At4g38680 | 2 |
| WRKY | WRKY6; At1g62300 | 9 |
| WRKY40; At1g80840 | ||
| WRKY4; At1g13960 | 8 | |
| WRKY60; At2g25000 | ||
| WRKY21; At2g30590 | ||
| WRKY54; At2g40750 | ||
| WRKY70; At3g56400 | ||
| WRKY53; At4g23810 | ||
| WRKY18; At4g31800 | ||
| WRKY26; At5g07100 | ||
| WRKY38; At5g22570 | ||
| WRKY30; At5g24110 | ||
| Primer Name | Sequence | Product Size [bp] |
|---|---|---|
| Confirmation of Transformation | ||
| Kanamycin_F | TGATCTCGTCGTGACCCAT | 234 |
| Kanamycin_R | AGAAGGCGATAGAAGGCGA | |
| Real-Time PCR | ||
| HMGR4_F | CTCAACCTGCTTGGCGTAA | 185 |
| HMGR4_R | AGTCTCGTGATGTCCCTGCT | |
| ACT7_F | TCCGTCTTGATCTTGCTGGT | 170 |
| ACT7_R | CGTCTTTGCAGTTTCGAGCT | |
| Analyte | Retention Time [min] | Standard Curve | R2 |
|---|---|---|---|
| DHTI | 6.025 | y = 190.163x − 16.690 | 0.99973 |
| CT | 10.009 | y = 146.106x − 10.926 | 0.99992 |
| TI | 10.938 | y = 241.276x − 21.816 | 0.99989 |
| TIIA | 14.808 | y = 327.209x − 20.014 | 0.99996 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majewska, M.; Szymczyk, P.; Gomulski, J.; Jeleń, A.; Grąbkowska, R.; Balcerczak, E.; Kuźma, Ł. The Expression Profiles of the Salvia miltiorrhiza 3-Hydroxy-3-methylglutaryl-coenzyme A Reductase 4 Gene and Its Influence on the Biosynthesis of Tanshinones. Molecules 2022, 27, 4354. https://doi.org/10.3390/molecules27144354
Majewska M, Szymczyk P, Gomulski J, Jeleń A, Grąbkowska R, Balcerczak E, Kuźma Ł. The Expression Profiles of the Salvia miltiorrhiza 3-Hydroxy-3-methylglutaryl-coenzyme A Reductase 4 Gene and Its Influence on the Biosynthesis of Tanshinones. Molecules. 2022; 27(14):4354. https://doi.org/10.3390/molecules27144354
Chicago/Turabian StyleMajewska, Małgorzata, Piotr Szymczyk, Jan Gomulski, Agnieszka Jeleń, Renata Grąbkowska, Ewa Balcerczak, and Łukasz Kuźma. 2022. "The Expression Profiles of the Salvia miltiorrhiza 3-Hydroxy-3-methylglutaryl-coenzyme A Reductase 4 Gene and Its Influence on the Biosynthesis of Tanshinones" Molecules 27, no. 14: 4354. https://doi.org/10.3390/molecules27144354







