Impact of Antibiotics as Waste, Physical, Chemical, and Enzymatical Degradation: Use of Laccases
Abstract
:1. Introduction
2. Antibiotics
3. Antibiotic Use in Livestock Farming
3.1. Antibiotics Use in Livestock Farming and Foodborne Diseases (FBD)
3.2. Waste from the Livestock Industry
4. Antibiotic Use by Humans
5. Presence of Antibiotics in Wastewaters
6. Global Problem of Antibiotic Resistance
7. Overviews of Degradation of Antibiotics
7.1. Biotic Degradation of Antibiotics
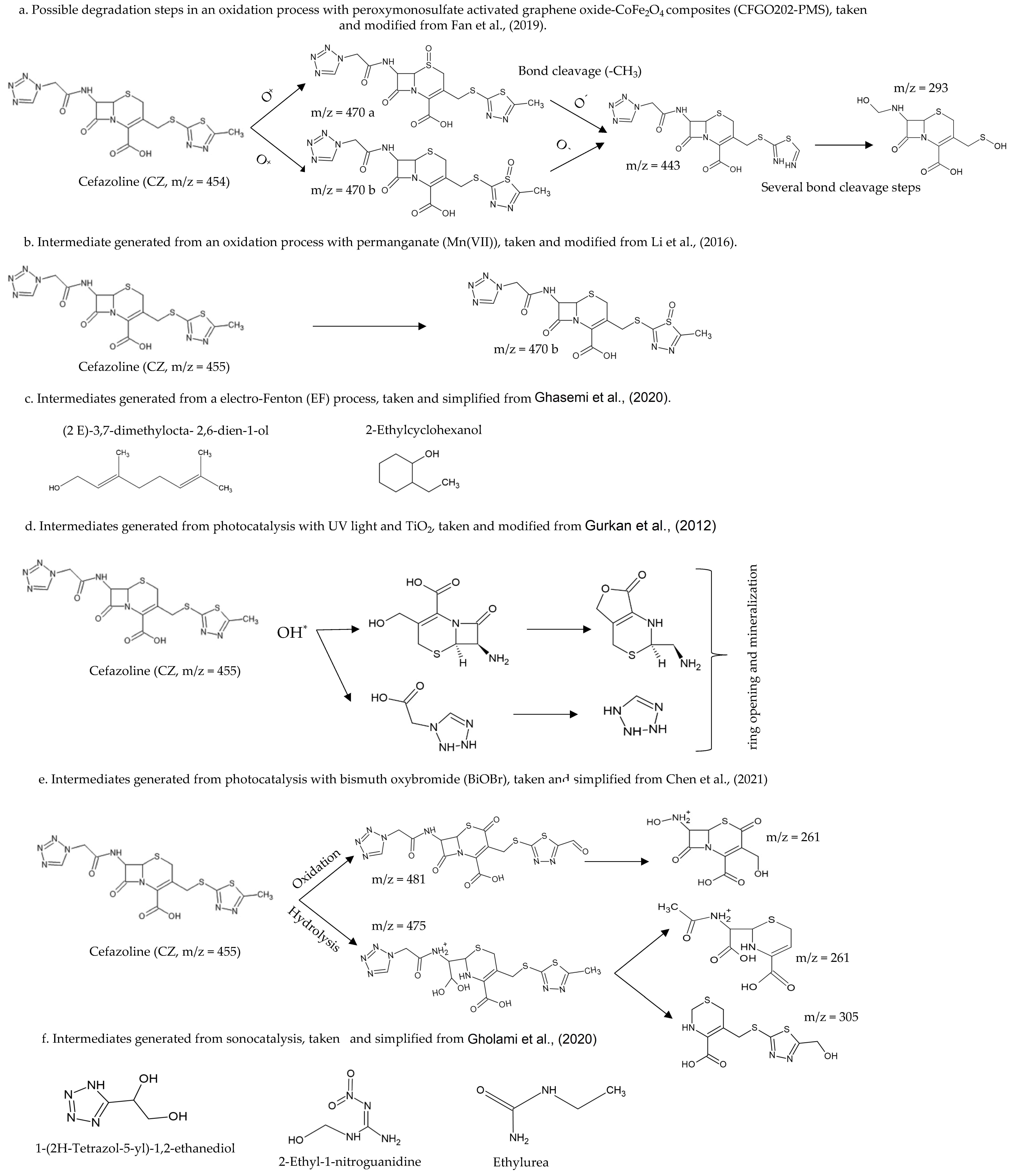
7.2. In Situ Chemical Oxidation (ISCO) of Antibiotics
7.3. Photocatalytic Advanced Oxidation Processes (AOP) for Heterogeneous Degradation of Antibiotics
7.3.1. Photodegradation of Antibiotics Using TiO2 Based Heterogeneous Semiconductors
7.3.2. Photodegradation of Antibiotics Using Non-TiO2 Based Semiconducting Catalysts
7.3.3. Photo-Fenton or Electro-Fenton of Antibiotics Removal
7.4. Degradation of Antibiotics in Water by Plasma Treatment
7.5. Cathodic Degradation of Antibiotics
7.6. Temperature Degradation of Antibiotics
7.7. Sonocatalytic Degradation of Antibiotics
8. Enzymatic Degradation of Antibiotics
8.1. Laccases
8.1.1. Laccase and the Degradation of Antibiotics
Laboratory Studies
Computational Studies for Antimicrobial Degradation Using Laccases
8.2. Ultrasound (Sonolysis) Combined with Enzymatic Degradation for the Degradation of Antibiotics
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aminov, R.I. A brief history of the antibiotic era: Lessons learned and challenges for the future. Front. Microbiol. 2010, 1, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spagnolo, F.; Trujillo, M.; Dennehya, J.J. Why do antibiotics exist? mBio 2021, 12, e01966-21. [Google Scholar] [CrossRef] [PubMed]
- Checa Artos, M.; Sosa del Castillo, D.; Ruiz Barzola, O.; Barcos-Arias, M. Presencia de productos farmacéuticos en el agua y su impacto en el ambiente. Bionat 2021, 6, 1618–1627. [Google Scholar] [CrossRef]
- Becker, D.; Varela Della Giustina, S.; Rodriguez-Mozaz, S.; Schoevaart, R.; Barcelo, D.; de Cazes, M.; Belleville, M.P.; Sanchez-Marcano, J.; de Gunzburg, J.; Couillerot, O.; et al. Removal of antibiotics in wastewater by enzymatic treatment with fungal laccase—Degradation of compounds does not always eliminate toxicity. Bioresour. Technol. 2016, 219, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Silbergeld, E.K.; Graham, J.; Price, L.B. Industrial food animal production, antimicrobial resistance, and human health. Annu. Rev. Public Health 2008, 29, 151–169. [Google Scholar] [CrossRef]
- Taylor, P.; Reeder, R. Antibiotic use on crops in low and middle-income countries based on recommendations made by agricultural advisors. CABI Agric. Biosci. 2020, 1, 1. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration (FDA). Summary Report on Antimicrobials Sold or Distributed for Use in Food-Producing Animals; U.S. Food and Drug Administration (FDA): Washington, DC, USA, 2019; p. 49.
- Sarmah, A.K.; Meyer, M.T.; Boxall, A.B. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 2006, 65, 725–759. [Google Scholar] [CrossRef]
- Osei Sekyere, J. Antibiotic Types and Handling Practices in Disease Management among Pig Farms in Ashanti Region, Ghana. J. Vet. Med. 2014, 2014, 531952. [Google Scholar] [CrossRef]
- Ferrari, B.t.; Paxéus, N.; Giudice, R.L.; Pollio, A.; Garric, J. Ecotoxicological impact of pharmaceuticals found in treated wastewaters: Study of carbamazepine, clofibric acid, and diclofenac. Ecotoxicol. Environ. Safe 2003, 55, 359–370. [Google Scholar] [CrossRef]
- Arenas, N.E.; Moreno-Melo, V. Producción pecuaria y emergencia de antibiótico resistencia en Colombia: Revisión sistemática. Infection 2018, 22, 110–119. [Google Scholar] [CrossRef]
- Mobarki, N.; Almerabi, B.; Hattan, A. Antibiotic resistance crisis. Int. J. Med. Devel. Count. 2019, 40, 561–564. [Google Scholar] [CrossRef] [Green Version]
- Leder, C.; Suk, M.; Lorenz, S.; Rastogi, T.; Peifer, C.; Kietzmann, M.; Jonas, D.; Buck, M.; Pahl, A.; Kümmerer, K. Reducing Environmental Pollution by Antibiotics through Design for Environmental Degradation. ACS Sustain. Chem. Eng. 2021, 9, 9358–9368. [Google Scholar] [CrossRef]
- Sundararaman, S.; Aravind Kumar, J.; Deivasigamani, P.; Devarajan, Y. Emerging pharma residue contaminants: Occurrence, monitoring, risk and fate assessment—A challenge to water resource management. Sci. Total. Environ. 2022, 825, 153897. [Google Scholar] [CrossRef]
- Szymańska, U.; Wiergowski, M.; Sołtyszewski, I.; Kuzemko, J.; Wiergowska, G.; Woźniak, M.K. Presence of antibiotics in the aquatic environment in Europe and their analytical monitoring: Recent trends and perspectives. Microchem. J. 2019, 147, 729–740. [Google Scholar] [CrossRef]
- Bedner, M.; Maccrehan, W.A. Transformation of acetaminophen by chlorination produces the toxicants 1,4-Benzoquinone and N-Acetyl-p-benzoquinone Imine. Environ. Sci. Technol. 2006, 40, 516–522. [Google Scholar] [CrossRef]
- Poutou, R.A.; Sánchez, L.; Díaz, K.; Máttar, S. Mecanismos de resistencia a los antibióticos betha-lactámicos. Med. UIS 1999, 13, 172–177. [Google Scholar]
- Denyer, S.P.; Hodges, N.A.; Gorman, S.P. Hugo and Russell’s Pharmaceutical Microbiology; Blackwell Science, Inc.: Oxford, UK, 2004. [Google Scholar]
- European Centre for Disease Prevention and Control (ECDC). Antimicrobial Consumption in the EU/EEA; Annual Epidemiological Report for 2019; European Centre for Disease Prevention and Control (ECDC): Stockholm, Sweden, 2019; p. 25. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing (CLSI M100-S17); Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2007; p. 182. [Google Scholar]
- World Health Organization (WHO). WHO Report on Surveillance of Antibiotic Consumption: 2016–2018 Early Implementation; World Health Organization (WHO): Geneva, Switzerland, 2018; p. 127. [Google Scholar]
- World Health Organization. Critically important antimicrobials for human medicine. In Ranking of Medically Important Antimicrobials for Risk Management of Antimicrobial Resistance due to Non-Human Use, 6th Revision 2018; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Forge, A.; Schacht, J. Aminoglycoside antibiotics. Audiol. Neurootol. 2000, 5, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Vigliarolo, L.O.; Di Pinto, P.M.; Suárez, M.C.; Lopardo, H.A.; Viegas Caetano, J.A. Antibióticos: Clasificación, Estructura, Mecanismos de Acción y Resistencia; Lopardo, H.A., Ed.; EDULP: Buenos Aires, Argentina, 2020; p. 191. [Google Scholar]
- Suarez, C.; Gudiol, F. Beta-lactam antibiotics. Enferm. Infecc. Microbiol. Clin. 2009, 27, 116–129. [Google Scholar]
- El-Gamal, M.I.; Brahim, I.; Hisham, N.; Aladdin, R.; Mohammed, H.; Bahaaeldin, A. Recent updates of carbapenem antibiotics. Eur. J. Med. Chem. 2017, 131, 185–195. [Google Scholar] [CrossRef]
- El-Shaboury, S.R.; Saleh, G.A.; Mohamed, F.A.; Rageh, A.H. Analysis of cephalosporin antibiotics. J. Pharm. Biomed. Anal. 2007, 45, 1–19. [Google Scholar] [CrossRef]
- Brewer, N.S.; Hellinger, W.C. The Monobactams. Mayo. Clin. Proc. 1991, 66, 1152–1157. [Google Scholar] [CrossRef]
- Garcia-Quetglas, E.; Azanza, J.R.; Sadaba, B.; Gil-Aldea, I. Farmacología de antimicrobianos utilizados en el tratamiento de las infecciones graves por bacterias Grampositivas. Rev. Esp. Quimioterap. 2003, 16, 277–288. [Google Scholar]
- Burdette, S.D.; Trotman, R. Tedizolid: The First Once-Daily Oxazolidinone Class Antibiotic. Clin. Infect. Dis. 2015, 61, 1315–1321. [Google Scholar] [PubMed] [Green Version]
- Calvo, J.; Martinez-Martinez, L. Mecanismos de acción de los antimicrobianos. Enfem. Infecc. Microbiol. Clín. 2009, 27, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Flamm, R.K.; Farrell, D.J.; Rhomberg, P.R.; Scangarella-Oman, N.E.; Sader, H.S. Gepotidacin (GSK2140944) In Vitro Activity against Gram-Positive and Gram-Negative Bacteria. Antimicrob. Agents Chemother. 2017, 61, e00468-17. [Google Scholar] [CrossRef] [Green Version]
- Cholo, M.C.; Steel, H.C.; Fourie, P.B.; Germishuizen, W.A.; Anderson, R. Clofazimine: Current status and future prospects. J. Antimicrob. Chemother. 2012, 67, 290–298. [Google Scholar] [CrossRef]
- García-Rodríguez, J.A.; Gutiérrez Zufiaurre, N.; Muñoz Bellido, L.J. Ácido fusídico. Rev. Esp. Quimioterap. 2003, 16, 161–171. [Google Scholar]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [Green Version]
- Qian, H.; Li, J.; Pan, X.; Sun, Z.; Ye, C.; Jin, G.; Fu, Z. Effects of streptomycin on growth of algae Chlorella vulgaris and Microcystis aeruginosa. Environ. Toxicol. 2012, 27, 229–237. [Google Scholar] [CrossRef]
- Wollenberger, L.; Halling-Sorensen, B.; Kusk, K.O. Acute and chronic toxicity of veterinary antibiotics to Daphnia magna. Chemosphere 2000, 40, 723–730. [Google Scholar] [CrossRef]
- Liljebjelke, K.A.; Hofacre, C.L.; White, D.G.; Ayers, S.; Lee, M.D.; Maurer, J.J. Diversity of Antimicrobial Resistance Phenotypes in Salmonella Isolated from Commercial Poultry Farms. Front. Vet. Sci. 2017, 4, 96. [Google Scholar] [CrossRef] [PubMed]
- Luczkiewicz, A.; Jankowska, K.; Fudala-Ksiazek, S.; Olanczuk-Neyman, K. Antimicrobial resistance of fecal indicators in municipal wastewater treatment plant. Water Res. 2010, 44, 5089–5097. [Google Scholar] [CrossRef] [PubMed]
- Avent, M.L.; Rogers, B.A.; Cheng, A.C.; Paterson, D.L. Current use of aminoglycosides: Indications, pharmacokinetics and monitoring for toxicity. Intern. Med. J. 2011, 41, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Turnidge, J. Pharmacodynamics and dosing of aminoglycosides. Infect. Dis. Clin. N. Am. 2003, 17, 503–528. [Google Scholar] [CrossRef]
- Duarte, J.L.d.S.; Solano, A.M.S.; Arguelho, M.L.P.M.; Tonholo, J.; Martínez-Huitle, C.A.; Zanta, C.L.d.P.e.S. Evaluation of treatment of effluents contaminated with rifampicin by Fenton, electrochemical and associated processes. J. Water Proc. Eng. 2018, 22, 250–257. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Global Antimicrobial Resistance Surveillance System (GLASS) Report: Early Implementation 2016–2017; Licence: CC BY-NC-SA 3.0 IGO; World Health Organization: Geneva, Switzerland, 2017; 164p. [Google Scholar]
- Havelkova, B.; Beklova, M.; Kovacova, V.; Hlavkova, D.; Pikula, J. Ecotoxicity of selected antibiotics for organisms of aquatic and terrestrial ecosystems. Neuroendocrinol. Lett. 2016, 37, 38–44. [Google Scholar]
- Ranjbar, R.; Sami, M. Genetic Investigation of Beta-Lactam Associated Antibiotic Resistance Among Escherichia Coli Strains Isolated from Water Sources. Open Microbiol. J. 2017, 11, 203–210. [Google Scholar] [CrossRef]
- Hoelle, J.; Johnson, J.R.; Johnston, B.D.; Kinkle, B.; Boczek, L.; Ryu, H.; Hayes, S. Survey of US wastewater for carbapenem-resistant Enterobacteriaceae. J. Water Health 2019, 17, 219–226. [Google Scholar] [CrossRef]
- Yilmaz, C.; Ozcengiz, G. Antibiotics: Pharmacokinetics, toxicity, resistance and multidrug efflux pumps. Biochem. Pharmacol. 2017, 133, 43–62. [Google Scholar] [CrossRef]
- Das, N.; Madhavan, J.; Selvi, A.; Das, D. An overview of cephalosporin antibiotics as emerging contaminants: A serious environmental concern. 3 Biotech. 2019, 9, 231. [Google Scholar] [CrossRef]
- Barriere, S.L. Pharmacology and Pharmacokinetics of Cefprozil. Clin. Infect. Dis. 1992, 14, S184–S188. [Google Scholar] [CrossRef] [PubMed]
- Selvi, A.; Salam, J.A.; Das, N. Biodegradation of cefdinir by a novel yeast strain, Ustilago sp. SMN03 isolated from pharmaceutical wastewater. World J. Microbiol. Biotechnol. 2014, 30, 2839–2850. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Tang, X.; Zuo, J.; Zhang, M.; Chen, L.; Li, Z. Distribution and persistence of cephalosporins in cephalosporin producing wastewater using SPE and UPLC-MS/MS method. Sci. Total Environ. 2016, 569–570, 23–30. [Google Scholar] [CrossRef]
- Bruyndonckx, R.; Adriaenssens, N.; Hens, N.; Versporten, A.; Monnet, D.L.; Molenberghs, G.; Goossens, H.; Weist, K.; Coenen, S.; Group, E.S.-N.s. Consumption of penicillins in the community, European Union/European Economic Area, 1997–2017. J. Antimicrob. Chemother. 2021, 76, ii14–ii21. [Google Scholar] [CrossRef] [PubMed]
- Bodey, G.P.; Nance, J. Amoxicillin: In Vitro and Pharmacological Studies. Antimicrob. Age Chemother. 1972, 1, 358–362. [Google Scholar] [CrossRef] [Green Version]
- Ighalo, J.O.; Igwegbe, C.A.; Aniagor, C.O.; Oba, S.N. A review of methods for the removal of penicillins from water. J. Water Prc. Eng. 2021, 39, 101886. [Google Scholar] [CrossRef]
- Arslan-Alaton, I.; Caglayan, A.E. Toxicity and biodegradability assessment of raw and ozonated procaine penicillin G formulation effluent. Ecotoxicol. Environ. Saf. 2006, 63, 131–140. [Google Scholar] [CrossRef]
- Szekeres, E.; Baricz, A.; Chiriac, C.M.; Farkas, A.; Opris, O.; Soran, M.L.; Andrei, A.S.; Rudi, K.; Balcazar, J.L.; Dragos, N.; et al. Abundance of antibiotics, antibiotic resistance genes and bacterial community composition in wastewater effluents from different Romanian hospitals. Environ. Pollut. 2017, 225, 304–315. [Google Scholar] [CrossRef]
- Felis, E.; Kalka, J.; Sochacki, A.; Kowalska, K.; Bajkacz, S.; Harnisz, M.; Korzeniewska, E. Antimicrobial pharmaceuticals in the aquatic environment—Occurrence and environmental implications. Eur. J. Pharmacol. 2020, 866, 172813. [Google Scholar] [CrossRef]
- McGuinness, W.A.; Malachowa, N.; DeLeo, F.R. Vancomycin Resistance in Staphylococcus aureus. Yale J. Biol. Med. 2017, 90, 269–281. [Google Scholar]
- Stein, G.E.; Babinchak, T. Tigecycline: An update. Diagn. Microbiol. Infect. Dis. 2013, 75, 331–336. [Google Scholar] [CrossRef] [PubMed]
- El-Azazy, M.; El-Shafie, A.S.; Al-Meer, S.; Al-Saad, K.A. Eco-structured Adsorptive Removal of Tigecycline from Wastewater: Date Pits’ Biochar versus the Magnetic Biochar. Nanomaterials 2020, 11, 30. [Google Scholar] [CrossRef] [PubMed]
- Zurfuh, K.; Poirel, L.; Nordmann, P.; Nuësch-Inderbinen, M.; Chler, H.H.; Stephan, R. Occurrence of the plasmid-borne mcr-1 colistin resistance gene in extended-spectrum-lactamase-producing Enterobacteriaceae in river water and imported vegetable samples in Switzerland. Antimicrob. Age Chemother. 2016, 60, 2594–2595. [Google Scholar] [CrossRef] [Green Version]
- Hembach, N.; Schmid, F.; Alexander, J.; Hiller, C.; Rogall, E.T.; Schwartz, T. Occurrence of the mcr-1 Colistin Resistance Gene and other Clinically Relevant Antibiotic Resistance Genes in Microbial Populations at Different Municipal Wastewater Treatment Plants in Germany. Front. Microbiol. 2017, 8, 1282. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Turiel, E.; He, L.; Martin-Esteban, A. Synthesis of Molecularly Imprinted Polymers for the Selective Extraction of Polymyxins from Environmental Water Samples. Polymers 2020, 12, 131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rincón-Gamboa, S.M.; Poutou-Piñales, R.A.; Carrascal-Camacho, A.K. Antimicrobial resistance of Non-Typhoid Salmonella in meat and meat products. Foods 2021, 10, 1731. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, R.; Ternes, T.; Haberer, K.; Kratz, K.-L. Occurrence of antibiotics in the aquatic environment. Sci. Total Environ. 1999, 225, 109–118. [Google Scholar] [CrossRef]
- Managaki, S.; Murata, A.; Takada, H.; Tuyen, B.C.; Chiem, N.H. Distribution of Macrolides, Sulfonamides, and Trimethoprim in Tropical Waters: Ubiquitous Occurrence of Veterinary Antibiotics in the Mekong Delta. Environ. Sci. Technol. 2007, 41, 8004–8010. [Google Scholar] [CrossRef]
- Zhang, C.; You, S.; Zhang, J.; Qi, W.; Su, R.; He, Z. An effective in-situ method for laccase immobilization: Excellent activity, effective antibiotic removal rate and low potential ecological risk for degradation products. Bioresour. Technol. 2020, 308, 123271. [Google Scholar] [CrossRef]
- Almeida, L.M.; Gaca, A.; Bispo, P.M.; Lebreton, F.; Saavedra, J.T.; Silva, R.A.; Basilio-Junior, I.D.; Zorzi, F.M.; Filsner, P.H.; Moreno, A.M.; et al. Coexistence of the Oxazolidinone Resistance-Associated Genes cfr and optrA in Enterococcus faecalis From a Healthy Piglet in Brazil. Front. Public Health 2020, 8, 518. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, J.; Schwarz, S.; Zhang, S.; Tao, J.; Fan, R.; Walsh, T.R.; Wu, C.; Wang, Y. Mobile oxazolidinone/phenicol resistance gene optrA in chicken Clostridium perfringens. J. Antimicrob. Chemother. 2020, 75, 3067–3069. [Google Scholar] [CrossRef] [PubMed]
- Timm, A.; Abendschon, P.; Tolgyesi, L.; Horn, H.; Borowska, E. Solar-mediated degradation of linezolid and tedizolid under simulated environmental conditions: Kinetics, transformation and toxicity. Chemosphere 2020, 241, 125111. [Google Scholar] [CrossRef] [PubMed]
- Shaw, K.J.; Barbachyn, M.R. The oxazolidinones: Past, present, and future. Ann. N. Y. Acad. Sci. 2011, 1241, 48–70. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Model List of Essential Medicines—22nd List; World Health Organization: Geneva, Switzerland, 2021; 66p. [Google Scholar]
- Merazi, Y.; Hammadi, K.; Fedoul, F.F. An investigation of the practices of veterinarians and breeders in the prevalence of antibiotic resistance in poultry farms in Algeria. Nat. Technol. 2021, 13, 14–33. [Google Scholar]
- Keating, G.M. Fosfomycin trometamol: A review of its use as a single-dose oral treatment for patients with acute lower urinary tract infections and pregnant women with asymptomatic bacteriuria. Drugs 2013, 73, 1951–1966. [Google Scholar] [CrossRef]
- Takahata, S.; Ida, T.; Hiraishi, T.; Sakakibara, S.; Maebashi, K.; Terada, S.; Muratani, T.; Matsumoto, T.; Nakahama, C.; Tomono, K. Molecular mechanisms of fosfomycin resistance in clinical isolates of Escherichia coli. Int. J. Antimicrob. Agents 2010, 35, 333–337. [Google Scholar] [CrossRef]
- Falagas, M.E.; Athanasaki, F.; Voulgaris, G.L.; Triarides, N.A.; Vardakas, K.Z. Resistance to fosfomycin: Mechanisms, Frequency and Clinical Consequences. Int. J. Antimicrob. Agents 2019, 53, 22–28. [Google Scholar] [CrossRef]
- Zeng, P.; Xie, X.; Song, Y.; Liu, R.; Zhu, C.; Galarneau, A.; Pic, J.S. Ion chromatography as highly suitable method for rapid and accurate determination of antibiotic fosfomycin in pharmaceutical wastewater. Water Sci. Technol. 2014, 69, 2014–2022. [Google Scholar] [CrossRef]
- Golet, E.M.; Xifra, i.; Siegrist, H.; Alder, A.C.; Giger, W. Environmental exposure Assessment of fluoroquinolone antibacterial agents from sewage to soil. Environ. Sci. Technol. 2003, 37, 3243–3249. [Google Scholar] [CrossRef]
- Zhang, Z.; Cheng, H. Recent Development in Sample Preparation and Analytical Techniques for Determination of Quinolone Residues in Food Products. Crit. Rev. Anal. Chem. 2017, 47, 223–250. [Google Scholar] [CrossRef]
- Castrignano, E.; Kannan, A.M.; Proctor, K.; Petrie, B.; Hodgen, S.; Feil, E.J.; Lewis, S.E.; Lopardo, L.; Camacho-Munoz, D.; Rice, J.; et al. (Fluoro)quinolones and quinolone resistance genes in the aquatic environment: A river catchment perspective. Water Res. 2020, 182, 116015. [Google Scholar] [CrossRef] [PubMed]
- Kergaravat, S.V.; Hernandez, S.R.; Gagneten, A.M. Second-, third- and fourth-generation quinolones: Ecotoxicity effects on Daphnia and Ceriodaphnia species. Chemosphere 2021, 262, 127823. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.D.M.; Ziora, Z.M.; Blaskovich, M.A.T. Quinolone antibiotics. Med. Chem. Commun. 2019, 10, 1719–1739. [Google Scholar] [CrossRef]
- Schwarz, S.; Kehrenberg, C.; Doublet, B.; Cloeckaert, A. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 2004, 28, 519–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira Toledo, S.L.; Silveira Silva, R.M.; Rodrigues dos Santos, I.C.; Lima, W.G.; Rodrigues Ferreira, L.G.; Paiva, M.C. Domestic wastewater treatment plants as sources of macrolide-lincosamide-streptogramin B- and penicillin-resistant Staphylococcus aureus in Brazil. Rev. Colomb. Cien. Quím.-Farm. 2020, 49, 267–279. [Google Scholar] [CrossRef]
- Guo, X.; Yan, Z.; Zhang, Y.; Xu, W.; Kong, D.; Shan, Z.; Wang, N. Behavior of antibiotic resistance genes under extremely high-level antibiotic selection pressures in pharmaceutical wastewater treatment plants. Sci. Total Environ. 2018, 612, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Dadashi, M.; Hajikhani, B.; Darban-Sarokhalil, D.; van Belkum, A.; Goudarzi, M. Mupirocin resistance in Staphylococcus aureus: A systematic review and meta-analysis. J. Glob. Antimicrob. Resist. 2020, 20, 238–247. [Google Scholar] [CrossRef]
- Holdiness, M.R. Clinical pharmacokinetics of clofazimine: A review. Clin. Pharmacokinet. 1989, 16, 74–85. [Google Scholar] [CrossRef]
- Williamson, D.A.; Monecke, S.; Heffernan, H.; Ritchie, S.R.; Roberts, S.A.; Upton, A.; Thomas, M.G.; Fraser, J.D. High usage of topical fusidic acid and rapid clonal expansion of fusidic acid-resistant Staphylococcus aureus: A cautionary tale. Clin. Infect. Dis. 2014, 59, 1451–1454. [Google Scholar] [CrossRef] [Green Version]
- Hruska, K.; Franek, M. Sulfonamides in the environment: A review and a case report. Vet. Med. 2012, 57, 1–35. [Google Scholar] [CrossRef] [Green Version]
- Vila-Costa, M.; Gioia, R.; Acena, J.; Perez, S.; Casamayor, E.O.; Dachs, J. Degradation of sulfonamides as a microbial resistance mechanism. Water Res. 2017, 115, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Scholar, E. Tetracycline. In X Pharm: The Comprehensive Pharmacology Reference; Dowd, F.J., Murrin, L.C., Ralevic, V., Scholar, E.M., Summers, R.J., Tew, K.D., Wecker, L., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2008; pp. 1–4. [Google Scholar]
- Pulicharla, R.; Brar, S.K.; Rouissi, T.; Auger, S.; Drogui, P.; Verma, M.; Surampalli, R.Y. Degradation of chlortetracycline in wastewater sludge by ultrasonication, Fenton oxidation, and ferro-sonication. Ultrason. Sonochem. 2017, 34, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhang, H.; Xiong, P.; Zhu, Q.; Liao, C.; Jiang, G. Occurrence, fate, and risk assessment of typical tetracycline antibiotics in the aquatic environment: A review. Sci. Total Environ. 2021, 753, 141975. [Google Scholar] [CrossRef]
- Page, S.W.; Gautier, P. Use of antimicrobial agents in livestock. Rev. Sci. Tech. 2012, 31, 145–188. [Google Scholar] [CrossRef]
- El Parlamento Europeo; Consejo de la Unión Europea. Reglamento (UE) 2019/6 del Parlamento Europeo y del Consejo de 11de Diciembre de 2018; El Parlamento Europeo: Washington, DC, USA, 2019; p. 125. [Google Scholar]
- World Health Organization. Initiative to Estimate the Global Burden of Foodborne Diseases: Fourth Formal Meeting of the Foodborne Disease Burden Epidemiology Reference Group (FERG): Sharing New Results, Making Future Plans, and Preparing Ground for the Countries; World Health Organization: Geneva, Switzerland, 2014; p. 108. [Google Scholar]
- Destro, M.T.; Ribeiro, V.B. Foodborne Zoonoses. In Encyclopedia of Meat Sciences; Devine, C., Dikeman, M., Eds.; Academic Press: Cambridge, MA, USA, 2014; pp. 17–21. [Google Scholar]
- Gebeyehu, D.T. Antibiotic Resistance Development in Animal Production: A Cross-Sectional Study. Vet. Med. Res. Rep. 2021, 12, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, S.A.; Ramachandran, A.; Perron, G.G. Antibiotic Pollution in the Environment: From Microbial Ecology to Public Policy. Microorganisms 2019, 7, 180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Checcucci, A.; Trevisi, P.; Luise, D.; Modesto, M.; Blasioli, S.; Braschi, I.; Mattarelli, P. Exploring the Animal Waste Resistome: The Spread of Antimicrobial Resistance Genes Through the Use of Livestock Manure. Front. Microbiol. 2020, 11, 1416. [Google Scholar] [CrossRef]
- Domenech, E.; Jimenez-Belenguer, A.; Amoros, J.A.; Ferrus, M.A.; Escriche, I. Prevalence and antimicrobial resistance of Listeria monocytogenes and Salmonella strains isolated in ready-to-eat foods in Eastern Spain. Food Cont. 2015, 47, 120–125. [Google Scholar] [CrossRef]
- Sallam, K.I.; Mohammed, M.A.; Hassan, M.A.; Tamura, T. Prevalence, molecular identification and antimicrobial resistance profile of Salmonella serovars isolated from retail beef products in Mansoura, Egypt. Food Cont. 2014, 38, 209–214. [Google Scholar] [CrossRef]
- Abd-Elghany, S.M.; Sallam, K.I.; Abd-Elkhalek, A.; Tamura, T. Occurrence, genetic characterization and antimicrobial resistance of Salmonella isolated from chicken meat and giblets. Epidemiol. Infect. 2015, 143, 997–1003. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, D.T.A.; Kanki, M.; Nguyen, P.D.; Le, H.T.; Ngo, P.T.; Tran, D.N.M.; Le, N.H.; Van Dang, C.; Kawai, T.; Kawahara, R.; et al. Prevalence, antibiotic resistance, and extended-spectrum and AmpC β-lactamase productivity of Salmonella isolates from raw meat and seafood samples in Ho Chi Minh City, Vietnam. Int. J. Food Microbiol. 2016, 236, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Ayala-Romero, C.; Ballen-Parada, C.; Rico-Gaitán, M.; Chamorro-Tobar, I.; Zambrano-Moreno, D.; Poutou-Piñales, R.; Carrascal-Camacho, A. Prevalence of Salmonella spp., in mesenteric pig’s ganglia at Colombian benefit plants. Rev. MVZ-Córdoba 2018, 23, 6474–6486. [Google Scholar] [CrossRef] [Green Version]
- Löfström, C.; Hansen, T.; Maurischat, S.; Malorny, B. Salmonella: Salmonellosis. In Encyclopedia of Food and Health; Caballero, B., Finglas, P., Toldra, F., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2015; pp. 701–705. [Google Scholar]
- Sánchez-Vargas, F.M.; Abu-El-Haija, M.A.; Gómez-Duarte, O.G. Salmonella infections: An update on epidemiology, management, and prevention. Travel. Med. Infect. Dis. 2011, 9, 263–277. [Google Scholar] [CrossRef] [PubMed]
- De Jong, A.; Bywater, R.; Butty, P.; Deroover, E.; Godinho, K.; Klein, U.; Marion, H.; Simjee, S.; Smets, K.; Thomas, V.; et al. A pan-European survey of antimicrobial susceptibility towards human-use antimicrobial drugs among zoonotic and commensal enteric bacteria isolated from healthy food-producing animals. J. Antimicrob. Chemother. 2009, 63, 733–744. [Google Scholar] [CrossRef] [Green Version]
- Antunes, P.; Mourão, J.; Campos, J.; Peixe, L. Salmonellosis: The role of poultry meat. Clin. Microbiol. Infect. 2016, 22, 110–121. [Google Scholar] [CrossRef] [Green Version]
- Marquis, H.; Drevets, D.A.; Bronze, M.S.; Kathariou, S.; Golos, T.G.; Iruretagoyena, J.I. Pathogenesis of Listeria Monocytogenes in Humans. In HUMAN Emerging and Re-emerging Infections: Viral and Parasitic Infections; Singh, S.K., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 749–772. [Google Scholar]
- Torres, K.J.; Sierra, S.C.; Poutou, R.A.; Vera, H.; Carrascal, A.K.; Mercado, M. Incidencia y diagnóstico de Listeria monocytogenes; microorganismo zoonótico emergente en la industria de alimentos. Rev. UDCA Act. Divulg. Cient 2004, 7, 25–57. [Google Scholar]
- Torres, K.J.; Sierra, S.C.; Poutou, R.A.; Carrascal, A.K.; Mercado, M. Patogénesis de Listeria monocytogenes, microorganismo zoonótico emergente. Rev. MVZ-Córdoba 2005, 10, 511–543. [Google Scholar] [CrossRef]
- Belalcazar, M.E.; Poutou, R.A.; Torres, K.J.; Gallegos, J.M.; Torres, O.; Carrascal, A.K. Listeria monocytogenes y listeriosis animal. Rev. UDCA Act. Inv. Cient. 2005, 8, 3–16. [Google Scholar]
- Ruiz-Bolivar, Z.; Neuque-Rico, M.C.; Poutou-Piñales, R.A.; Carrascal-Camacho, A.K.; Máttar-Velilla, S. Antimicrobial susceptibility of L. monocytogenes food-isolates from different cities of Colombia. Foodborne. Path Dis. 2011, 8, 913–919. [Google Scholar] [CrossRef] [Green Version]
- Allen, K.J.; Wałecka-Zacharska, E.; Chen, J.C.; Kosek-Paszkowska, K.; Devlieghere, F.; Van Meervenne, E.; Osek, J.; Wieczorek, K.; Bania, J. Listeria monocytogenes—An examination of food chain factors potentially contributing to antimicrobial resistance. Food Microbiol. 2016, 54, 178–189. [Google Scholar] [CrossRef]
- Tenover, F.C. Mechanisms of antimicrobial resistance in bacteria. Am. J. Infect. Control. 2006, 34, s3–s10. [Google Scholar] [CrossRef] [PubMed]
- Morejón García, M. Betalactamasas de espectro extendido. Rev. Cubana. Med. 2013, 52, 272–280. [Google Scholar]
- Sanseverino, I.; Navarro Cuenca, A.; Loos, R.; Marinov, D.; Lettieri, T. State of the Art on the Contribution of Water to Antimicrobial Resistance, EUR 29592; Publications Office of the European Union: Luxembourg, 2018; 110p, ISBN 978-92-79-98478-5. [Google Scholar] [CrossRef]
- Grohmann, E.; Muth, G.; Espinosa, M. Conjugative plasmid transfer in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2003, 67, 277–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poutou, R.A.; Mattar, S. Genética Molecular Bacteriana. In Bacteriología Clínica: Estudio Etiológico de las Enfermedades Infecciosas de Origen Bacteriano; Editorial: Universidad de Córdoba Vicerrectoría de Investigación y Extensiones; Asociación Colombiana de Infectología (ACIN): Córdoba, Spain, 2002. [Google Scholar]
- Arutyunov, D.; Frost, L.S. F conjugation: Back to the beginning. Plasmid 2013, 70, 18–32. [Google Scholar] [CrossRef] [PubMed]
- Kohler, V.; Keller, W.; Grohmann, E. Regulation of Gram-Positive Conjugation. Front. Microbiol. 2019, 10, 1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Y.; Xu, L.; Rysz, M.; Wang, Y.; Zhang, H.; Alvarez, P.J. Occurrence and transport of tetracycline, sulfonamide, quinolone, and macrolide antibiotics in the Haihe River Basin, China. Environ. Sci. Technol. 2011, 45, 1827–1833. [Google Scholar] [CrossRef]
- Ramírez, G.D.; Vélez, G.; Rondón, I.S. Determinación de residuos de antibióticos y tiempo de retiro en leche proveniente del municipio de Cartago (Valle del Cauca). Rev. Col. Cienc. Anim. 2012, 5, 25–31. [Google Scholar]
- Xu, D.; Xiao, Y.; Pan, H.; Mei, Y. Toxic effects of tetracycline and its degradation products on freshwater green algae. Ecotoxicol. Environ. Saf. 2019, 174, 43–47. [Google Scholar] [CrossRef]
- Song, Y.; Han, Z.; Song, K.; Zhen, T. Antibiotic Consumption Trends in China: Evidence From Six-Year Surveillance Sales Records in Shandong Province. Front. Pharmacol. 2020, 11, 491. [Google Scholar] [CrossRef]
- Anh, H.Q.; Le, T.P.Q.; Da Le, N.; Lu, X.X.; Duong, T.T.; Garnier, J.; Rochelle-Newall, E.; Zhang, S.; Oh, N.H.; Oeurng, C.; et al. Antibiotics in surface water of East and Southeast Asian countries: A focused review on contamination status, pollution sources, potential risks, and future perspectives. Sci. Total Environ. 2021, 764, 142865. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (Ed.) Antibiotic Use in the United States, 2021 Update: Progress and Opportunities; US Department of Health and Human Services: Atlanta, GA, USA, 2021; 26p.
- Instituto Nacional de Salud (INS). Consumo de Antibióticos en el Ambito Hospitalario, Colombia, 2017; Instituto Nacional de Salud (INS): Singapore, 2017; 16p.
- Pallares, C.J.; Martínez, E. Implementación de un programa de uso regulado de antibióticos en 2 unidades de cuidado intensivo medico-quirúrgico en un hospital universitario de tercer nivel en Colombia. Infection 2012, 16, 192–198. [Google Scholar] [CrossRef]
- Villalobos, A.P.; Barrero, L.I.; Rivera, S.M.; Ovalle, M.V.; Valera, D. Vigilancia de infecciones asociadas a la atención en salud, resistencia bacteriana y consumo de antibióticos en hospitales de alta complejidad, Colombia, 2011. Biomédica 2013, 34, 67. [Google Scholar] [CrossRef] [Green Version]
- Diwan, V.; Tamhankar, A.J.; Khandal, R.K.; Sen, S.; Aggarwal, M.; Marothi, Y.; Iyer, R.V.; Sundblad-Tonderski, K.; Stålsby-Lundborg, C. Aesneatrcihb airtoiclteics and antibiotic-resistant bacteria in waters associated with a hospital in Ujjain, India. BMC Public Health 2010, 10, 414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lacorte, S.; Gomez-Canela, C.; Calas-Blanchard, C. Pharmaceutical residues in senior residences wastewaters: High loads, emerging risks. Molecules 2021, 26, 5047. [Google Scholar] [CrossRef] [PubMed]
- Lacorte, S.; Luis, S.; Gomez-Canela, C.; Sala-Comorera, T.; Courtier, A.; Roig, B.; Oliveira-Brett, A.M.; Joannis-Cassan, C.; Aragones, J.I.; Poggio, L.; et al. Pharmaceuticals released from senior residences: Occurrence and risk evaluation. Environ. Sci. Pollut. Res. 2018, 25, 6095–6106. [Google Scholar] [CrossRef] [Green Version]
- Kummerer, K. Significance of antibiotics in the environment. J. Antimicrob. Chemother. 2003, 52, 5–7. [Google Scholar] [CrossRef] [Green Version]
- Larsson, D.G. Antibiotics in the environment. Ups. J. Med. Sci. 2014, 119, 108–112. [Google Scholar] [CrossRef]
- Li, S.; Shi, W.; Li, H.; Xu, N.; Zhang, R.; Chen, X.; Sun, W.; Wen, D.; He, S.; Pan, J.; et al. Antibiotics in water and sediments of rivers and coastal area of Zhuhai City, Pearl River estuary, south China. Sci. Total Environ. 2018, 636, 1009–1019. [Google Scholar] [CrossRef]
- Yang, S.; Carlson, K.H. Solid-phase extraction-high-performance liquid chromatography-ion trap mass spectrometry for analysis of trace concentrations of macrolide antibiotics in natural and waste water matrices. J. Chromatogr. A 2004, 1038, 141–155. [Google Scholar] [CrossRef]
- Lien, L.T.; Hoa, N.Q.; Chuc, N.T.; Thoa, N.T.; Phuc, H.D.; Diwan, V.; Dat, N.T.; Tamhankar, A.J.; Lundborg, C.S. Antibiotics in wastewater of a rural and an urban hospital before and after wastewater treatment, and the relationship with antibiotic use-A one year study from Vietnam. Int. J. Environ. Res. Public Health 2016, 13, 588. [Google Scholar] [CrossRef] [Green Version]
- Serna-Galvis, E.; Martínez-Mena, Y.L.; Porras, J.; Torres-Palma, R.A. Antibióticos de alto consumo en Colombia, excreción en orina y presencia en aguas residuales—Una revisión bibliográfica. Ing. Compet. 2021, 24, e30711267. [Google Scholar] [CrossRef]
- Deak, D.; Outterson, K.; Powers, J.H.; Kesselheim, A.S. Progress in the Fight Against Multidrug-Resistant Bacteria? A Review of U.S. Food and Drug Administration-Approved Antibiotics, 2010–2015. Ann. Intern. Med. 2016, 165, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Andrei, S.; Droc, G.; Stefan, G. FDA approved antibacterial drugs: 2018–2019. Discoveries 2019, 7, e102. [Google Scholar] [CrossRef] [PubMed]
- Malenfant, J.H.; Brewer, T.F. Rifampicin Mono-Resistant Tuberculosis-A Review of an Uncommon But Growing Challenge for Global Tuberculosis Control. Open Forum. Infect. Dis. 2021, 8, ofab018. [Google Scholar] [CrossRef]
- Pan, M.; Chu, L.M. Adsorption and degradation of five selected antibiotics in agricultural soil. Sci. Total Environ. 2016, 545–546, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Cycon, M.; Mrozik, A.; Piotrowska-Seget, Z. Antibiotics in the Soil Environment-Degradation and Their Impact on Microbial Activity and Diversity. Front. Microbiol. 2019, 10, 338. [Google Scholar] [CrossRef]
- Ezzariai, A.; Hafidi, M.; Khadra, A.; Aemig, Q.; El Fels, L.; Barret, M.; Merlina, G.; Patureau, D.; Pinelli, E. Human and veterinary antibiotics during composting of sludge or manure: Global perspectives on persistence, degradation, and resistance genes. J. Hazard. Mater. 2018, 359, 465–481. [Google Scholar] [CrossRef]
- Wu, S.; Hu, Y.H. A comprehensive review on catalysts for electrocatalytic and photoelectrocatalytic degradation of antibiotics. Chem. Eng. J. 2021, 409, 127739. [Google Scholar] [CrossRef]
- Homem, V.; Santos, L. Degradation and removal methods of antibiotics from aqueous matrices—A review. J. Env. Manag. 2011, 92, 2304–2347. [Google Scholar] [CrossRef]
- Abdurahman, M.H.; Abdullah, A.Z.; Shoparwe, N.F. A comprehensive review on sonocatalytic, photocatalytic, and sonophotocatalytic processes for the degradation of antibiotics in water: Synergistic mechanism and degradation pathway. Chem. Eng. J. 2021, 413, 127412. [Google Scholar] [CrossRef]
- Loftin, K.A.; Adams, C.D.; Meyer, M.T.; Surampalli, R. Effects of ionic strength, temperature, and pH on degradation of selected antibiotics. J. Environ. Qual. 2008, 37, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Calcio Gaudino, E.; Canova, E.; Liu, P.; Wu, Z.; Cravotto, G. Degradation of Antibiotics in Wastewater: New Advances in Cavitational Treatments. Molecules 2021, 26, 617. [Google Scholar] [CrossRef] [PubMed]
- Zeghioud, H.; Kamagate, M.; Coulibaly, L.S.; Rtimi, S.; Assadi, A.A. Photocatalytic degradation of binary and ternary mixtures of antibiotics: Reactive species investigation in pilot scale. Chem. Eng. Res. Des. 2019, 144, 300–309. [Google Scholar] [CrossRef] [Green Version]
- Dorival-Garcia, N.; Zafra-Gomez, A.; Navalon, A.; Gonzalez-Lopez, J.; Hontoria, E.; Vilchez, J.L. Removal and degradation characteristics of quinolone antibiotics in laboratory-scale activated sludge reactors under aerobic, nitrifying and anoxic conditions. J. Environ. Manag. 2013, 120, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Gan, J. Sorption and degradation of wastewater-associated non-steroidal anti-inflammatory drugs and antibiotics in soils. Chemosphere 2011, 83, 240–246. [Google Scholar] [CrossRef]
- Ji, Y.; Ferronato, C.; Salvador, A.; Yang, X.; Chovelon, J.M. Degradation of ciprofloxacin and sulfamethoxazole by ferrous-activated persulfate: Implications for remediation of groundwater contaminated by antibiotics. Sci. Total Environ. 2014, 472, 800–808. [Google Scholar] [CrossRef]
- Jiang, M.; Wang, L.; Ji, R. Biotic and abiotic degradation of four cephalosporin antibiotics in a lake surface water and sediment. Chemosphere 2010, 80, 1399–1405. [Google Scholar] [CrossRef]
- Braschi, I.; Blasioli, S.; Fellet, C.; Lorenzini, R.; Garelli, A.; Pori, M.; Giacomini, D. Persistence and degradation of new beta-lactam antibiotics in the soil and water environment. Chemosphere 2013, 93, 152–159. [Google Scholar] [CrossRef]
- Ahmad, F.; Zhu, D.; Sun, J. Environmental fate of tetracycline antibiotics: Degradation pathway mechanisms, challenges, and perspectives. Environ. Sci. Eur. 2021, 33, 64. [Google Scholar] [CrossRef]
- Ruan, Y.; Kumar Awasthi, M.; Cai, L.; Lu, H.; Xu, X.; Li, W. Simultaneous aerobic denitrification and antibiotics degradation by strain Marinobacter hydrocarbonoclasticus RAD-2. Bioresour. Technol. 2020, 313, 123609. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, J.; Zhong, Z.; Guo, C.; Li, L.; He, Y.; Fan, W.; Chen, Y. Degradation of sulfonamides antibiotics in lake water and sediment. Environ. Sci. Pollut. Rese. Int. 2013, 20, 2372–2380. [Google Scholar] [CrossRef] [PubMed]
- Nnenna, F.-P.; Lekiah, P.; Obemeata, O. Degradation of antibiotics by bacteria and fungi from the aquatic environment. J. Toxicol. Environ. Health Sci. 2011, 30, 275–285. [Google Scholar]
- Alexy, R.; Kumpel, T.; Kummerer, K. Assessment of degradation of 18 antibiotics in the Closed Bottle Test. Chemosphere 2004, 57, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.B.; Zakaria, M.P.; Latif, P.A.; Saari, N. Degradation of veterinary antibiotics and hormone during broiler manure composting. Bioresour. Technol. 2013, 131, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Shi, Y.; Dong, W.; Wen, X.; Jiang, M.; Lu, J. Thermo-activated persulfate oxidation system for tetracycline antibiotics degradation in aqueous solution. Chem. Eng. J. 2016, 298, 225–233. [Google Scholar] [CrossRef] [Green Version]
- Yin, R.; Guo, W.; Wang, H.; Du, J.; Zhou, X.; Wu, Q.; Zheng, H.; Chang, J.; Ren, N. Selective degradation of sulfonamide antibiotics by peroxymonosulfate alone: Direct oxidation and nonradical mechanisms. Chem. Eng. J. 2018, 334, 2539–2546. [Google Scholar] [CrossRef]
- Lange, F.; Cornelissen, S.; Kubac, D.; Sein, M.M.; von Sonntag, J.; Hannich, C.B.; Golloch, A.; Heipieper, H.J.; Moder, M.; von Sonntag, C. Degradation of macrolide antibiotics by ozone: A mechanistic case study with clarithromycin. Chemosphere 2006, 65, 17–23. [Google Scholar] [CrossRef]
- Pelalak, R.; Alizadeh, R.; Ghareshabani, E.; Heidari, Z. Degradation of sulfonamide antibiotics using ozone-based advanced oxidation process: Experimental, modeling, transformation mechanism and DFT study. Sci. Total Environ. 2020, 734, 139446. [Google Scholar] [CrossRef]
- Wu, X.; Zhao, W.; Huang, Y.; Zhang, G. A mechanistic study of amorphous CoSx cages as advanced oxidation catalysts for excellent peroxymonosulfate activation towards antibiotics degradation. Chem. Eng. J. 2020, 381, 122768. [Google Scholar] [CrossRef]
- Guo, W.; Su, S.; Yi, C.; Ma, Z. Degradation of antibiotics amoxicillin by Co3O4-catalyzed peroxymonosulfate system. Environ. Prog. Sustain. Energ. 2013, 32, 193–197. [Google Scholar] [CrossRef]
- Calvete, M.J.F.; Piccirillo, G.; Vinagreiro, C.S.; Pereira, M.M. Hybrid materials for heterogeneous photocatalytic degradation of antibiotics. Coordin. Chem. Rev. 2019, 395, 63–85. [Google Scholar] [CrossRef]
- Rokesh, K.; Sakar, M.; Do, T.O. Emerging Hybrid Nanocomposite Photocatalysts for the Degradation of Antibiotics: Insights into Their Designs and Mechanisms. Nanomaterials 2021, 11, 572. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhuan, R. Degradation of antibiotics by advanced oxidation processes: An overview. Sci. Total Environ. 2020, 701, 135023. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Li, B.; Pu, Q.; Chen, X.; Wen, G.; Li, Z. Preparation of Ag-AgVO3/g-C3N4 composite photo-catalyst and degradation characteristics of antibiotics. J. Hazard. Mater. 2019, 373, 303–312. [Google Scholar] [CrossRef]
- Wang, J.; Zhuan, R.; Chu, L. The occurrence, distribution and degradation of antibiotics by ionizing radiation: An overview. Sci. Total Environ. 2019, 646, 1385–1397. [Google Scholar] [CrossRef]
- He, X.; Mezyk, S.P.; Michael, I.; Fatta-Kassinos, D.; Dionysiou, D.D. Degradation kinetics and mechanism of beta-lactam antibiotics by the activation of H2O2 and Na2S2O8 under UV-254nm irradiation. J. Hazard. Mater. 2014, 279, 375–383. [Google Scholar] [CrossRef]
- Jeong, J.; Song, W.; Cooper, W.J.; Jung, J.; Greaves, J. Degradation of tetracycline antibiotics: Mechanisms and kinetic studies for advanced oxidation/reduction processes. Chemosphere 2010, 78, 533–540. [Google Scholar] [CrossRef]
- Saidulu, D.; Gupta, B.; Gupta, A.K.; Ghosal, P.S. A review on occurrences, eco-toxic effects, and remediation of emerging contaminants from wastewater: Special emphasis on biological treatment based hybrid systems. J. Environ. Chem. Eng. 2021, 9, 105282. [Google Scholar] [CrossRef]
- Li, D.; Shi, W. Recent developments in visible-light photocatalytic degradation of antibiotics. Chin. J. Catal. 2016, 37, 792–799. [Google Scholar] [CrossRef]
- Elmolla, E.S.; Chaudhuri, M. Photocatalytic degradation of amoxicillin, ampicillin and cloxacillin antibiotics in aqueous solution using UV/TiO2 and UV/H2O2/TiO2 photocatalysis. Desalination 2010, 252, 46–52. [Google Scholar] [CrossRef]
- Elmolla, E.S.; Chaudhuri, M. Degradation of amoxicillin, ampicillin and cloxacillin antibiotics in aqueous solution by the UV/ZnO photocatalytic process. J. Hazard. Mater. 2010, 173, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Xu, S.; Wang, Y.; Sun, X.; Gao, Y.; Gao, B. Enhanced degradation of ciprofloxacin by graphitized mesoporous carbon (GMC)-TiO2 nanocomposite: Strong synergy of adsorption-photocatalysis and antibiotics degradation mechanism. J. Colloid. Interface Sci. 2018, 527, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Abellán, M.N.; Giménez, J.; Esplugas, S. Photocatalytic degradation of antibiotics: The case of sulfamethoxazole and trimethoprim. Catal. Today 2009, 144, 131–136. [Google Scholar] [CrossRef]
- Ding, J.; Dai, Z.; Qin, F.; Zhao, H.; Zhao, S.; Chen, R. Z-scheme BiO1-xBr/Bi2O2CO3 photocatalyst with rich oxygen vacancy as electron mediator for highly efficient degradation of antibiotics. Appl. Catal. B Environ. 2017, 205, 281–291. [Google Scholar] [CrossRef]
- Wang, K.; Li, Y.; Zhang, G.; Li, J.; Wu, X. 0D Bi nanodots/2D Bi3NbO7 nanosheets heterojunctions for efficient visible light photocatalytic degradation of antibiotics: Enhanced molecular oxygen activation and mechanism insight. Appl. Catal. B Environ. 2019, 240, 39–49. [Google Scholar] [CrossRef]
- Yan, W.; Yan, L.; Jing, C. Impact of doped metals on urea-derived g-C3N4 for photocatalytic degradation of antibiotics: Structure, photoactivity and degradation mechanisms. Appl. Catal. B Environ. 2019, 244, 475–485. [Google Scholar] [CrossRef]
- Wang, A.; Zheng, Z.; Wang, H.; Chen, Y.; Luo, C.; Liang, D.; Hu, B.; Qiu, R.; Yan, K. 3D hierarchical H2-reduced Mn-doped CeO2 microflowers assembled from nanotubes as a high-performance Fenton-like photocatalyst for tetracycline antibiotics degradation. Appl. Catal. B Environ. 2020, 277, 119171. [Google Scholar] [CrossRef]
- Shi, W.; Li, M.; Huang, X.; Ren, H.; Guo, F.; Tang, Y.; Lu, C. Construction of CuBi2O4/Bi2MoO6 p-n heterojunction with nanosheets-on-microrods structure for improved photocatalytic activity towards broad-spectrum antibiotics degradation. Chem. Eng. J. 2020, 394, 125009. [Google Scholar] [CrossRef]
- Li, S.; Wang, C.; Liu, Y.; Xue, B.; Jiang, W.; Liu, Y.; Mo, L.; Chen, X. Photocatalytic degradation of antibiotics using a novel Ag/Ag2S/Bi2MoO6 plasmonic p-n heterojunction photocatalyst: Mineralization activity, degradation pathways and boosted charge separation mechanism. Chem. Eng. J. 2021, 415, 128991. [Google Scholar] [CrossRef]
- Che, H.; Che, G.; Jiang, E.; Liu, C.; Dong, H.; Li, C. A novel Z-Scheme CdS/Bi3O4Cl heterostructure for photocatalytic degradation of antibiotics: Mineralization activity, degradation pathways and mechanism insight. J. Taiwan Inst. Chem. Eng. 2018, 91, 224–234. [Google Scholar] [CrossRef]
- Li, H.; Li, T.; He, S.; Zhou, J.; Wang, T.; Zhu, L. Efficient degradation of antibiotics by non-thermal discharge plasma: Highlight the impacts of molecular structures and degradation pathways. Chem. Eng. J. 2020, 395, 125091. [Google Scholar] [CrossRef]
- Magureanu, M.; Piroi, D.; Mandache, N.B.; David, V.; Medvedovici, A.; Bradu, C.; Parvulescu, V.I. Degradation of antibiotics in water by non-thermal plasma treatment. Water Res. 2011, 45, 3407–3416. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-S.; Yang, C.-S.; Mok, Y.S. Degradation of veterinary antibiotics by dielectric barrier discharge plasma. Chem. Eng. J. 2013, 219, 19–27. [Google Scholar] [CrossRef]
- Sarangapani, C.; Ziuzina, D.; Behan, P.; Boehm, D.; Gilmore, B.F.; Cullen, P.J.; Bourke, P. Degradation kinetics of cold plasma-treated antibiotics and their antimicrobial activity. Sci. Rep. 2019, 9, 3955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, D.; Liang, B.; Yun, H.; Cheng, H.; Ma, J.; Cui, M.; Wang, A.; Ren, N. Cathodic degradation of antibiotics: Characterization and pathway analysis. Water Res. 2015, 72, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Zhang, J.; Chen, H.; Wang, J.; Sun, W.; Zhang, X.; Yang, Y.; Wang, Q.; Ma, J. Effect of temperature on sulfonamide antibiotics degradation, and on antibiotic resistance determinants and hosts in animal manures. Sci. Total Environ. 2017, 607–608, 725–732. [Google Scholar] [CrossRef]
- Chakma, S.; Dikshit, P.K.; Galodiya, M.N.; Giri, A.S.; Moholkar, V.S. The role of ultrasound in enzymatic degradation mechanism. J. Taiwan Inst. Chem. Eng. 2020, 107, 54–71. [Google Scholar] [CrossRef]
- Adewuyi, Y.G. Sonochemistry in environmental remediation. 2. Heterogeneous sonophotocatalytic oxidation processes for the treatment of pollutants in water. Environ. Sci. Technol. 2005, 39, 8557–8570. [Google Scholar] [CrossRef]
- De Bel, E.; Dewulf, J.; Witte, B.D.; Van Langenhove, H.; Janssen, C. Influence of pH on the sonolysis of ciprofloxacin: Biodegradability, ecotoxicity and antibiotic activity of its degradation products. Chemosphere 2009, 77, 291–295. [Google Scholar] [CrossRef]
- Liu, P.; Wu, Z.; Abramova, A.V.; Cravotto, G. Sonochemical processes for the degradation of antibiotics in aqueous solutions: A review. Ultrason. Sonochem. 2021, 74, 105566. [Google Scholar] [CrossRef]
- Li, L.; Wei, D.; Wei, G.; Du, Y. Oxidation of cefazolin by potassium permanganate: Transformation products and plausible pathways. Chemosphere 2016, 149, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Konstantinou, I.K.; Albanis, T.A. TiO2-assisted photocatalytic degradation of azo dyes in aqueous solution: Kinetic and mechanistic investigations. Appl. Catal. B Environ. 2004, 49, 1–14. [Google Scholar] [CrossRef]
- Fan, Y.; Zhou, Z.; Feng, Y.; Zhou, Y.; Wen, L.; Shih, K. Degradation mechanisms of ofloxacin and cefazolin using peroxymonosulfate activated by reduced graphene oxide-CoFe2O4 composites. Chem. Eng. J. 2020, 383, 123056. [Google Scholar] [CrossRef]
- Ghasemi, M.; Khataee, A.; Gholami, P.; Soltani, R.D.C.; Hassani, A.; Orooji, Y. In-situ electro-generation and activation of hydrogen peroxide using a CuFeNLDH-CNTs modified graphite cathode for degradation of cefazolin. J. Environ. Manag. 2020, 267, 110629. [Google Scholar] [CrossRef] [PubMed]
- Gurkan, Y.Y.; Turkten, N.; Hatipoglu, A.; Cinar, Z. Photocatalytic degradation of cefazolin over N-doped TiO2 under UV and sunlight irradiation: Prediction of the reaction paths via conceptual DFT. Chem. Eng. J. 2012, 184, 113–124. [Google Scholar] [CrossRef]
- Chen, Y.; Tian, H.; Zhu, W.; Zhang, X.; Li, R.; Chen, C.; Huang, Y. l-Cysteine directing synthesis of BiOBr nanosheets for efficient cefazolin photodegradation: The pivotal role of thiol. J. Hazard. Mater. 2021, 414, 125544. [Google Scholar] [CrossRef]
- Gholami, P.; Dinpazhoh, L.; Khataee, A.; Hassani, A.; Bhatnagar, A. Facile hydrothermal synthesis of novel Fe-Cu layered double hydroxide/biochar nanocomposite with enhanced sonocatalytic activity for degradation of cefazolin sodium. J. Hazard. Mater. 2020, 381, 120742. [Google Scholar] [CrossRef]
- Liu, J.; Li, Z.; Wang, M.; Jin, C.; Kang, J.; Tang, Y.; Li, S. Eu2O3/Co3O4 nanosheets for levofloxacin removal via peroxymonosulfate activation: Performance, mechanism and degradation pathway. Separat. Pur. Technol. 2021, 274, 118666. [Google Scholar] [CrossRef]
- He, Z.; Zheng, W.; Li, M.; Liu, W.; Zhang, Y.; Wang, Y. Fe2P/biocarbon composite derived from a phosphorus-containing biomass for levofloxacin removal through peroxymonosulfate activation. Chem. Eng. J. 2022, 427, 130928. [Google Scholar] [CrossRef]
- Xu, K.; Ben, W.; Ling, W.; Zhang, Y.; Qu, J.; Qiang, Z. Impact of humic acid on the degradation of levofloxacin by aqueous permanganate: Kinetics and mechanism. Water Res. 2017, 123, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Gao, Y.; Jiang, J.; Shen, Y.M.; Pang, S.Y.; Song, Y.; Guo, Q. A comparison study of levofloxacin degradation by peroxymonosulfate and permanganate: Kinetics, products and effect of quinone group. J. Hazard. Mater. 2021, 403, 123834. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Hou, Y.; Zhu, H.; Xiong, J.; Huang, W.; Yu, Z.; Wang, S. Levofloxacin degradation performance and mechanism in the novel electro-Fenton system constructed with vanadium oxide electrodes under neutral pH. Chem. Eng. J. 2022, 433, 133574. [Google Scholar] [CrossRef]
- Zhang, Y.; Hua, S.; Sun, X.; Liu, Z.; Dang, Y.; Zhang, L.; Zhou, Y. A novel electrochemical cathode based on sea urchin-like NiO/Co3O4 composite inducing efficient Fenton-like process for levofloxacin degradation. Appl. Catal. A 2021, 628, 118403. [Google Scholar] [CrossRef]
- Wang, A.; Chen, Z.; Zheng, Z.; Xu, H.; Wang, H.; Hu, K.; Yan, K. Remarkably enhanced sulfate radical-based photo-Fenton-like degradation of levofloxacin using the reduced mesoporous MnO@MnOx microspheres. Chem. Eng. J. 2020, 379, 122340. [Google Scholar] [CrossRef]
- Gong, Y.; Li, J.; Zhang, Y.; Zhang, M.; Tian, X.; Wang, A. Partial degradation of levofloxacin for biodegradability improvement by electro-Fenton process using an activated carbon fiber felt cathode. J. Hazard. Mater. 2016, 304, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Ge, M.; Guo, C. Efficient removal of levofloxacin from different water matrices via simultaneous adsorption and photocatalysis using a magnetic Ag3PO4/rGO/CoFe2O4 catalyst. Chemosphere 2021, 268, 128834. [Google Scholar] [CrossRef]
- Rong, F.; Xue, Y.; Tang, W.; Lu, Q.; Wei, M.; Guo, E.; Pang, Y. Visible-light-active 1D Ag-CoWO4/CdWO4 plasmonic photocatalysts boosting levofloxacin conversion. J. Taiwan Inst. Chem. Eng. 2022, 133, 104267. [Google Scholar] [CrossRef]
- Wei, H.; Hu, D.; Su, J.; Li, K. Intensification of levofloxacin sono-degradation in a US/H2O2 system with Fe3O4 magnetic nanoparticles. Chin. J. Chem. Eng. 2015, 23, 296–302. [Google Scholar] [CrossRef]
- Conde-Cid, M.; Fernandez-Calvino, D.; Novoa-Munoz, J.C.; Arias-Estevez, M.; Diaz-Ravina, M.; Fernandez-Sanjurjo, M.J.; Nunez-Delgado, A.; Alvarez-Rodriguez, E. Biotic and abiotic dissipation of tetracyclines using simulated sunlight and in the dark. Sci. Total Environ. 2018, 635, 1520–1529. [Google Scholar] [CrossRef]
- Norzaee, S.; Taghavi, M.; Djahed, B.; Kord Mostafapour, F. Degradation of Penicillin G by heat activated persulfate in aqueous solution. J. Environ. Manag. 2018, 215, 316–323. [Google Scholar] [CrossRef]
- Zhu, L.; Santiago-Schubel, B.; Xiao, H.; Hollert, H.; Kueppers, S. Electrochemical oxidation of fluoroquinolone antibiotics: Mechanism, residual antibacterial activity and toxicity change. Water Res. 2016, 102, 52–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Ran, J.; Mao, K.; Yang, X.; Zhong, L.; Yang, C.; Feng, X.; Zhang, H. Recent progress in Fenton/Fenton-like reactions for the removal of antibiotics in aqueous environments. Ecotoxicol. Environ. Saf. 2022, 236, 113464. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Kou, L.; Fan, Q.; Jiang, K.; Wang, J. Simultaneous recovery of phosphate and degradation of antibiotics by waste sludge-derived biochar. Chemosphere 2022, 291, 132832. [Google Scholar] [CrossRef] [PubMed]
- Al-Jubouri, S.M.; Al-Jendeel, H.A.; Rashid, S.A.; Al-Batty, S. Antibiotics adsorption from contaminated water by composites of ZSM-5 zeolite nanocrystals coated carbon. J. Water. Proc. Eng. 2022, 47, 102745. [Google Scholar] [CrossRef]
- Bekkali, C.E.; Bouyarmane, H.; Karbane, M.E.; Masse, S.; Saoiabi, A.; Coradin, T.; Laghzizil, A. Zinc oxide-hydroxyapatite nanocomposite photocatalysts for the degradation of ciprofloxacin and ofloxacin antibiotics. Colloids. Surf. A Physicochem. Eng. Aspects. 2018, 539, 364–370. [Google Scholar] [CrossRef] [Green Version]
- Felis, E.; Buta-Hubeny, M.; Zielinski, W.; Hubeny, J.; Harnisz, M.; Bajkacz, S.; Korzeniewska, E. Solar-light driven photodegradation of antimicrobials, their transformation by-products and antibiotic resistance determinants in treated wastewater. Sci. Total Environ. 2022, 836, 155447. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, Y.; Li, C. Photocatalytic degradation of ofloxacin on Gd2Ti2O7 supported on quartz spheres. J. Phys. Chem. Solids. 2018, 118, 144–149. [Google Scholar] [CrossRef]
- Pal, S.; Ahamed, Z.; Pal, P. Removal of antibiotics and pharmaceutically active compounds from water Environment: Experiments towards industrial scale up. Separat. Purif. Technol. 2022, 295, 121249. [Google Scholar] [CrossRef]
- Reis, A.C.; Kolvenbach, B.A.; Nunes, O.C.; Corvini, P.F.X. Biodegradation of antibiotics: The new resistance determinants—Part I. N Biotechnol. 2020, 54, 34–51. [Google Scholar] [CrossRef]
- Chaturvedi, P.; Giri, B.S.; Shukla, P.; Gupta, P. Recent advancement in remediation of synthetic organic antibiotics from environmental matrices: Challenges and perspective. Bioresour. Technol. 2021, 319, 124161. [Google Scholar] [CrossRef]
- Oberoi, A.S.; Jia, Y.; Zhang, H.; Khanal, S.K.; Lu, H. Insights into the Fate and Removal of Antibiotics in Engineered Biological Treatment Systems: A Critical Review. Environ. Sci. Technol. 2019, 53, 7234–7264. [Google Scholar] [CrossRef] [PubMed]
- Langbehn, R.K.; Michels, C.; Soares, H.M. Antibiotics in wastewater: From its occurrence to the biological removal by environmentally conscious technologies. Environ. Pollut. 2021, 275, 116603. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Zhang, W.; Cai, Y. Laccase immobilization for water purification: A comprehensive review. Chem. Eng. J. 2021, 403, 126272. [Google Scholar] [CrossRef]
- Bilal, M.; Ashraf, S.S.; Barcelo, D.; Iqbal, H.M.N. Biocatalytic degradation/redefining "removal" fate of pharmaceutically active compounds and antibiotics in the aquatic environment. Sci. Total Environ. 2019, 691, 1190–1211. [Google Scholar] [CrossRef]
- Park, H.; Choung, Y.-K. Degradation of Antibiotics (Tetracycline, Sulfathiazole, Ampicillin) Using Enzymes of Glutathion S-Transferase. Human. Ecol. Risk Assess. Int. J. 2007, 13, 1147–1155. [Google Scholar] [CrossRef]
- Wen, X.; Jia, Y.; Li, J. Enzymatic degradation of tetracycline and oxytetracycline by crude manganese peroxidase prepared from Phanerochaete chrysosporium. J. Hazard. Mater. 2010, 177, 924–928. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Zeng, Z.; Du, C.; Huang, D.; Zeng, G.; Xiao, R.; Lai, C.; Xu, P.; Zhang, C.; Wan, J.; et al. Immobilized laccase on bentonite-derived mesoporous materials for removal of tetracycline. Chemosphere 2019, 222, 865–871. [Google Scholar] [CrossRef]
- Tian, Q.; Dou, X.; Huang, L.; Wang, L.; Meng, D.; Zhai, L.; Shen, Y.; You, C.; Guan, Z.; Liao, X. Characterization of a robust cold-adapted and thermostable laccase from Pycnoporus sp. SYBC-L10 with a strong ability for the degradation of tetracycline and oxytetracycline by laccase-mediated oxidation. J. Hazard. Mater. 2020, 382, 121084. [Google Scholar] [CrossRef]
- Copete-Pertuz, L.S.; Plácido, J.; Serna-Galvis, E.A.; Torres-Palma, R.A.; Mora, A. Elimination of Isoxazolyl-Penicillins antibiotics in waters by the ligninolytic native Colombian strain Leptosphaerulina sp. considerations on biodegradation process and antimicrobial activity removal. Sci. Total Environ. 2018, 630, 1195–1204. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Leng, Y.; Wan, D.; Chang, F.; Huang, Y.; Li, Z.; Xiong, W.; Wang, J. Transformation of Tetracycline by Manganese Peroxidase from Phanerochaete chrysosporium. Molecules 2021, 26. [Google Scholar]
- Weng, S.S.; Ku, K.L.; Lai, H.T. The implication of mediators for enhancement of laccase oxidation of sulfonamide antibiotics. Bioresour. Technol. 2012, 113, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Hakulinen, N.; Rouvinen, J. Three-dimensional structures of laccases. CMLS 2015, 72, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, U.N.; Singh, P.; Pandey, V.P.; Kumar, A. Structure–function relationship among bacterial, fungal and plant laccases. J. Mol. Catal. B Enz. 2011, 68, 117–128. [Google Scholar] [CrossRef]
- Rivera-Hoyos, C.M.; Morales-Álvarez, E.D.; Poutou-Piñales, R.A.; Pedroza-Rodríguez, A.M.; Rodríguez-Vázquez, R.; Delgado-Boada, J.M. Fungal laccases. Fung. Biol. Rev. 2013, 27, 67–82. [Google Scholar] [CrossRef]
- Shraddha; Shekher, R.; Sehgal, S.; Kamthania, M.; Kumar, A. Laccase:microbial sources, production, purification, and potential biotechnological applications. Enz. Res. 2011, 2011, 217861. [Google Scholar]
- Christopher, L.P.; Yao, B.; Ji, Y. Lignin biodegradation with laccase-mediator systems. Front. Ener. Res. 2014, 2, 12. [Google Scholar] [CrossRef]
- Falade, A.O.; Nwodo, U.U.; Iweriebor, B.C.; Green, E.; Mabinya, L.V.; Okoh, A.I. Lignin peroxidase functionalities and prospective applications. Microbiol. Open 2017, 6, e00394. [Google Scholar] [CrossRef] [Green Version]
- Wesenberg, D.; Kyriakides, I.; Agathos, S.N. White-rot fungi and their enzymes for the treatment of industrial dye effluents. Biotechnol. Adv. 2003, 22, 161–187. [Google Scholar] [CrossRef]
- Kong, W.; Fu, X.; Wang, L.; Alhujaily, A.; Zhang, J.; Ma, F.; Zhang, X.; Yu, H. A novel and efficient fungal delignification strategy based on versatile peroxidase for lignocellulose bioconversion. Biotechnol. Biofuels. 2017, 10, 218. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez Couto, S.; Toca Herrera, J.L. Industrial and biotechnological applications of laccases: A review. Biotechnol. Adv. 2006, 24, 500–513. [Google Scholar] [CrossRef]
- Forgacs, E.; Cserháti, T.; Oros, G. Removal of synthetic dyes from wastewaters: A review. Environ. Int. 2004, 30, 953–971. [Google Scholar] [CrossRef] [PubMed]
- Majeau, J.-A.; Brar, S.K.; Tyagi, R.D. Laccases for removal of recalcitrant and emerging pollutants. Biores. Technol. 2010, 101, 2331–2350. [Google Scholar] [CrossRef] [PubMed]
- Ardila-Leal, L.D.; Poutou-Piñales, R.A.; Pedroza-Rodríguez, A.M.; Quevedo-Hidalgo, B.E. A brief history of colour, the environmental impact of synthetic dyes and removal by using laccases. Molecules 2021, 26, 3813. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, W.; Ng, T.B.; Deng, X.; Lin, J.; Ye, X. Laccases: Production, expression regulation, and applications in pharmaceutical biodegradation. Front. Microbiol. 2017, 8, 832. [Google Scholar] [CrossRef]
- Navada, K.K.; Kulal, A. Enzymatic degradation of chloramphenicol by laccase from Trametes hirsuta and comparison among mediators. Int. Biodet. Biodegrad. 2019, 138, 63–69. [Google Scholar] [CrossRef]
- Najafabadipour, N.; Mojtabavi, S.; Jafari-Nodoushan, H.; Samadi, N.; Faramarzi, M.A. High efficiency of osmotically stable laccase for biotransformation and micro-detoxification of levofloxacin in the urea-containing solution: Catalytic performance and mechanism. Colloids. Surf. B Biointerf. 2021, 207, 112022. [Google Scholar] [CrossRef]
- Kelbert, M.; Pereira, C.S.; Daronch, N.A.; Cesca, K.; Michels, C.; de Oliveira, D.; Soares, H.M. Laccase as an efficacious approach to remove anticancer drugs: A study of doxorubicin degradation, kinetic parameters, and toxicity assessment. J. Hazard. Mater. 2021, 409, 124520. [Google Scholar] [CrossRef]
- Rivera-Hoyos, C.M.; Morales-Álvarez, E.D.; Poveda-Cuevas, S.A.; Reyes-Guzmán, E.A.; Poutou-Piñales, R.A.; Reyes-Montaño, E.A.; Pedroza-Rodríguez, A.M.; Rodríguez-Vázquez, R.; Cardozo-Bernal, Á.M. Computational analysis and low-scale constitutive expression of laccases synthetic genes GlLCC1 from Ganoderma lucidum and POXA 1B from Pleurotus ostreatus in Pichia pastoris. PLoS ONE 2015, 10, e0116524. [Google Scholar] [CrossRef]
- Sáenz-Suárez, H.; Chávez-Zobbel, A.; Lareo, L.R.; Oribio-Quinto, C.; Martínez-Mendoza, J. Predicción computacional de estructura terciaria de las proteínas humanas Hsp27, aB-cristalina y HspB8. Univ. Sci. 2011, 16, 15–28. [Google Scholar] [CrossRef]
- Zárate-Bonilla, L.J.; del Portillo, P.; Sáenz-Suárez, H.; Janneth, G.-S.; Barreto-Sampaio, G.E.; Poutou-Piñales, R.A.; Felipe Rey, A.; Rey, J.G. Computational modeling and preliminary iroN, fepA, cirA gene expression in Salmonella Enteritidis under iron deficiency induced conditions. Poult. Sci. 2014, 93, 221–230. [Google Scholar] [CrossRef]
- Sáenz-Suárez, H.; Rivera-Hoyos, C.; Morales-Álvarez, E.; Poutou-Piñales, R.; Sáenz-Moreno, J.; Pedroza-Rodríguez, A. Modelación computacional preliminar de la estructura 3D de dos lacasas fúngicas. Salud. Arte. Cuidado. 2014, 7, 5–16. [Google Scholar]
- Sáenz-Suárez, H.; Poutou-Piñales, R.A.; González-Santos, J.; Barreto, G.E.; Prieto-Navarrera, L.P.; Sáenz-Moreno, J.A.; Landázuri, P.; Barrera-Avellaneda, L.A. Prediction of glycation sites: New insights from protein structural analysis. Turk. J. Biol. 2016, 40, 12–25. [Google Scholar] [CrossRef]
- Niño-Gómez, D.C.; Rivera-Hoyos, C.M.; Morales-Álvarez, E.D.; Reyes-Montaño, E.A.; Vargas-Alejo, N.E.; Ramírez-Casallas, I.N.; Erkan Turkmen, K.; Sáenz-Suárez, H.; Sáenz-Moreno, J.A.; Poutou-Piñales, R.A.; et al. “In silico” characterization of 3-phytase A and 3-phytase B from Aspergillus niger. Enz. Res. 2017, 2017, 9746191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sáenz, H.; Lareo, L.; Poutou, R.A.; Sosa, C.; Barrera, L.A. Predicción computacional de la estructura terciaria de la iduronato 2-sulfato sulfatasa humana. Biomédica 2007, 27, 7–20. [Google Scholar] [CrossRef] [Green Version]
- Ardila-Leal, L.D.; Monterey-Gutiérrez, P.A.; Poutou-Piñales, R.A.; Quevedo-Hidalgo, B.E.; Galindo, J.F.; Pedroza-Rodríguez, A.M. Recombinant laccase rPOXA 1B real-time and accelerated stability studies supported by molecular dynamics. BMC Biotechnol. 2021, 21, 37. [Google Scholar] [CrossRef]
- Pagadala, N.S.; Syed, K.; Tuszynski, J. Software for molecular docking: A review. Biophys. Rev. 2017, 9, 91–102. [Google Scholar] [CrossRef]
- Ardila-Leal, L.D. Producción a escala piloto (10L) y caracterización de un concentrado enzimático de rPOXA 1B para la remoción de colorantes. In Microbiology; Pontificia Universidad Javeriana: Bogotá D.C., Colombia, 2021; p. 361. [Google Scholar]
- Sarkar, S.; Banerjee, A.; Chakraborty, N.; Soren, K.; Chakraborty, P.; Bandopadhyay, R. Structural-functional analyses of textile dye degrading azoreductase, laccase and peroxidase: A comparative in silico study. Elect. J. Biotechnol. 2020, 43, 48–54. [Google Scholar] [CrossRef]
- Singh, A.; Kumari, S.; Pal, T.K. In silico analysis for laccase-mediated bioremediation of the emerging pharmaceutical pollutants. Int. J. Bioautom. 2015, 19, 423–432. [Google Scholar]
- Yue, S.-Y. Distance-constrained molecular docking by simulated annealing. Prot. Eng. 1990, 4, 177–184. [Google Scholar] [CrossRef]
- Reva, B.A.; Finkelstein, A.V.; Skolnick, J. What is the probability of a chance prediction of a protein structure with an rmsd of 6 å? Fold Des. 1998, 3, 141–147. [Google Scholar] [CrossRef] [Green Version]
- Cárdenas-Moreno, Y.; Espinosa, L.A.; Vieyto, J.C.; González-Durruthy, M.; del Monte-Martinez, A.; Guerra-Rivera, G.; Sánchez López, M.I. Theoretical study on binding interactions of laccase-enzyme from Ganoderma weberianum with multiples ligand substrates with environmental impact. Ann. Prot. Bioinf. 2019, 3, 001–009. [Google Scholar]
- Sutar, R.S.; Rathod, V.K. Ultrasound assisted Laccase catalyzed degradation of Ciprofloxacin hydrochloride. J. Ind. Eng. Chem. 2015, 31, 276–282. [Google Scholar] [CrossRef]


| Structure | Target | Antibiotic | Abbreviations in This Paper | Use | |
|---|---|---|---|---|---|
| CLSI | H | A | |||
| Critical Importance Antibiotics | |||||
| Aminoglycosides | |||||
| Aminoglycosides consist of several cyclitol rings in their structure; usually, three and five sugars are linked by glycosidic bonds. Amino and hydroxyl groups are attached to the rings providing the chemical properties of the compound [23]. | They alter cell membrane permeability and also inhibit protein synthesis by binding to the 30s ribosomal subunit [23]. | Aminoglycosides + 2 Deoxystreptamine | |||
| Amikacin | AN | X | |||
| Paromomycin | PLZ | X | |||
| Streptomycin | STR | X | X | ||
| Gentamicin | GM | X | X | ||
| Kanamycin | K | X | X | ||
| Netilmicine | NET | X | |||
| Tobramycin | TM | X | X | ||
| Apramycin | APR | X | |||
| Dihydrostreptomycin | DST | X | |||
| Plazomycin | PLZ | X | |||
| Neomycin | NEO | X | X | ||
| Isepamycin | ISE | X | |||
| Arbekacin | ABK | X | |||
| Fortimicin A | FM-A | X | |||
| Bekanamycin | AKM | X | |||
| Dibekacin | DKB | X | |||
| Ribostamycin | RIB | X | |||
| Ansamycins | |||||
| Contain an aliphatic chain connecting the two ends of a naphthoquinone core [24] | Binds to the β subunit of RNA polymerase, inhibiting its activity [24]. | Rifampicins | |||
| Rifaximin | RFP | X | |||
| Rifampicin | RIF | X | |||
| Rifapentine | RPT | X | |||
| Rifabutin | RFB | X | |||
| β-lactams | |||||
| The β-lactam ring chemically defines this class of antibiotics. This ring is bound to other radicals. The association of different types of linear chains modifies the properties of the compound and the different groups of β-lactam antibiotics are formed [17,25]. Their structure consists of a five-membered unsaturated ring fused to an β-lactam ring. Carbapenems differ from penicillins by the C2-C3 double bond and the carbon in place of the sulphur at C1. Additionally, carbapenems have a trans-1-hydroxyethyl substituent in place of the acylamino substituent on the β-lactam ring [26]. | They act by two mechanisms: inhibition of wall synthesis and induction of bacterial autolysis. Transpeptidase enzymes (E.C. 3.4.16.4) are involved in the last stage of wall synthesis, linking the bonds of the peptidoglycan chains. The β-lactam ring is structurally similar to the region of the pentapeptide to which transpeptidases bind, thus the ring binds to the enzymes inhibiting cell wall formation. In addition, β-lactam activate an endogenous autolysin that degrades the peptidoglycan [17,25]. | Carbapenems | |||
| Doripenem | DOR | X | |||
| Ertapenem | ETP | X | |||
| Meropenem | MEM | X | |||
| Imipenem | IPM | X | |||
| Panipenem | PAPM | X | |||
| Faropenem | FRPM | X | |||
| Biapenem | BPM | X | |||
| The chemical structure of cephalosporins comes from 7-cephalosporanic acid. Cephalosporins structure are a fusion of a two-ring system of -lactam-3-dihydrothiazine, known as 7-aminocephalosporanic acid (7-ACA), and vary in their side-chain substitutions at C3 (R2) and C7 [27]. | 1, 2, 3,4 and 5 Generation Cephalosporins | ||||
| Cefditoren | CDN | X | |||
| Cefmenoxime | CMX | X | |||
| Cephpyrome | CPO | X | |||
| Ceftriaxone | CRO | X | |||
| Cefoperazone | CFP | X | |||
| Cefquinome | CEQ | X | |||
| Cefotaxime | CTX | X | |||
| Ceftazidime | CAZ | X | |||
| Cefetameta | CAT | X | |||
| Cefpodoxime | CPD | X | |||
| Ceftibuten | CBT | X | |||
| Cefdinir | CDR | X | |||
| Cefepime | FEP | X | |||
| Cefixime | CFM | X | |||
| Ceftaroline fosamil | CPT | X | |||
| Ceftiofur | CFT | X | |||
| Cefovecin | CFO | X | |||
| Cefaclor | CEC | X | |||
| Cefadroxil | CFR | X | X | ||
| Cephalexin | CN | X | X | ||
| Cephalonium | CFL | X | |||
| Cephapirin | CAP | X | X | ||
| Cephalotin | CF | X | X | ||
| Cefazolin | CZ | X | X | ||
| Cefoxitin | FOX | X | |||
| Cefprozil | CPR | X | |||
| Cefuroxime | CXM | X | X | ||
| Loracarbef | LOR | X | |||
| Cefotetan | CTT | X | |||
| Cephradine | BAN | X | |||
| Tazobactam | TZB | X | |||
| Cefcapene | CFPM | X | |||
| Cefodizime | CDZM | X | |||
| Cefoselis | CFSL | X | |||
| Cefozopran | ZOP | X | |||
| Cefsulodin | CFS | X | |||
| Ceftizoxime | ZOX | X | |||
| Ceftobiprole | BPR | X | |||
| Ceftozolane | CTZ | X | |||
| Latamoxef | LMOX | X | |||
| Cephacetrile | CEC | X | |||
| Cephaloridine | CPH | X | |||
| Cefamandol | CFM | X | |||
| Cefatrizine | CTZ | X | |||
| Cefazedone | CFZD | X | |||
| Cefbuperazone | CFB | X | |||
| Cefmetazol | CMZ | X | |||
| Cefminox | CMNX | X | |||
| Cefonicid | CID | X | |||
| Ceforanide | CFR | X | |||
| Cefotiam | CTM | X | |||
| Cefroxadine | CXD | X | |||
| Ceftezole | CTZ | X | |||
| Flomoxef | FMOX | X | |||
| Monobactams have a sulphonic acid group on the nitrogen at the N-l position; the sulphonic acid activates the l3-lactam ring and thus acetylates the transpeptidase enzymes [28]. | Penicillin monobactam | ||||
| Aztreonam | ATM | X | |||
| Carumonam | CAR | X | |||
| The basic structure of penicillin (6-aminopenicillanic acid) consists of a thiazolidine ring, an attached p-lactam ring and a side-chain [17,25]. | Antipseudomonal penicillin | ||||
| Carbenicillin | CB | X | |||
| Ticarcillin | TIC | X | X | ||
| Piperacillin | PIP | X | |||
| Sulbenicillin | SBPC | X | |||
| Azlocillin | AZ | X | |||
| Carindacillin | CIPC | X | |||
| Mezlocilin | MEZ | X | |||
| Sultamicillin | SBTPC | X | |||
| Hetacillin | HET | X | |||
| Amdinocillin | MEC | X | |||
| Ampicillin | AM | X | |||
| Talampicillin | AMX | X | X | ||
| Azidocillin | AZD | X | |||
| Bacampicillin | B | X | |||
| Epicillin | EP | X | |||
| Temocillin | TEM | X | |||
| Methampicillin | MTP | X | |||
| Pivampicillin | PMPC | XX | |||
| Amoxicillin (Clavulanic acid) | AMC | X | X | ||
| Glycopeptides and Lipoglycopeptides | |||||
| Consist of a central heptapeptide core to which amino acid residues and sugars are attached [29]. | In Gram-positive bacteria, Glycopeptides and lipoglycopeptides bind to the D-alanyl-D-alanine terminus at the carboxy-terminal end of bacterial wall precursor peptides, thus blocking peptidoglycan synthesis [24,29]. | Telavancine | TLV | X | |
| Teicoplanin | TEC | X | |||
| Vancomycin | VA | X | |||
| Avoparcin | AV | X | |||
| Dalbavancine | DAL | X | |||
| Oritavancine | ORI | X | |||
| Ramoplanin | RAM | X | |||
| Glycyliclinas | |||||
| Structurally similar to the tetracyclines, it has a central structure of four carbocyclic rings, with a t-butylglycylamide substitution at position 9 of the minocycline that confers a broad spectrum of activity [24]. | Inhibits protein synthesis by reversibly binding to the 30S subunit of the bacterial ribosome, blocking the entry of the aminoacyl tRNA into the A site of the ribosome, thus preventing amino acid incorporation and subsequent elongation of peptide chains [24]. | Tigecycline | TGC | X | |
| Lipopeptides | |||||
| A cyclic molecule with 13 amino acids, ten of which are part of the cyclic structure, and the remaining three make up a side chain with an N-decanoyl residue [24]. | They insert into the membrane bilayer causing its depolarisation, with a strong loss of potassium ion leading to cell death; a side-chain bearing an N-decanoyl residue [24]. | Daptomycin | DAP | X | |
| Colistin | CL | X | |||
| Polymyxin B | PB | X | X | ||
| Macrolides | |||||
| Are a lactonic ring with 14 to 16 carbons, bound to an aminated sugar [24]. | Binds to sequences of the 23S rRNA domain V, which is part of the 50S subunit, preventing elongation of the peptide chain by blocking the polypeptide exit tunnel and thus dissociating the peptidyl-RNA complex from the ribosome [24]. | Azithromycin | AZM | X | X |
| Gamithromycin | GM | X | X | ||
| Josamycin | JM | X | X | ||
| Tulathromycin | TUL | X | |||
| Tylvalosin | TVN | X | |||
| Tylosin | TLY | X | |||
| Tilmicosin | TMS | X | |||
| Midecamycin | MDM | X | |||
| Dirithromycin | DTM | X | |||
| Rokitamycin | RKM | X | |||
| Roxithromycin | RXT | X | |||
| Clarithromycin | CLR | X | |||
| Spiramycin | SP | X | |||
| Fidaxomicin | FDX | X | |||
| Erythromycin | E | X | |||
| Telithromycin | TEL | X | X | ||
| Fluoroerythromycin | X | ||||
| Kitasamycin | KIA | X | |||
| Oleandomycin | OL | X | |||
| Tildispyrosine | TD | X | |||
| Troleandomycin | TAO | X | |||
| Quinupristin-dalfopristin | SYN | X | |||
| Pristinamycin | PT | X | |||
| Virginiamycin | VM | X | |||
| Solithromycin | SOL | X | |||
| Cethromycin | CET | X | |||
| Oxazolidinones | |||||
| Oxazolidinone consists of a ring with three carbon atoms, an oxygen atom at position one and a nitrogen atom at position three [30]. | The antibiotic binds to the 50S subunit, affecting protein synthesis, and also inhibits the initiation complex by binding to the 70S subunit. The D-ring of the tedizolid contributes to the presence of additional hydrogen bonds that provide further interactions between the tedizolid and the bacterial ribosome; therefore, the drug is more potent [30]. | Radezolid | RAD | X | |
| Linezolid | LZD | X | |||
| Cadazolid | CDZ | X | |||
| Tedizolid | TZD | X | |||
| Phosphomycins | |||||
| It is cis-1,2-epoxypropylphosphonic acid (-), a simple, water-soluble molecule with only three carbon atoms and no nitrogen. The carbon atom is bonded to the phosphorus atom without an intermediate oxygen bridge. The antimicrobial activity is due to the epoxy bond [24]. | Fosfomycin enters the membrane via two permeases (E.C. 3.1.3.9); the inducible D-glucose-6-phosphate transport system and the L-α-glycerophosphate system. The antibiotic competes with the substrate of the enzyme UDP-N-acetylglucosamine-3-O-enolpyruvyltransferase (MurA) (E.C. 2.5.1.7), an enzyme that catalyses the first stage of peptidoglycan heteropolymer biosynthesis [24]. | Fosfomycin | FOS | X | X |
| Quinolones and Fluoroquinolones | |||||
| They interfere with DNA synthesis inducing cell death and penetrating the wall through porins to interact with two enzymes, the DNA gyrase (E.C. 5.6.2.2) and topoisomerase IV (E.C. 5.99.1.2), both responsible for DNA supercoiling [24]. | Quinolones and fluoroquinolones interfere with DNA synthesis, inducing cell death. They penetrate the wall through porins and interact with two enzymes; DNA gyrase (E.C. 5.6.2.2) and topoisomerase IV, (E.C. 5.99.1.2) responsible for DNA supercoiling. [24]. | Pefloxacin | PEF | X | |
| Besifloxacin | BES | X | |||
| Delafloxacin | DLX | X | |||
| Gemifloxacin | GEM | X | |||
| Nadifloxacin | X | ||||
| Nalidixic acid | NAL | X | |||
| Ozenoxacin | OZN | X | |||
| Oxolinic acid | OA | X | |||
| Enrofloxacin | ENR | X | |||
| Difloxacin | DIF | X | |||
| Pradofloxacin | PARA | X | |||
| Moxifloxacin | MXF | X | X | ||
| Levofloxacin | LVX | X | X | ||
| Ibafloxacin | X | ||||
| Flumequine | FLU | X | |||
| Marbofloxacin | MAR | X | |||
| Orbifloxacin | OBFX | X | |||
| Rufloxacin | RFX | X | X | ||
| Ciprofloxacin | CIP | X | X | ||
| Norfloxacin | NX | X | |||
| Gatifloxacin | GAT | X | |||
| Ofloxacin | OFL | X | |||
| Lomefloxacin | LOM | X | |||
| Enoxacin | GRN | X | |||
| Grepafloxacin | GRX | X | |||
| Pazufloxacin | PZFX | X | |||
| Pipemidic acid | HPPA | X | |||
| Prulifloxacin | PUFX | X | |||
| Rosoxacin | RSX | X | |||
| Sitafloxacin | STFX | X | |||
| Sparfloxacin | SPX | X | |||
| Termafloxacin | TEM | ||||
| Phenicols | |||||
| Phenicols feature a p-nitrophenyl group and an N-dichloroacetyl substituent attached to a three-carbon chain [24]. | Prevents peptide bond formation by binding to the L16 protein located in the 50S subunit of the ribosome, which mediates tRNA binding to peptidyl transferase (E.C. 2.3.2.12) [24]. | Chloramphenicol | CHL | X | X |
| Thiamphenicol | X | X | |||
| Florfenicol | FLO | X | |||
| Lincosamides | |||||
| Lincomycin consists of an amino acid attached to an amino sugar; clindamycin differs structurally due to the substitution of a chlorine atom by a hydroxyl group and the inversion of carbon [24]. | It binds to sequences of rRNA 23S domain V, which is part of the 50S subunit of the ribosome, preventing the elongation of the peptide chain by blocking the polypeptide exit tunnel, and thus the peptidyl-RNA complex dissociates from the ribosome [24]. | Clindamycin | CM | X | X |
| Lincomycin | LIN | X | X | ||
| Pirlimycin | PIR | X | |||
| Pseudomonic Acids | |||||
| It consists of a 9-hydroxy-nonanoic acid chain, some spatial similarity in structure to the amino acid isoleucine [31]. | DNA gyrase and topoisomerase IV inhibitor [32]. Binds to the enzyme isoleucyl-tRNA (E.C. 6.1.1.5), preventing the incorporation of isoleucine into proteins [31]. | Mupirocin | MUP | X | |
| Rhinophenazines | |||||
| It contains a phenazine core with an alkyl imino (R-imino) group at position two and phenyl substituents at positions 3 and 10 of the phenazine core. The alkyl imino group is essential for antimicrobial activity [33]. | Its target is the bacterial respiratory chain and ion transporters. Intracellular redox cycling, involving oxidation of reduced clofazimine, generates antimicrobial reactive oxygen species (ROS) and hydrogen peroxide (H2O2). Additionally, clofazimine interaction with membrane phospholipids promotes membrane dysfunction and interferes with K+ uptake. Both mechanisms result in interference with cellular energy metabolism by disrupting ATP production [33]. | Clofazimine | CFZ | X | |
| Steroidals | |||||
| They have a steroid structure (cyclopentanoperhydrophenanthrene ring), and are a tetracyclic triterpenoid [34]. | Inhibits protein synthesis by preventing translocation of elongation factor (EF-G) during protein synthesis [34]. | Fusidic acid | FUS | X | |
| Sulphonamides | |||||
| They have a similar structure to para-aminobenzoic acid. Contain an aromatic NH2 substituent at the para position of the benzene ring. Contain a substituent at the ortho- and meta-position of the benzene ring. Additionally, it contains a double substituent at position N1 and a sulphonamide group on the benzene ring [24]. | They are competitive p-aminobenzoic acid (PABA) antagonists, binding to the enzyme tetrahydropteroic acid synthetase (E.C 6.3.2.17) inhibiting folic acid synthesis [24]. | Sulfamethoxazole | SMX | X | X |
| Sulphadimethoxine | SDM | X | |||
| Sulphadiazine | SDZ | X | X | ||
| Sulfamerazine | SMZ | X | |||
| Sulphanilamide | SA | X | |||
| Sulfathiazole | STZ | X | X | ||
| Sulfadimidine = Sulfamethazine | SM2 | X | |||
| Sulfamethizole | SMZ | X | X | ||
| Sulfisoxazole | FIS | X | |||
| Trimethoprim | SXT | X | X | ||
| Brodimoprim | BDM | X | |||
| Formosulfathiazole | X | ||||
| Iclaprim | ICL | X | |||
| Phthalylsulfathiazole | PA | X | |||
| Sulphaisodimidine | SU | X | |||
| Sulphalene | X | ||||
| Sulfamazone | SZO | X | |||
| Sulfamethoxypyridazine | SP | X | |||
| Sulphamethomidine | SM | X | |||
| Sulfamethoxydiazine | SMD | X | |||
| Sulphametrol | SMT | X | |||
| Sulfamoxol | SMO | X | |||
| Sulfaperin | SFL | X | |||
| Sulfafenazol | SPZ | X | |||
| Sulphapyridine | SP | X | |||
| Sulfatiourea | SFTu | X | |||
| Tetroxoprim | TXP | X | |||
| Tetracyclines | |||||
| Composed of a linear fused tetracyclic fused core to which several functional groups are attached (Chlorine (Cl), Hydrogen (H) dimethylamine (N(CH3)2, methyl (CH2), methylenes (CH3) hydroxyl (OH) [35]. | It binds to the 30S subunit of the bacterial ribosome and prevents the binding of the aminoacyl tRNA, disrupting the incorporation of amino acids into the growing peptide [35]. | Chlortetracycline | CTC | X | X |
| Demeclocycline | DMC | X | |||
| Omadacycline | OMC | X | |||
| Lymecycline | LYME | X | |||
| Eravacycline | ERV | X | |||
| Doxycycline | DO | X | X | ||
| Oxytetracycline | OXT | X | X | ||
| Rolitetracycline | RTC | X | |||
| Tetracycline | TE | X | X | ||
| Minocycline | MI | X | |||
| Clomocycline | CLM | X | |||
| Penimepicycline | PNM | X | |||
| Methacycline | ME | X | |||
| Types of Antibiotic Degradation | Techniques | Process or Materials | Antibiotics Reported | Degradation/Removal | References |
|---|---|---|---|---|---|
| Biotic | Hydrolysis | Lake sediment. | Cephradine (BAN), Cefuroxime (CXM), Ceftriaxone (CRO), Cefepime (FEP). | Reporting removal of Ceftriaxone disodium (CRO) of 3% and Cefotiam dihydrochloride (CTM) of 7% in 28 d. | [155] |
| Microbial degradation | Bacterial suspensions of poultry manure and soil that produce humic acids. | Tetracycline (TE), Oxytetracycline (OXT), Chlortetracycline (CTC). | Removal between 88 and 75% in 15 min. | [218] | |
| Degradation attributed to a co-metabolic process | The strain was isolated from a reactor used for the treatment of aquaculture effluent. | Oxytetracycline (OXT), Ciprofloxacin (CIP). | Oxytetracycline (OXT) between 90.3 and 97.4 and Ciprofloxacin (CIP) were unable to degrade. | [159] | |
| Chemical | Heat activated of Persulfate | Aqueous solution at different pH. | Penicillin G (PG). | At pH 5 was removal 82.6 and at higher pH, the removal decreased. | [219] |
| Electrochemical oxidation | EC reactor with a built-in platinum counter electrode and Roxy potentiostat. | Ciprofloxacin (CIP), Norfloxacin (NX), Ofloxacin (OFL). | Reporting removal of Ciprofloxacin (CIP) of 90%, Norfloxacin (NX) of 62% and Ofloxacin 97.3%. | [220] | |
| Fenton process | The reaction was improving with H2O2, promoting catalytic production. | Thiazole sulphate, Tylosin (TLY), Ciprofloxacin (CIP), Amoxicillin (AMC) Cloxacillin (CLO), Tetracycline (TE). | Reporting removal between 77 and 97.1%. | [221] | |
| Physical | Adsorption | Biochar of waste sludge. | Tetracycline (TE), Sulfamethazine (SM2) | Weak adsorption using biochar could be overcome with the use of peroxymonosulfate. | [222] |
| ZSM-5 zeolite and zeolites nanocrystals. | Ciprofloxacin (CIP). | Removal between 54 and 90% according to material, time and antibiotic concentration. | [223] | ||
| Temperature | Increased temperature with incubation | Sulphonamide (SUL). | Minimum temperature of 60% removing between 78.1 and 98.3 on different sulphonamide antibiotics. | [195] | |
| Photodegradation | It was evaluated under a 125 W UV A-B-C (200–600 nm) irradiation. | Ciprofloxacin (CIP), Ofloxacin (OFL). | There is no high evidence of degradation of antibiotics by exposure to UV light. | [224] | |
| Physico-chemical | Plasma treatment | Plasma reactor in coaxial configuration operated in pulsed mode. | Oxacillin (OX), Amoxicillin (AMC), Ampicillin (AM). | Oxacillin and Amoxicillin (AMC) had >90% conversion and Ampicillin (AM) was 29%. | [191] |
| Photocatalysis | Solarbox 1500 photoreactor produced by an air-cooled xenon lamp. | Ampicillin (AM), Enrofloxacin (ENR), Tylosin (TLY), Vancomycin (VA), Clindamycin (CM), Trimethoprim (SXT), Zetronidazole, Sulfadiazine (SD), Doxycycline (DO), Oxytetracycline (OXT). | The removal of antibiotics by photocatalytic oxidation was between 40 and 100% with the exception of Clindamycin (CM) which was not removed. | [225] | |
| Quartz reactor with a 20 W lamp and irradiation of 2300 μW/cm2. | Ciprofloxacin (CIP), Ofloxacin (OFL). | Removal of ~70%. | [226] | ||
| Advanced oxidation processes (AOP) | Oxidation with agents such as hydrogen peroxide, ozone, titanium dioxide and UV light using semiconducting materials, such as TiO2, SnO2, CeO2, ZnO as catalysts. | Penicillins, Sulphonamides, Phenicols, β-lactams, Tetracyclines, Fluoroquinolones | Reporting removal with different efficiency ranges between 78 and 100%. | [227] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mora-Gamboa, M.P.C.; Rincón-Gamboa, S.M.; Ardila-Leal, L.D.; Poutou-Piñales, R.A.; Pedroza-Rodríguez, A.M.; Quevedo-Hidalgo, B.E. Impact of Antibiotics as Waste, Physical, Chemical, and Enzymatical Degradation: Use of Laccases. Molecules 2022, 27, 4436. https://doi.org/10.3390/molecules27144436
Mora-Gamboa MPC, Rincón-Gamboa SM, Ardila-Leal LD, Poutou-Piñales RA, Pedroza-Rodríguez AM, Quevedo-Hidalgo BE. Impact of Antibiotics as Waste, Physical, Chemical, and Enzymatical Degradation: Use of Laccases. Molecules. 2022; 27(14):4436. https://doi.org/10.3390/molecules27144436
Chicago/Turabian StyleMora-Gamboa, María P. C., Sandra M. Rincón-Gamboa, Leidy D. Ardila-Leal, Raúl A. Poutou-Piñales, Aura M. Pedroza-Rodríguez, and Balkys E. Quevedo-Hidalgo. 2022. "Impact of Antibiotics as Waste, Physical, Chemical, and Enzymatical Degradation: Use of Laccases" Molecules 27, no. 14: 4436. https://doi.org/10.3390/molecules27144436
APA StyleMora-Gamboa, M. P. C., Rincón-Gamboa, S. M., Ardila-Leal, L. D., Poutou-Piñales, R. A., Pedroza-Rodríguez, A. M., & Quevedo-Hidalgo, B. E. (2022). Impact of Antibiotics as Waste, Physical, Chemical, and Enzymatical Degradation: Use of Laccases. Molecules, 27(14), 4436. https://doi.org/10.3390/molecules27144436







