Metabolic Profiling and Transcriptional Analysis of Carotenoid Accumulation in a Red-Fleshed Mutant of Pummelo (Citrus grandis)
Abstract
:1. Introduction
2. Results
2.1. Carotenoid Composition and Content
2.2. Quantification of Flavones and Coumarins
2.3. Identification of Volatile Compounds
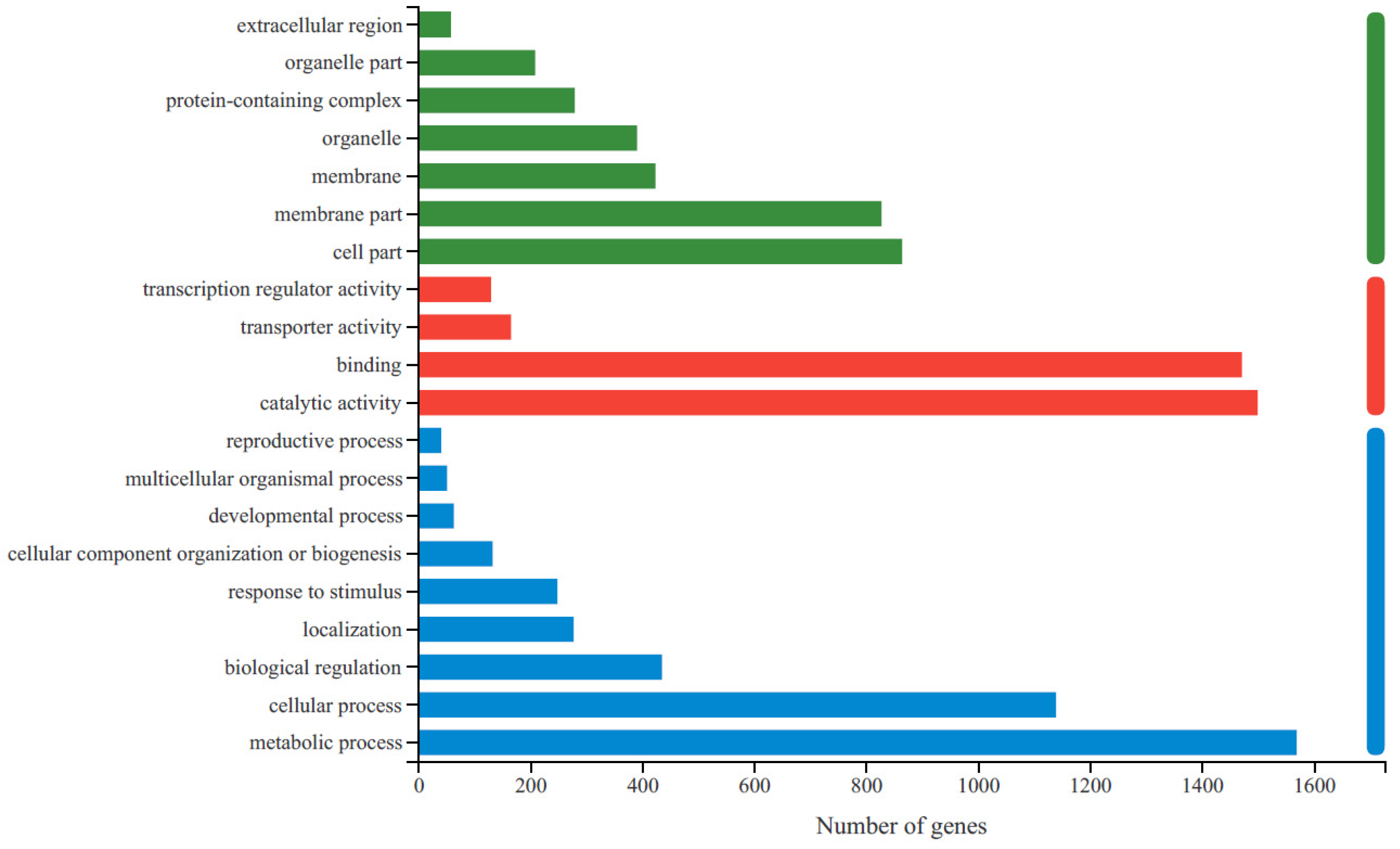
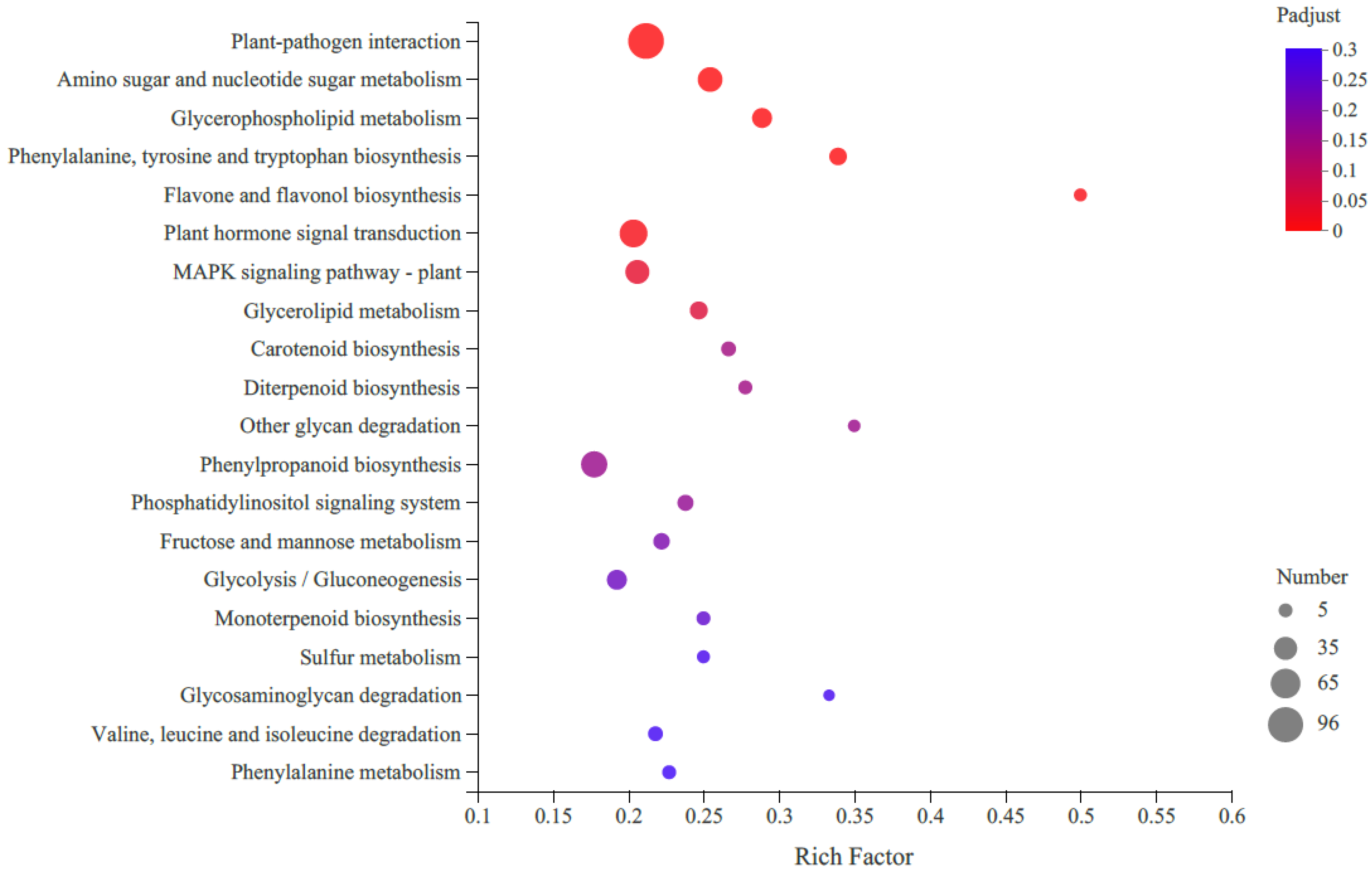
2.4. RNA-Seq Analysis
2.5. DEGs Related to Carotenoid Accumulation
3. Discussion
4. Materials and methods
4.1. Plant Materials
4.2. Carotenoid Extraction, Identification and Quantification
4.3. Analysis of Phytochemical Compounds by UPLC-DAD
4.4. Volatile Organic Compounds Identified by HS-SPME and GC/MS
4.5. RNA Extraction, Library Construction, and RNA-Seq
4.6. Transcriptome Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Purkiewicz, A.; Pietrzak-Fiećko, R. Antioxidant properties of fruit and vegetable whey beverages and fruit and vegetable mousses. Molecules 2021, 26, 3126. [Google Scholar] [CrossRef] [PubMed]
- Chayut, N.; Yuan, H.; Saar, Y.; Zheng, Y.; Sun, T.; Zhou, X.; Hermanns, A.; Oren, E.; Faigenboim, A.; Hui, M.; et al. Comparative transcriptome analyses shed light on carotenoid production and plastid development in melon fruit. Hortic. Res. 2021, 8, 112. [Google Scholar] [CrossRef] [PubMed]
- Niyogi, K.K.; Shih, C.; Soon, C.W.; Pogson, B.J.; Dellapenna, D.; Björkman, O. Photoprotection in a zeaxanthin- and lutein-deficient double mutant of Arabidopsis. Photosynth. Res. 2001, 67, 139–145. [Google Scholar] [CrossRef]
- Cazzonelli, C.; Pogson, B. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef]
- Cezare-Gomes, E.A.; Mejia-da-Silva, L.d.C.; Pérez-Mora, L.S.; Matsudo, M.C.; Ferreira-Camargo, L.S.; Singh, A.K.; de Carvalho, J.C.M. Potential of microalgae carotenoids for industrial application. Appl. Biochem. Biotechnol. 2019, 188, 602–634. [Google Scholar] [CrossRef]
- Xu, C.J.; Fraser, P.D.; Wang, W.J.; Bramley, P.M. Differences in the carotenoid content of ordinary citrus and lycopene-accumulating mutants. J. Agric. Food Chem. 2006, 54, 5474–5481. [Google Scholar] [CrossRef]
- Guo, F.; Yu, H.W.; Xu, Q.; Deng, X.X. Transcriptomic analysis of differentially expressed genes in an orange-pericarp mutant and wild type in pummel (Citrus grandis). BMC Plant Biol. 2015, 15, 435. [Google Scholar] [CrossRef] [Green Version]
- Alquezar, B.; Rodrigo, M.J.; Zacarías, L. Regulation of carotenoid biosynthesis during fruit maturation in the red-fleshed orange mutant Cara Cara. Phytochemistry 2008, 69, 1997–2007. [Google Scholar] [CrossRef]
- Lu, Q.; Huang, X.J.; Lv, S.Y.; Pan, S.Y. Carotenoid profiling of red navel orange “Cara Cara” harvested from five regions in China. Food Chem. 2017, 232, 788–798. [Google Scholar] [CrossRef]
- Liu, Q.; Xu, J.; Liu, Y.Z.; Zhao, X.L.; Deng, X.X.; Guo, L.L.; Gu, J.Q. A novel bud mutation that confers abnormal patterns of lycopene accumulation in sweet orange fruit (Citrus sinensis L. Osbeck). J. Exp. Bot. 2007, 58, 4161–4171. [Google Scholar] [CrossRef]
- Zhang, M.; Duan, C.; Zang, Y.; Huang, Z.; Liu, G. The flavonoid composition of flavedo and juice from the pummelo cultivar (Citrus grandis (L.) Osbeck) and the grapefruit cultivar (Citrus paradisi) from China. Food Chem. 2011, 129, 1530–1536. [Google Scholar] [CrossRef]
- Tocmo, R.; Pena-Fronteras, J.; Calumba, K.F.; Mendoza, M.; Johnson, J.J. Valorization of pomelo (Citrus grandis Osbeck) peel: A review of current utilization, phytochemistry, bioactivities, and mechanisms of action. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1969–2012. [Google Scholar] [CrossRef]
- Chinese Pharmacopeia Commission. Pharmacopeia of the People’s Republic of China; China Medical Science Publisher: Beijing, China, 2020; Volume 1, pp. 76–77.
- Jiang, K.; Song, Q.; Wang, L.; Xie, T.Z.; Wu, X.; Wang, P.; Yin, G.; Ye, W.C.; Wang, T.J. Antitussive, expectorant and anti-inflammatory activities of different extracts from Exocarpium Citri grandis. J. Ethnopharmacol. 2014, 156, 97–101. [Google Scholar] [CrossRef]
- Zeng, X.; Su, W.W.; Zheng, Y.Y.; Liu, H.; Li, P.L.; Zhang, W.J.; Liang, Y.T.; Bai, Y.; Peng, W.; Yao, H.L. UFLC-Q-TOF-MS/MS-based screening and identification of flavonoids and derived metabolites in human urine after oral administration of Exocarpium Citri Grandis Extract. Molecules 2018, 23, 895. [Google Scholar] [CrossRef] [Green Version]
- Kuo, P.C.; Liao, Y.R.; Hung, H.Y.; Chuang, C.W.; Hwang, T.L.; Huang, S.C.; Wu, T.S. Anti-inflammatory and neuroprotective constituents from the peels of Citrus grandis. Molecules 2017, 22, 967. [Google Scholar] [CrossRef] [Green Version]
- Tian, D.; Wang, F.; Duan, M.; Cao, L.; Zhang, Y.; Yao, X.; Tang, J. Coumarin analogues from the Citrus grandis (L.) Osbeck and their hepatoprotective activity. J. Agric. Food Chem. 2019, 67, 1937–1947. [Google Scholar] [CrossRef]
- Jiang, C.C.; Zhang, Y.F.; Lin, Y.J.; Chen, Y.; Lu, X.K. Illumina® Sequencing Reveals Candidate Genes of Carotenoid Metabolism in Three Pummelo Cultivars (Citrus Maxima) with Different Pulp Color. Int. J. Mol. Sci. 2019, 20, 2246. [Google Scholar] [CrossRef] [Green Version]
- Li, P.L.; Liu, M.H.; Hu, J.H.; Su, W.W. Systematic Chemical Profiling of Citrus Grandis ‘tomentosa’ by Ultrafast Liquid chromatography/diode-array detector/quadrupole Time-of-flight Tandem Mass Spectrometry. J. Pharm. Biomed. Anal. 2014, 90, 167–179. [Google Scholar] [CrossRef]
- Wetzel, C.M.; Jiang, C.Z.; Meehan, L.J.; Voytas, D.F.; Rodermel, S.R. Nuclear-organelle interactions: The immutans variegation mutant of Arabidopsis is plastid autonomous and impaired in carotenoid biosynthesis. Plant J. 1994, 61, 161–175. [Google Scholar] [CrossRef]
- Liu, C.H.; He, M.; Wang, Z.; Xu, J. Integrative Analysis of terpenoid profiles and hormones from fruits of red-flesh citrus mutants and their wild types. Molecules 2019, 24, 3456. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.H.; Zhang, H.P.; He, M.; Liu, X.; Chen, S.L.; He, Z.Y.; Ye, J.L.; Xu, J. Lycopene accumulation in Cara Cara red-flesh navel orange is correlated with weak abscisic acid catabolism. J. Agric. Food Chem. 2021, 69, 8236–8246. [Google Scholar] [CrossRef]
- Ikoma, Y.; Komatsu, A.; Kita, M.; Ogawa, K.; Omura, M.; Yano, M.; Moriguchi, T. Expression of a phytoene synthase gene and characteristic carotenoid accumulation during citrus fruit development. Physiol. Plantarum. 2001, 111, 232–238. [Google Scholar] [CrossRef]
- Kim, I.J.; Ko, K.C.; Kim, C.S.; Chung, W.I. Isolation and expression patterns of a cDNA encoding phytoene synthase in Citrus. J. Plant Physiol. 2001, 58, 795–800. [Google Scholar] [CrossRef]
- Bruno, M.; Koschmieder, J.; Wuest, F.; Schaub, P.; Fehling-Kaschek, M.; Timmer, J.; Beyer, P.; Al-Babili, S. Enzymatic study on AtCCD4 and AtCCD7 and their potential to form acyclic regulatory metabolites. J. Exp. Bot. 2016, 67, 5993–6005. [Google Scholar] [CrossRef] [Green Version]
- Hou, X.; Rivers, J.; León, P.; McQuinn, R.P.; Pogson, B.J. Synthesis and function of apocarotenoid signals in plants. Trends Plant Sci. 2016, 21, 792–803. [Google Scholar] [CrossRef]
- Zhang, H.P.; Chen, J.J.; Peng, Z.X.; Shi, M.Y.; Liu, X.; Wen, H.; Jiang, Y.W.; Cheng, Y.J.; Xu, J.; Zhang, H.Y. Integrated Transcriptomic and Metabolomic analysis reveals a transcriptional regulation network for the biosynthesis of carotenoids and flavonoids in ‘Cara cara’ navel Orange. BMC Plant Biol. 2021, 21, 29. [Google Scholar] [CrossRef]
- Yu, K.Q.; Xu, Q.; Da, X.L.; Guo, F.; Ding, Y.D.; Deng, X.X. Transcriptome changes during fruit development and ripening of sweet orange (Citrus sinensis). BMC Genom. 2012, 13, 10. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.W.; Zhang, Y.; Zheng, X.J.; Zhu, K.J.; Xu, Q.; Deng, X.X. Molecular characterization, critical amino acid identification, and promoter analysis of a lycopene β-cyclase gene from citrus. Tree Genet. Genomes 2016, 12, 106. [Google Scholar] [CrossRef]
- Tatmala, N.; Ma, G.; Zhang, L.C.; Kato, M.; Kaewsuksaeng, S. Characterization of carotenoid accumulation and carotenogenic gene expression during fruit ripening in red colored pulp of ‘Siam Red Ruby’ pumelo (Citrus grandis) cultivated in Thailand. Horticult. J. 2020, 89, 237–243. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.Y.; Lu, X.N.; Tang, Z.L.; Zhang, X.N.; Lei, F.J.; Hou, L.P.; Li, M.L. Combined analysis of carotenoid metabolites and the transcriptome to reveal the molecular mechanism underlying fruit colouration in zucchini (Cucurbita pepo L.). Food Chem. 2021, 2, 100021. [Google Scholar] [CrossRef]
- Wang, W.; Liu, D.X.; Qin, M.; Xie, Z.B.; Chen, R.Y.; Zhang, Y.T. Effects of supplemental lighting on potassium transport and fruit coloring of tomatoes grown in hydroponics. Int. J. Mol. Sci. 2021, 22, 2687. [Google Scholar] [CrossRef]
- Xie, Z.S.; Liu, Q.D.; Liang, Z.K.; Zhao, M.Q.; Yu, X.X.; Yang, D.P.; Xu, X.J. The GC/MS analysis of volatile components extracted by different methods from Exocarpium Citri Grandis. J. Anal. Methods Chem. 2013, 2013, 918406. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Mao, X.Z.; Huang, J.J.; Ding, Y.; Wu, J.M.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L.P. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [Green Version]

| Sample | Total Carotenoids | Β-Carotene | Lutein | Zeaxanthin | Lycopene | Β-Cryptoxanthin | Γ-Carotene | Phytoene |
|---|---|---|---|---|---|---|---|---|
| R-HJH Albedo | 15.553 ± 0.100 | 1.347 ± 0.096 | 1.193 ± 0.188 | 0.197 ± 0.017 | 10.407 ± 0.315 | 0.109 ± 0.031 | 0.268 ± 0.026 | 1.166 ± 0.259 |
| HJH Albedo | 3.864 ± 0.289 | 0.235 ± 0.035 | 2.593 ± 0.097 | 0.169 ± 0.016 | 0.294 ± 0.189 | 0.002 ± 0.001 | 0.076 ± 0.005 | - |
| R-HJH juice sacs | 19.432 ± 1.104 | 1.890 ± 0.191 | 1.600 ± 0.047 | 0.689 ± 0.084 | 11.333 ± 0.636 | 0.075 ± 0.008 | 0.341 ± 0.024 | 3.017 ± 0.158 |
| HJH juice sacs | 6.720 ± 0.153 | 0.200 ± 0.007 | 1.667 ± 0.058 | 1.313 ± 0.030 | 3.207 ± 0.061 | 0.003 ± 0.001 | 0.110 ± 0.003 | - |
| HJH | R-HJH | |
|---|---|---|
| naringin | 55.81 ± 2.48 | 38.76 ± 1.82 |
| rhoifolin | 10.37 ± 0.29 | 5.72 ± 0.15 |
| isoimperatorin | 0.95 ± 0.01 | 0.35 ± 0.01 |
| bergapten | 0.33 | 0.24 ± 0.01 |
| No | Name | CAS# | RI | Exocarp | Pulp | ||
|---|---|---|---|---|---|---|---|
| No. | HJU | R-HJH | HJU | R-HJH | |||
| 1 | α-Pinene | 80-56-8 | 7.103 | - | - | 5.58 ± 0.29 | 2.53 ± 0.18 |
| 2 | β-Myrcene | 123-35-3 | 8.613 | 813.58 ± 74.28 | 268.86 ± 42.92 | 169.82 ± 7.53 | |
| 3 | α-Phellandrene | 99-83-2 | 8.993 | 173.41 ± 14.46 | 78.72 ± 8.32 | 11.28 ± 1.26 | 6.59 ± 0.63 |
| 4 | D-Limonene | 5989-27-5 | 9.757 | 8594.44 ± 700.36 | 5721.00 ± 827.98 | 774.94 ± 43.00 | 398.28 ± 22.36 |
| 5 | γ-Terpinene | 99-85-4 | 10.537 | 917.96 ± 78.94 | 124.34 ± 18.61 | 103.85 ± 4.20 | 42.47 ± 2.61 |
| 6 | Linalool | 78-70-6 | 11.679 | 1189.20 ± 26.86 | 915.71 ± 22.37 | 76.03 ± 2.76 | 37.68 ± 0.65 |
| 7 | Nonanal | 124-19-6 | 11.791 | 454.39 ± 371.06 | 63.48 ± 8.13 | 6.74 ± 0.66 | 4.58 ± 0.38 |
| 8 | α-Terpineol | 10482-56-1 | 14.302 | 467.10 ± 8.22 | 231.96 ± 4.00 | 26.97 ± 1.24 | 16.59 ± 0.55 |
| 9 | Neral | 106-26-3 | 15.676 | 722.94 ± 17.79 | 430.55 ± 18.60 | 18.52 ± 0.68 | 9.59 ± 0.29 |
| 10 | Geraniol | 106-24-1 | 16.017 | 95.81 ± 1.86 | 63.39 ± 1.40 | 6.76 ± 0.30 | 7.18 ± 0.47 |
| 11 | Citral | 5392-40-5 | 16.485 | 1036.55 ± 24.90 | 620.00 ± 26.35 | 29.05 ± 1.41 | 18.27 ± 0.58 |
| 12 | Dodecanal | 112-54-9 | 20.102 | 91.55 ± 7.81 | 88.59 ± 17.96 | 4.73 ± 0.48 | 1.72 ± 0.03 |
| 13 | Caryophyllene | 87-44-5 | 20.532 | 324.47 ± 33.88 | 134.35 ± 36.49 | 12.14 ± 1.31 | 2.16 ± 0.21 |
| 14 | γ-Muurolene | 30021-74-0 | 21.925 | 223.37 ± 25.60 | 127.79 ± 33.25 | 9.73 ± 1.07 | - |
| 15 | Germacrene D | 23986-74-5 | 22.070 | 1085.83 ± 110.57 | 591.64 ± 182.15 | 32.36 ± 3.29 | 4.17 ± 0.37 |
| 16 | α-Muurolene | 10208-80-7 | 23.372 | 111.13 ± 10.70 | 65.07 ± 15.59 | 4.31 ± 0.46 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, C.; Peng, C.; Qiu, D.; Zeng, J. Metabolic Profiling and Transcriptional Analysis of Carotenoid Accumulation in a Red-Fleshed Mutant of Pummelo (Citrus grandis). Molecules 2022, 27, 4595. https://doi.org/10.3390/molecules27144595
Zhu C, Peng C, Qiu D, Zeng J. Metabolic Profiling and Transcriptional Analysis of Carotenoid Accumulation in a Red-Fleshed Mutant of Pummelo (Citrus grandis). Molecules. 2022; 27(14):4595. https://doi.org/10.3390/molecules27144595
Chicago/Turabian StyleZhu, Congyi, Cheng Peng, Diyang Qiu, and Jiwu Zeng. 2022. "Metabolic Profiling and Transcriptional Analysis of Carotenoid Accumulation in a Red-Fleshed Mutant of Pummelo (Citrus grandis)" Molecules 27, no. 14: 4595. https://doi.org/10.3390/molecules27144595
APA StyleZhu, C., Peng, C., Qiu, D., & Zeng, J. (2022). Metabolic Profiling and Transcriptional Analysis of Carotenoid Accumulation in a Red-Fleshed Mutant of Pummelo (Citrus grandis). Molecules, 27(14), 4595. https://doi.org/10.3390/molecules27144595





