Transcriptome Analysis of the Anti-Proliferative Effects of Ginsenoside Rh3 on HCT116 Colorectal Cancer Cells
Abstract
1. Introduction
2. Results
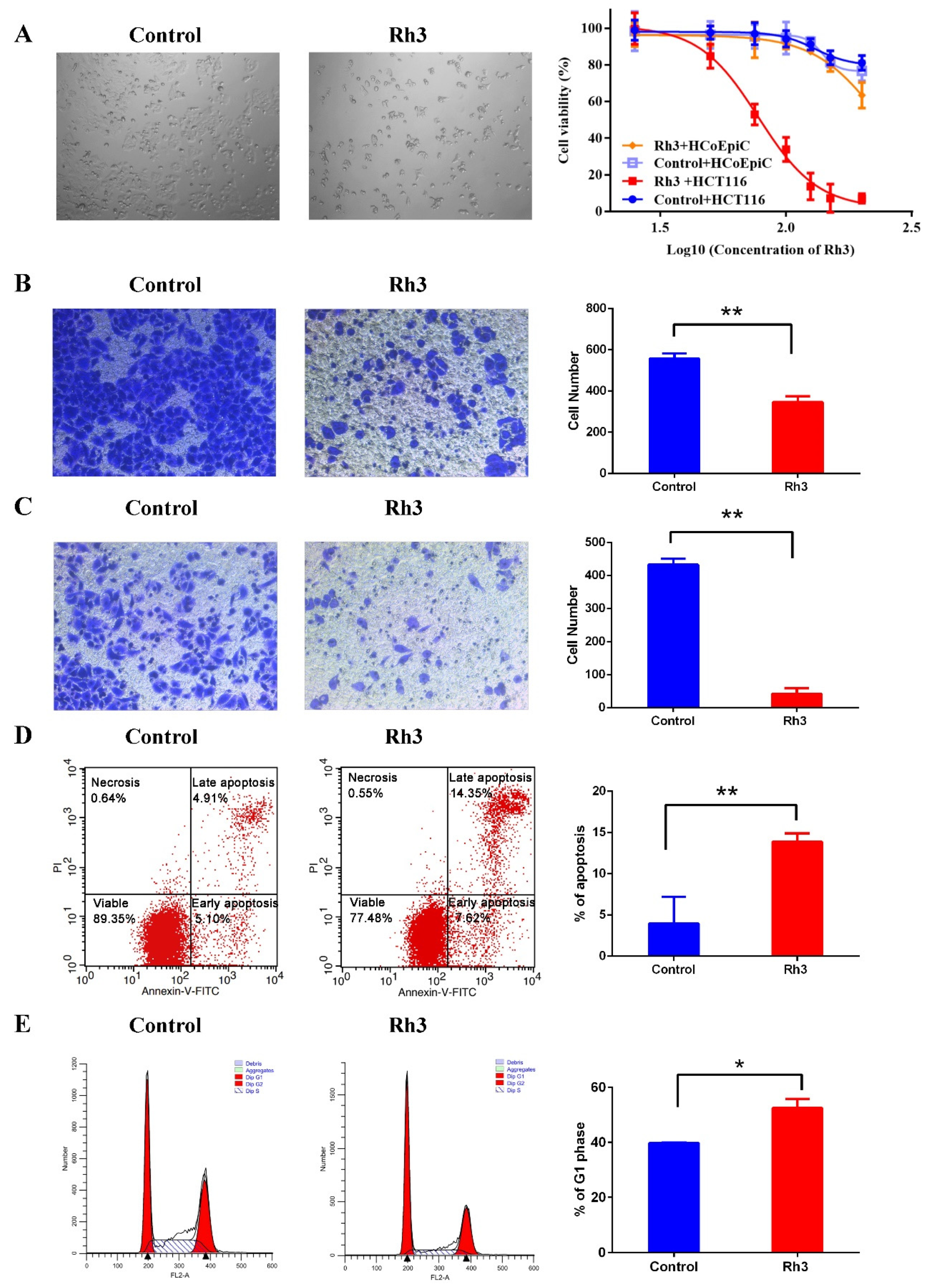
2.1. Effect of Rh3 on Viability, Migration, Invasion, and Cell Cycle Progression in HCT116 Cells
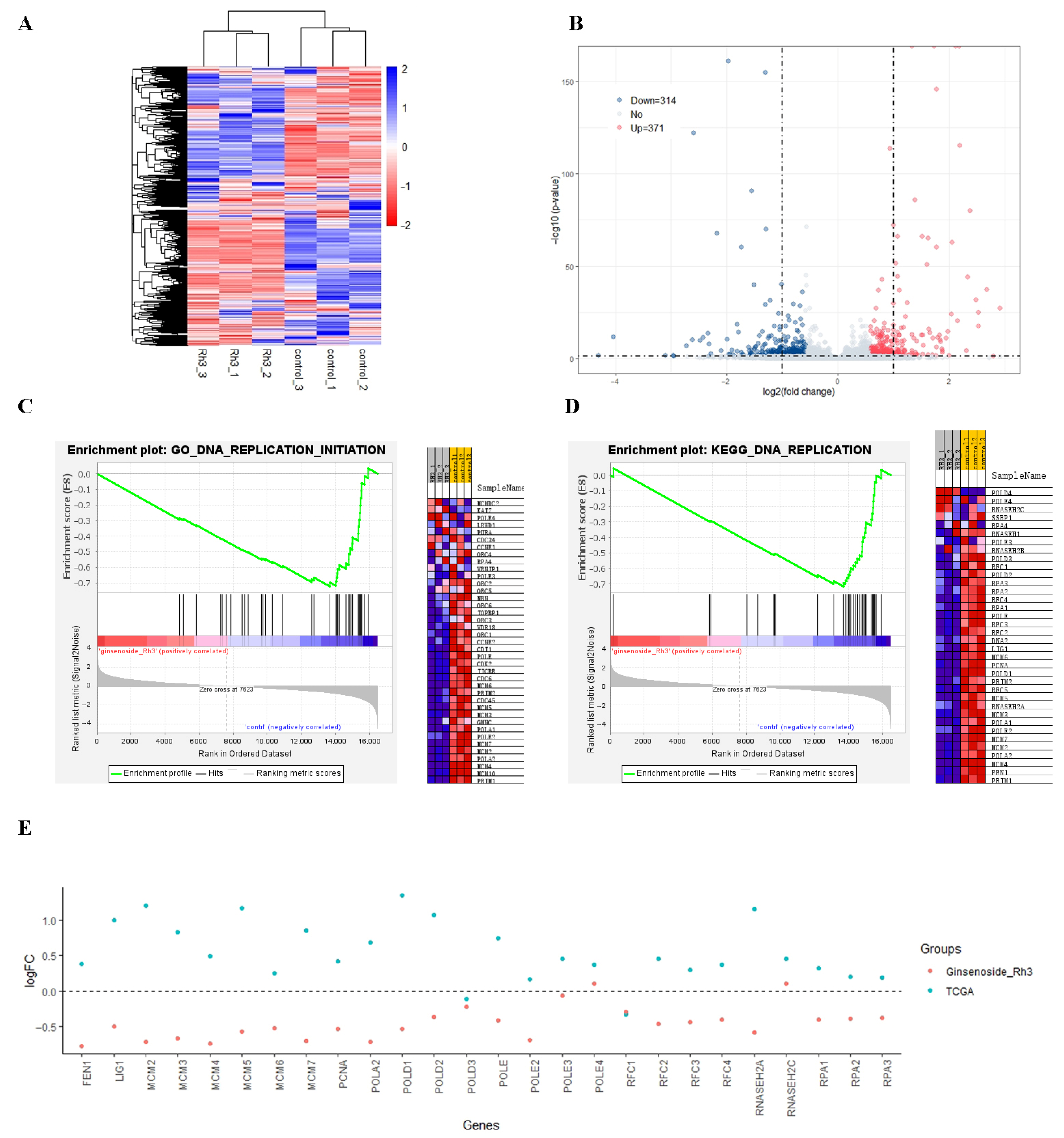
2.2. Effect of Rh3 on Gene Transcription in HCT116 Cells
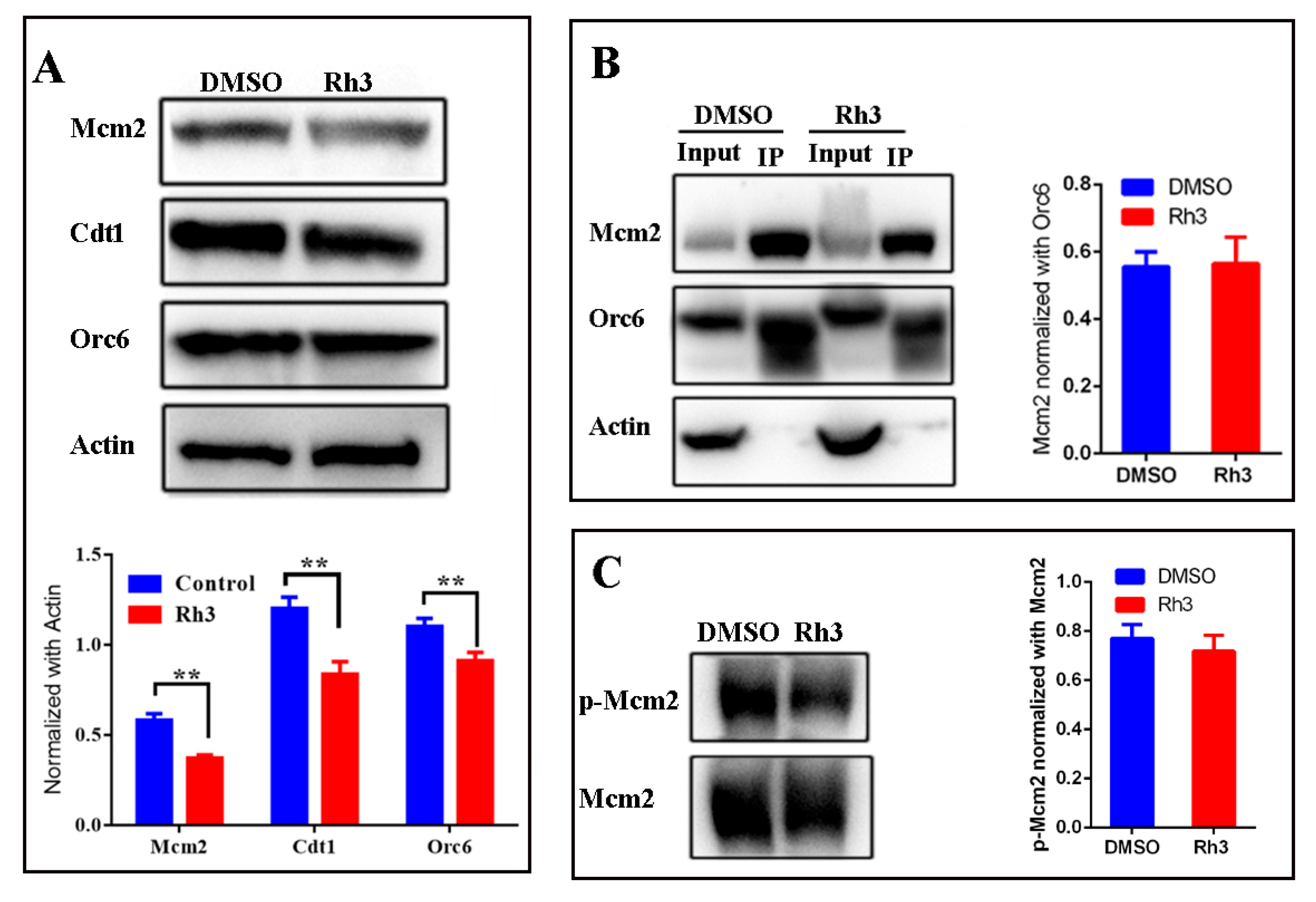
2.3. Effect of Rh3 on Expression of Proteins Related to the Initiation of DNA Replication
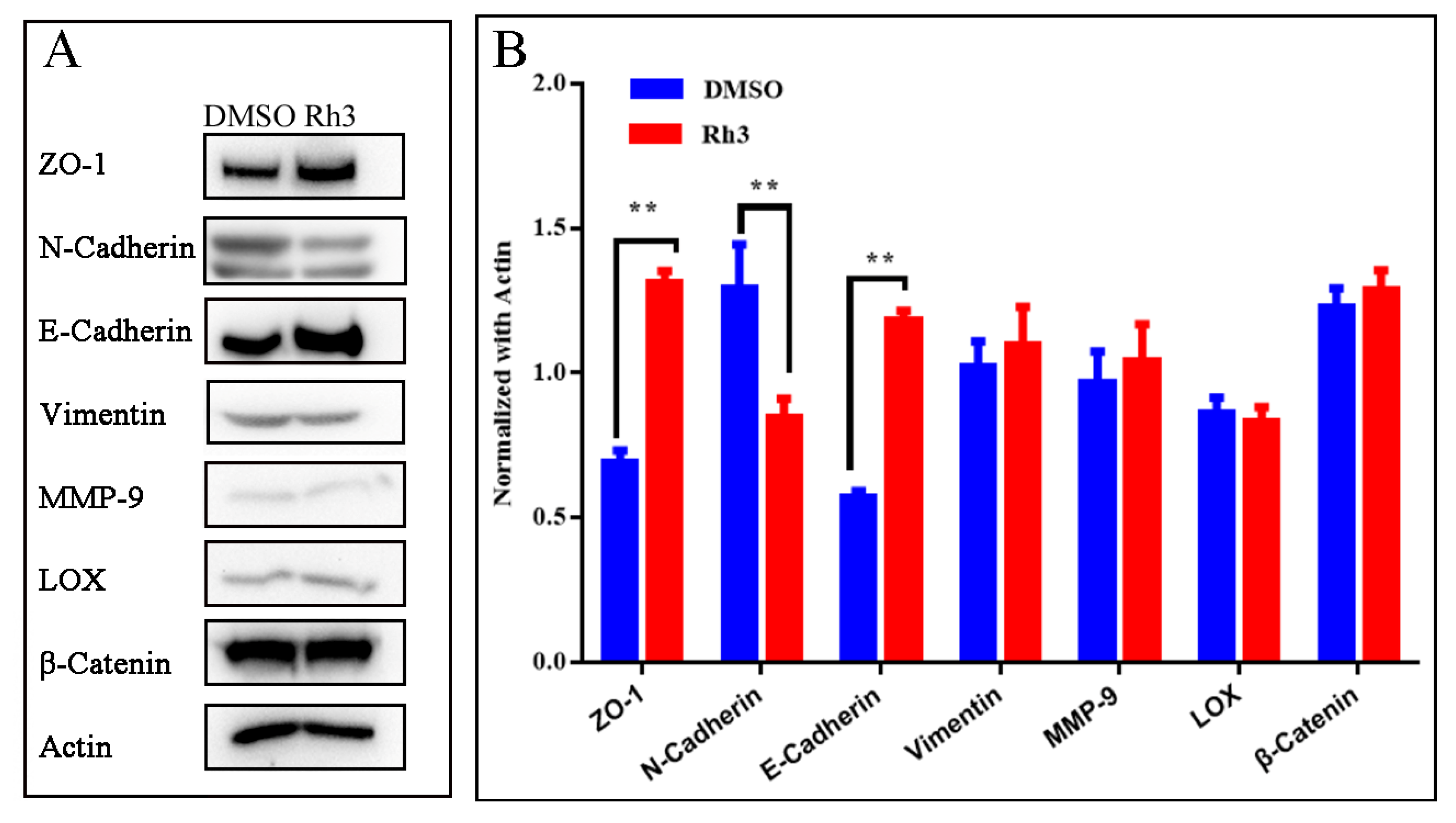
2.4. Effect of Rh3 on Expression of Proteins Related to Cell Migration and Invasion
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Reagents
4.2. Cell Proliferation Assay
4.3. Selectivity Index (SI)
4.4. Cell Migration and Invasion Assay
4.5. Cell Apoptosis and Cell Cycle Analyses
4.6. RNA-Seq
4.7. Analysis of RNA-Seq Data
4.8. Colon Cancer Data Acquisition and Analysis
4.9. Western Blotting and Co-IP
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Dong, X.; Li, H.; Cao, M.; Sun, D.; He, S.; Yang, F.; Yan, X.; Zhang, S.; Li, N.; et al. Cancer statistics in China and United States, 2022: Profiles, trends, and determinants. Chin. Med. J. 2022, 135, 584–590. [Google Scholar] [CrossRef]
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal cancer. Lancet 2019, 394, 1467–1480. [Google Scholar] [CrossRef]
- Baldo, B.A.; Pham, N.H. Adverse reactions to targeted and non-targeted chemotherapeutic drugs with emphasis on hypersensitivity responses and the invasive metastatic switch. Cancer Metastasis Rev. 2013, 32, 723–761. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Yu, X.; Huangpu, H.; Yao, F. Ginsenoside Rb3 protects cardiomyocytes against hypoxia/reoxygenation injury via activating the antioxidation signaling pathway of PERK/Nrf2/HMOX1. Biomed. Pharm. 2019, 109, 254–261. [Google Scholar] [CrossRef]
- Lee, J.O.; Choi, E.; Shin, K.K.; Hong, Y.H.; Kim, H.G.; Jeong, D.; Hossain, M.A.; Kim, H.S.; Yi, Y.S.; Kim, D.; et al. Compound K, a ginsenoside metabolite, plays an antiinflammatory role in macrophages by targeting the AKT1-mediated signaling pathway. J. Ginseng Res. 2019, 43, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Ji, H.W.; Kim, H.W.; Yun, S.H.; Park, J.E.; Kim, S.J. Ginsenoside Rg3 prevents oncogenic long noncoding RNA ATXN8OS from inhibiting tumor-suppressive microRNA-424-5p in breast cancer cells. Biomolecules 2021, 11, 118. [Google Scholar] [CrossRef]
- Han, J.; Oh, J.P.; Yoo, M.; Cui, C.H.; Jeon, B.M.; Kim, S.C.; Han, J.H. Minor ginsenoside F1 improves memory in APP/PS1 mice. Mol. Brain 2019, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Quan, K.; Liu, Q.; Wan, J.Y.; Zhao, Y.J.; Guo, R.Z.; Alolga, R.N.; Li, P.; Qi, L.W. Rapid preparation of rare ginsenosides by acid transformation and their structure-activity relationships against cancer cells. Sci. Rep. 2015, 5, 8598. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Gao, Y.; Zhu, J.; Zhang, J.; Dong, M.; Mao, Y. Protective action of the ginsenoside Rh3 in a rat myocardial ischemia-reperfusion injury model by inhibition of apoptosis induced via p38 mitogen-activated protein kinase/caspase-3 signaling. J. Int. Med. Res. 2020, 48, 300060520969090. [Google Scholar] [CrossRef]
- Wang, X.M.; She, C.; Li, Q.; Zhang, D.; Xu, J.X.; Li, M.H.; Li, P.; Xu, H.B. Ginsenoside Rh3 activates Nrf2 signaling and protects endometrial cells from oxygen and glucose deprivation-reoxygenation. Aging 2020, 12, 6109–6119. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.S.; Lee, J.E.; Park, J.I.; Myung, C.H.; Lim, Y.H.; Park, C.K.; Hwang, J.S. Inhibitory mechanism of ginsenoside Rh3 on granulocyte-macrophage colony-stimulating factor expression in UV-B-irradiated murine SP-1 keratinocytes. J. Ginseng Res. 2020, 44, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Chung, I.; Lee, J.; Park, Y.S.; Lim, Y.; Chang, D.H.; Park, J.; Hwang, J.S. Inhibitory mechanism of Korean Red Ginseng on GM-CSF expression in UVB-irradiated keratinocytes. J. Ginseng Res. 2015, 39, 322–330. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.Z.; Li, K.R.; Yu, Q.; Jiang, Q.; Yao, J.; Cao, C. Activation of Nrf2 by Ginsenoside Rh3 protects retinal pigment epithelium cells and retinal ganglion cells from UV. Free Radic. Biol. Med. 2018, 117, 238–246. [Google Scholar] [CrossRef]
- Lee, Y.Y.; Park, J.S.; Lee, E.J.; Lee, S.Y.; Kim, D.H.; Kang, J.L.; Kim, H.S. Anti-inflammatory mechanism of ginseng saponin metabolite Rh3 in lipopolysaccharide-stimulated microglia: Critical role of 5′-adenosine monophosphate-activated protein kinase signaling pathway. J. Agric. Food Chem. 2015, 63, 3472–3480. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.L.; Kang, K.S. Protective effect of ginsenoside Rh3 against anticancer drug-induced apoptosis in LLC-PK1 kidney cells. J. Ginseng Res. 2017, 41, 227–231. [Google Scholar] [CrossRef]
- Cong, Z.; Zhao, Q.; Yang, B.; Cong, D.; Zhou, Y.; Lei, X.; Zhang, X. Ginsenoside Rh3 inhibits proliferation and induces apoptosis of colorectal cancer cells. Pharmacology 2020, 105, 329–338. [Google Scholar] [CrossRef]
- Li, N.; Lam, W.H.; Zhai, Y.; Cheng, J.; Cheng, E.; Zhao, Y.; Gao, N.; Tye, B.K. Structure of the origin recognition complex bound to DNA replication origin. Nature 2018, 559, 217–222. [Google Scholar] [CrossRef]
- Takara, T.J.; Bell, S.P. Multiple Cdt1 molecules act at each origin to load replication-competent Mcm2-7 helicases. EMBO J. 2011, 30, 4885–4896. [Google Scholar] [CrossRef]
- Liu, C.; Wu, R.; Zhou, B.; Wang, J.; Wei, Z.; Tye, B.K.; Liang, C.; Zhu, G. Structural insights into the Cdt1-mediated MCM2-7 chromatin loading. Nucleic Acids Res. 2012, 40, 3208–3217. [Google Scholar] [CrossRef]
- Yuan, Z.; Riera, A.; Bai, L.; Sun, J.; Nandi, S.; Spanos, C.; Chen, Z.A.; Barbon, M.; Rappsilber, J.; Stillman, B.; et al. Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1. Nat. Struct. Mol. Biol. 2017, 24, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Lei, M.; Kawasaki, Y.; Young, M.R.; Kihara, M.; Sugino, A.; Tye, B.K. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997, 11, 3365–3374. [Google Scholar] [CrossRef]
- Zhu, S.; Liu, X.; Xue, M.; Li, Y.; Cai, D.; Wang, S.; Zhang, L. 20(S)-ginsenoside Rh2 induces caspase-dependent promyelocytic leukemia-retinoic acid receptor A degradation in NB4 cells via Akt/Bax/caspase9 and TNF-alpha/caspase8 signaling cascades. J. Ginseng Res. 2021, 45, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Xiao-Ling, S.; Yaliu, S.; Ming-Ming, L.; Xue, F.; Xian-Sheng, M.; Li, F. Comparative pharmacokinetics of ginsenoside Rg3 and ginsenoside Rh2 after oral administration of ginsenoside Rg3 in normal and walker 256 tumor-bearing rats. Pharmacogn. Mag. 2016, 12, 21–24. [Google Scholar] [PubMed]
- Wang, J.; Chen, Y.; Dai, C.; Shang, Y.; Xie, J. Ginsenoside Rh2 alleviates tumor-associated depression in a mouse model of colorectal carcinoma. Am. J. Transl. Res. 2016, 8, 2189–2195. [Google Scholar] [PubMed]
- Chen, Y.; Zhang, Y.; Song, W.; Zhang, Y.; Dong, X.; Tan, M. Ginsenoside Rh2 improves the cisplatin anti-tumor effect in lung adenocarcinoma A549 cells via superoxide and PD-L1. Anticancer Agents Med. Chem. 2020, 20, 495–503. [Google Scholar] [CrossRef]
- Zhao, L.; Sun, W.; Zheng, A.; Zhang, Y.; Fang, C.; Zhang, P. Ginsenoside Rg3 suppresses ovarian cancer cell proliferation and invasion by inhibiting the expression of lncRNA H19. Acta Biochim. Pol. 2021, 68, 575–582. [Google Scholar] [CrossRef]
- Liu, M.Y.; Liu, F.; Li, Y.J.; Yin, J.N.; Gao, Y.L.; Wang, X.Y.; Yang, C.; Liu, J.G.; Li, H.J. Ginsenoside Rg5 inhibits human osteosarcoma cell proliferation and induces cell apoptosis through PI3K/Akt/mTORC1-Related LC3 autophagy pathway. Oxidative Med. Cell. Longev. 2021, 2021, 5040326. [Google Scholar] [CrossRef]
- Sharma, A.; Lee, H.J. Ginsenoside compound k: Insights into recent studies on pharmacokinetics and health-promoting activities. Biomolecules 2020, 10, 1028. [Google Scholar] [CrossRef]
- Park, J.S.; Park, E.M.; Kim, D.H.; Jung, K.; Jung, J.S.; Lee, E.J.; Hyun, J.W.; Kang, J.L.; Kim, H.S. Anti-inflammatory mechanism of ginseng saponins in activated microglia. J. Neuroimmunol. 2009, 209, 40–49. [Google Scholar] [CrossRef]
- Kim, E.J.; Jung, I.H.; Van Le, T.K.; Jeong, J.J.; Kim, N.J.; Kim, D.H. Ginsenosides Rg5 and Rh3 protect scopolamine-induced memory deficits in mice. J. Ethnopharmacol. 2013, 146, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Martino-Echarri, E.; Henderson, B.R.; Brocardo, M.G. Targeting the DNA replication checkpoint by pharmacologic inhibition of Chk1 kinase: A strategy to sensitize APC mutant colon cancer cells to 5-fluorouracil chemotherapy. Oncotarget 2014, 5, 9889–9900. [Google Scholar] [CrossRef] [PubMed]
- Grant, G.D.; Cook, J.G. The temporal regulation of S phase proteins during G1. Adv. Exp. Med. Biol. 2017, 1042, 335–369. [Google Scholar] [PubMed]
- Lin, Y.C.; Prasanth, S.G. Replication initiation: Implications in genome integrity. DNA Repair 2021, 103, 103131. [Google Scholar] [CrossRef]
- Cox, T.R. The matrix in cancer. Nat. Rev. Cancer 2021, 21, 217–238. [Google Scholar] [CrossRef]
- Loh, C.Y.; Chai, J.Y.; Tang, T.F.; Wong, W.F.; Sethi, G.; Shanmugam, M.K.; Chong, P.P.; Looi, C.Y. The E-cadherin and N-cadherin switch in epithelial-to-mesenchymal transition: Signaling, therapeutic implications, and challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef]
- Murdocca, M.; De Masi, C.; Pucci, S.; Mango, R.; Novelli, G.; Di Natale, C.; Sangiuolo, F. LOX-1 and cancer: An indissoluble liaison. Cancer Gene Ther. 2021, 28, 1088–1098. [Google Scholar] [CrossRef]
- Buttacavoli, M.; Di Cara, G.; Roz, E.; Pucci-Minafra, I.; Feo, S.; Cancemi, P. Integrated multi-omics investigations of metalloproteinases in colon cancer: Focus on MMP2 and MMP9. Int. J. Mol. Sci. 2021, 22, 12389. [Google Scholar] [CrossRef]
- Zucker, S.; Vacirca, J. Role of matrix metalloproteinases (MMPs) in colorectal cancer. Cancer Metastasis Rev. 2004, 23, 101–117. [Google Scholar] [CrossRef]
- Chen, G.T.; Tifrea, D.F.; Murad, R.; Habowski, A.N.; Lyou, Y.; Duong, M.R.; Hosohama, L.; Mortazavi, A.; Edwards, R.A.; Waterman, M.L. Disruption of beta-catenin dependent Wnt signaling in colon cancer cells remodels the microenvironment to promote tumor invasion. Mol. Cancer Res. 2022, 20, 468–484. [Google Scholar] [CrossRef] [PubMed]
- Rosivatz, E.; Becker, I.; Bamba, M.; Schott, C.; Diebold, J.; Mayr, D.; Hofler, H.; Becker, K.F. Neoexpression of N-cadherin in E-cadherin positive colon cancers. Int. J. Cancer 2004, 111, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Polette, M.; Mestdagt, M.; Bindels, S.; Nawrocki-Raby, B.; Hunziker, W.; Foidart, J.M.; Birembaut, P.; Gilles, C. Beta-catenin and ZO-1: Shuttle molecules involved in tumor invasion-associated epithelial-mesenchymal transition processes. Cells Tissues Organs 2007, 185, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Badisa, R.B.; Darling-Reed, S.F.; Joseph, P.; Cooperwood, J.S.; Latinwo, L.M.; Goodman, C.B. Selective cytotoxic activities of two novel synthetic drugs on human breast carcinoma MCF-7 cells. Anticancer Res. 2009, 29, 2993–2996. [Google Scholar] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty Heatmaps, R Package Version 1.0.12. 2019. Available online: https://CRAN.R-project.org/package=pheatmap (accessed on 9 December 2020).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teng, S.; Lei, X.; Zhang, X.; Shen, D.; Liu, Q.; Sun, Y.; Wang, Y.; Cong, Z. Transcriptome Analysis of the Anti-Proliferative Effects of Ginsenoside Rh3 on HCT116 Colorectal Cancer Cells. Molecules 2022, 27, 5002. https://doi.org/10.3390/molecules27155002
Teng S, Lei X, Zhang X, Shen D, Liu Q, Sun Y, Wang Y, Cong Z. Transcriptome Analysis of the Anti-Proliferative Effects of Ginsenoside Rh3 on HCT116 Colorectal Cancer Cells. Molecules. 2022; 27(15):5002. https://doi.org/10.3390/molecules27155002
Chicago/Turabian StyleTeng, Siying, Xi Lei, Xinmin Zhang, Dongming Shen, Qiuyi Liu, Yingjie Sun, Yi Wang, and Zhongyi Cong. 2022. "Transcriptome Analysis of the Anti-Proliferative Effects of Ginsenoside Rh3 on HCT116 Colorectal Cancer Cells" Molecules 27, no. 15: 5002. https://doi.org/10.3390/molecules27155002
APA StyleTeng, S., Lei, X., Zhang, X., Shen, D., Liu, Q., Sun, Y., Wang, Y., & Cong, Z. (2022). Transcriptome Analysis of the Anti-Proliferative Effects of Ginsenoside Rh3 on HCT116 Colorectal Cancer Cells. Molecules, 27(15), 5002. https://doi.org/10.3390/molecules27155002





