4. Material and Methods
Chemistry General. Purity and characterization of compounds were established by a combination of MS, LC/MS, NMR, HPLC and TLC analytical techniques, as described below. 1H spectra were recorded on a Bruker Avance DPX-300 (300 MHz) spectrometer in chloroform-d (7.26 ppm) or methanol-d4 (3.31 ppm) with tetramethylsilane (0.00 ppm) or solvent peaks as the internal reference unless otherwise noted. 13C spectra were recorded on a JEOP 400YH (100 MHz). Chemical shifts are reported in ppm relative to the solvent signal and coupling constant (J) values are reported in hertz (Hz). TLC was performed on EMD precoated silica gel 60 F254 plates. TLC spots were visualized with UV light or I2 detection. Low-resolution mass spectra were obtained using a single quadrupole PE Sciex API 150EX (ESI). Unless stated otherwise, all test compounds were at least 95% pure as determined by HPLC. HPLC method: a Waters 2695 Separation Module equipped with a Waters 2996 Photodiode Array Detector and a Phenomenex Synergi 4 μm Hydro-RP 80A C18 250 × 4.6 mm column using a flow rate of 1 mL/min starting with 1 min at 5% solvent B, followed by a 15 min gradient of 5–95% solvent B, followed by 9 min at 95% solvent B (solvent A, water with 0.1% TFA; solvent B, acetonitrile with 0.1% TFA and 5% water; absorbance monitored at 220 and 280 nm). LC/MS Instrument: an Agilent InfinityLab MSD single quadrupole mass spectrometer equipped with an API-ES and an Agilent Infinity II 1260 HPLC equipped with an Agilent Infinity 1260 variable wavelength detector and an Agilent Poroshell 120 SB-C18 2.7 μm 50 × 4.8 mm column. HPLC Method: With a flow rate of 1 mL/min, a 4.0 min gradient of 5–95% solvent B, followed by 2.5 min at 95% solvent B (solvent A, water with 0.1% formic acid; solvent B, acetonitrile with 0.1% formic acid and 5% water; absorbance monitored at 220 and 280 nm). MS Method: using atmospheric pressure ionization–electrospray, positive and negative ions were monitored in the range of 70–700 or 300–2000 (for compounds with a MW > 700).
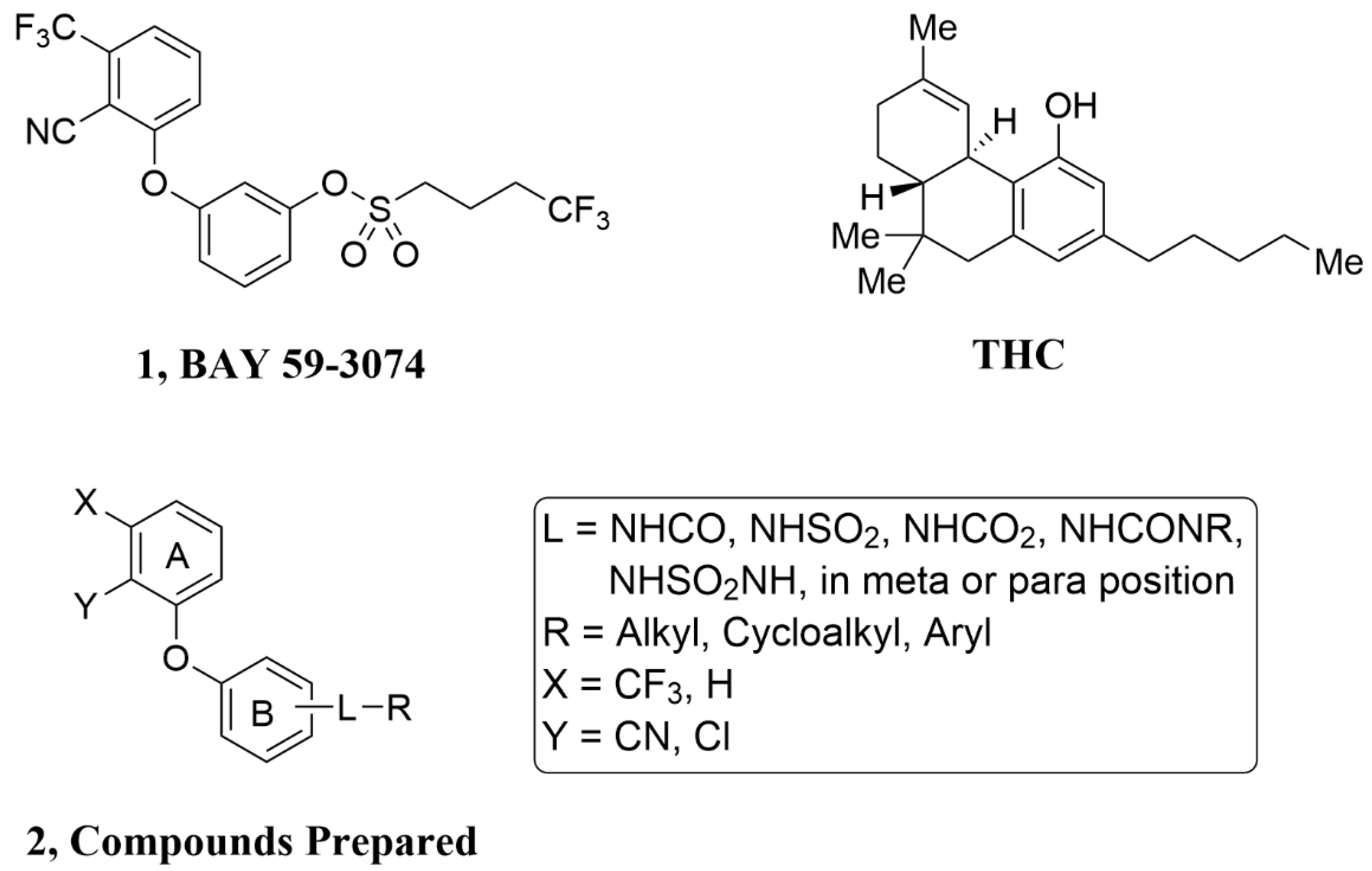
General Procedure A: Sulfonamides, Sulfamides, Carbamates, Amides and Ureas of 3 from Sulfonyl Chlorides, Sulfamoyl Chlorides, Carbamoyl Chlorides, Acid Chlorides and Isocyanates, Respectively. To a solution of 3 (0.2 mmol, 1 equiv.) in THF (1 mL) was added the sulfonyl chloride, sulfamoyl chloride, carbamoyl chloride, acid chloride or isocyanate (0.24 mmol, 1.2 equiv.), followed by triethylamine (0.26 mmol, 1.3 equiv.). The mixture was stirred at rt for 15 h. Water (0.4 mL), ethyl acetate (3 mL) and then saturated NaHCO3 solution (0.8 mL) were added. After 10 min, the aqueous layer was removed. Celite (600 mg) was added to the organic layer, and the solvent was evaporated. Flash chromatography using silica gel with an ethyl acetate/hexanes gradient provided the purified sulfonamide, sulfamide, carbamate, amide, or urea.
General Procedure B: Phenyl Ethers 3 from 2-Fluorobenzonitriles. A mixture of 2-fluoro-6-trifluoromethylbenzonitrile or 2-fluorobenzonitrile (2 mmol), 3-aminophenol or 4-aminophenol (2.2 mmol, 1.1 equiv.), K2CO3 (6 mmol, 3 equiv.) and DMF (4 mL) was stirred at rt for 15 min and then at 80 °C for 4 h. Ethyl acetate (20 mL) was added, followed by water (4 mL) and brine (10 mL). After 10 min, the aqueous layer was removed, and the organic layer was washed with 0.8 M NaHCO3 solution (2 × 6 mL). Celite (3 g) was added to the organic layer and the solvent evaporated. Flash chromatography using silica gel with an ethyl acetate/hexanes gradient provided the purified phenyl ether 3.
2-(3-Aminophenoxy)-6-trifluoromethylbenzonitrile(3A). The title compound was prepared by the general procedure B from 2-fluoro-6-trifluoromethylbenzonitrile (380 mg, 2.0 mmol), 3-aminophenol (240 mg, 1.1 equiv.), and K2CO3 (820 mg, 3 equiv.) to provide 550 mg (99%) of a colorless oil. Rf = 0.35 (40% ethyl acetate/hexanes; UV active). LC/MS (m/z) 279.0 (M+1), >95% at 4.36 min.
2-(4-Aminophenoxy)-6-trifluoromethylbenzonitrile(3B). The title compound was prepared by the general procedure B from 2-fluoro-6-trifluoromethylbenzonitrile (380 mg, 2.0 mmol), 4-aminophenol (240 mg, 1.1 equiv.), and K2CO3 (820 mg, 3 equiv.) to provide 451 mg (82%) of a tan crystalline solid. Rf = 0.24 (40% ethyl acetate/hexanes; UV active). LC/MS (m/z) 279.0 (M+1), 85% at 3.94 min.
2-(3-Aminophenoxy)benzonitrile(3C). The title compound was prepared by the general procedure B from 2-fluorobenzonitrile (0.17 mL, 1.5 mmol), 3-aminophenol (180 mg, 1.1 equiv.), and K2CO3 (620 mg, 3 equiv.) to provide 270 mg (86%) of a colorless oil. Rf = 0.40 (40% ethyl acetate/hexanes; UV active). LC/MS (m/z) 211.0 (M+1), >97% at 3.84 min.
3-[2-Chloro-3-(trifluoromethyl)phenoxy]benzenamine(3D). A mixture of 2-chloro-3-trifluoromethylphenol (0.27 mL, 2.0 mmol), 3-fluoronitrobenzene (0.26 mL, 1.2 equiv.), K2CO3 (820 mg, 3 equiv.) and DMA (4 mL) was stirred at rt for 15 min and then at 70 °C for 2 h. Ethyl acetate (20 mL) was added, followed by water (4 mL) and brine (10 mL). After 10 min, the aqueous layer was removed, and the organic layer was washed with brine (2 × 6 mL). Celite (3 g) was added to the organic layer and the solvent evaporated. Flash chromatography using silica gel with an ethyl acetate/hexanes gradient provided 620 mg of a yellow oil. Rf = 0.44 (20% ethyl acetate/hexanes; UV active). To the solution of the obtained oil in ethanol (6 mL) was added SnCl2 dihydrate (2.2 g, 5 equiv.), and the mixture was heated at 50 °C for 15 h. Most of the solvent was evaporated (rotary evaporator) and CH2Cl2 (20 mL) was added, followed by slow addition of saturated NaHCO3 solution (20 mL, vigorous gas evolution). Then, 6 N NaOH (1 mL) was added, and the mixture was stirred vigorously for 30 min. Water (20 mL) was added and the layers separated (a solid is removed with the aqueous layer). The aqueous layer was extracted with CH2Cl2 (2 × 10 mL). Celite (3 g) was added to the combined organic layers and the solvent evaporated. Flash chromatography using silica gel with an ethyl acetate/hexanes gradient provided 459 mg (80%) of a clear oil. Rf = 0.26 (20% ethyl acetate/hexanes; UV active). LC/MS (m/z) 288.0 (M+1), >95% at 4.69 min.
N-{3-[2-Cyano-3-(trifluoromethyl)phenoxy]phenyl}-4,4,4-trifluorobutane-1-sulfonamide(4). The title compound was prepared by the general procedure A from 3A (50 mg, 0.18 mmol), 4,4,4-trifluorobutane-1-sulfonyl chloride (46 mg, 1.2 equiv.), and pyridine (0.030 mL, 2 equiv.) to provide 42 mg (53%) of an off-white crystalline solid, mp 96–97 °C. Rf = 0.34 (40% ethyl acetate/hexanes; UV active). 1H NMR (300 MHz, CDCl3) δ 7.57–7.73 (m, 1H), 7.51 (d, J = 8.1 Hz, 1H), 7.41 (t, J = 8.4 Hz, 1H), 7.13 (d, J = 8.3 Hz, 1H), 7.04–7.08 (m, 2H), 6.89 (d, J = 8.5 Hz, 1H), 6.83 (s, 1H), 3.24 (t, J = 7.4 Hz, 2H), 2.21–2.43 (m, 2H), 2.05–2.20 (m, 2H). MS (m/z) 451.3 (M-1). HPLC 98% at 15.80 min.
N-{3-[2-Chloro-3-(trifluoromethyl)phenoxy]phenyl}-4,4,4-trifluorobutane-1-sulfonamide(5). The title compound was prepared by the general procedure A from 3D (44 mg, 0.15 mmol), 4,4,4-trifluorobutane-1-sulfonyl chloride (38 mg, 1.2 equiv.), and triethylamine (0.027 mL, 1.3 equiv.) to provide 43 mg (62%) of a colorless oil. Rf = 0.36 (30% ethyl acetate/hexanes; UV active). 1H NMR (300 MHz, CDCl3) δ 7.46–7.63 (m, 1H), 7.28–7.42 (m, 2H), 7.14–7.24 (m, 1H), 6.95 (d, J = 7.7 Hz, 1H), 6.86 (s, 1H), 6.75 (d, J = 7.7 Hz, 1H), 6.58 (s, 1H), 3.20 (br s, 2H), 2.18–2.41 (m, 2H), 2.06–2.17 (m, 2H). MS (m/z) 460.3 (M-1). HPLC 96% at 17.00 min.
N-[3-(2-Cyanophenoxy)phenyl]-4,4,4-trifluorobutane-1-sulfonamide(6). The title compound was prepared by the general procedure A from 3C (32 mg, 0.15 mmol), 4,4,4-trifluorobutane-1-sulfonyl chloride (38 mg, 1.2 equiv.), and triethylamine (0.027 mL, 1.3 equiv.) to provide 17 mg (30%) of a colorless oil. Rf = 0.25 (30% ethyl acetate/hexanes; UV active). 1H NMR (300 MHz, CDCl3) δ 7.68 (d, J = 7.7 Hz, 1H), 7.48–7.59 (m, 1H), 7.31–7.43 (m, 1H), 7.16–7.25 (m, 1H), 6.92–7.07 (m, 3H), 6.86 (d, J = 9.0 Hz, 1H), 6.62 (s, 1H), 3.22 (t, J = 7.4 Hz, 2H), 2.19–2.41 (m, 2H), 2.06–2.19 (m, 2H). MS (m/z) 383.3 (M-1). HPLC > 99% at 14.95 min.
N-{3-[2-Cyano-3-(trifluoromethyl)phenoxy]phenyl}pentane-1-sulfonamide(7). The title compound was prepared by the general procedure A from 3A (42 mg, 0.15 mmol), n-pentane-1-sulfonyl chloride (0.026 mL, 1.2 equiv.), and pyridine (0.027 mL, 2.2 equiv.) to provide 60 mg (97%) of a colorless oil. Rf = 0.49 (40% ethyl acetate/hexanes; UV active). 1H NMR (300 MHz, CDCl3) δ 7.55–7.70 (m, 1H), 7.50 (d, J = 8.1 Hz, 1H), 7.39 (t, J = 8.5 Hz, 1H), 7.12 (d, J = 8.1 Hz, 1H), 6.98–7.08 (m, 2H), 6.87 (d, J = 8.5 Hz, 1H), 6.75 (s, 1H), 3.15 (t, J = 7.5 Hz, 2H), 1.74–1.91 (m, 2H), 1.21–1.47 (m, 4H), 0.89 (t, J = 7.4 Hz, 3H). MS (m/z) 413.4 (M+1), 411.4 (M-1). HPLC > 99% at 16.46 min.
N-{3-[2-Cyano-3-(trifluoromethyl)phenoxy]phenyl}-1-phenylmethanesulfonamide(8). The title compound was prepared by the general procedure A from 3A (45 mg, 0.15 mmol), 1-phenylmethanesulfonyl chloride (35 mg, 1.2 equiv.), and triethylamine (0.027 mL, 1.3 equiv.) to provide 23 mg (36%) of an off-white crystalline solid, mp 131–132 °C. Rf = 0.48 (40% ethyl acetate/hexanes; UV active). 1H NMR (300 MHz, CDCl3) δ 7.62 (br s, 1H), 7.52 (br s, 1H), 7.36 (br s, 6H), 7.09 (d, J = 7.4 Hz, 1H), 6.89–6.98 (m, 1H), 6.84 (br s, 2H), 6.48 (s, 1H), 4.39 (s, 2H). MS (m/z) 431.2 (M-1). HPLC > 99% at 15.87 min.
N-{3-[2-Chloro-3-(trifluoromethyl)phenoxy]phenyl}-1-phenylmethanesulfonamide(9). The title compound was prepared by the general procedure A from 3D (44 mg, 0.15 mmol), 1-phenylmethanesulfonyl chloride (35 mg, 1.2 equiv.), and triethylamine (0.027 mL, 1.3 equiv.) to provide 23 mg (35%) of an off-white crystalline solid, mp 129–130 °C. Rf = 0.36 (30% ethyl acetate/hexanes; UV active). 1H NMR (300 MHz, CDCl3) δ 7.48–7.60 (m, 1H), 7.21–7.46 (m, 8H), 6.89 (d, J = 7.5 Hz, 1H), 6.69–6.77 (m, 2H), 6.32 (s, 1H), 4.36 (s, 2H). MS (m/z) 440.4 (M-1). HPLC > 99% at 17.25 min.
N-[3-(2-Cyanophenoxy)phenyl]-1-phenylmethanesulfonamide(10). The title compound was prepared by the general procedure A from 3C (32 mg, 0.15 mmol), 1-phenylmethanesulfonyl chloride (35 mg, 1.2 equiv.), and triethylamine (0.027 mL, 1.3 equiv.) to provide 12 mg (22%) of a colorless oil. Rf = 0.25 (30% ethyl acetate/hexanes; UV active). 1H NMR (300 MHz, CDCl3) δ 7.69 (d, J = 7.7 Hz, 1H), 7.48–7.59 (m, 1H), 7.27–7.41 (m, 6H), 7.15–7.24 (m, 1H), 6.89–7.00 (m, 2H), 6.79–6.88 (m, 2H), 6.34 (s, 1H), 4.38 (s, 2H). MS (m/z) 363.4 (M-1). HPLC > 98% at 15.00 min.
1-(2-Chlorophenyl)-N-{3-[2-cyano-3-(trifluoromethyl)phenoxy]phenyl}methanesulfonamide(11). The title compound was prepared by the general procedure A from 3A (42 mg, 0.15 mmol), 1-(2-chlorophenyl)methanesulfonyl chloride (41 mg, 1.2 equiv.), and pyridine (0.036 mL, 3 equiv.) to provide 43 mg (61%) of a white crystalline solid, mp 121–122 °C. Rf = 0.24 (30% ethyl acetate/hexanes; UV active). 1H NMR (300 MHz, CDCl3) δ 7.55–7.69 (m, 1H), 7.49 (d, J = 7.5 Hz, 1H), 7.38–7.46 (m, 1H), 7.24–7.37 (m, 3H), 7.06 (d, J = 8.5 Hz, 1H), 6.94 (d, J = 8.7 Hz, 1H), 6.91 (s, 1H), 6.82 (s, 1H), 6.80 (d, J = 8.7 Hz, 1H), 4.63 (s, 2H). MS (m/z) 465.2 (M-1). HPLC 96% at 16.24 min.
1-(3-Chlorophenyl)-N-{3-[2-cyano-3-(trifluoromethyl)phenoxy]phenyl}methanesulfonamide(12). The title compound was prepared by the general procedure A from 3A (42 mg, 0.15 mmol), 1-(3-chlorophenyl)methanesulfonyl chloride (41 mg, 1.2 equiv.), and pyridine (0.036 mL, 3 equiv.) to provide 46 mg (66%) of an off-white crystalline solid, mp 144–145 °C. Rf = 0.28 (30% ethyl acetate/hexanes; UV active). 1H NMR (300 MHz, CDCl3) δ 7.63 (t, J = 8.1 Hz, 1H), 7.51 (d, J = 7.9 Hz, 1H), 7.28–7.45 (m, 3H), 7.07–7.21 (m, 2H), 6.82–7.02 (m, 3H), 6.69 (s, 1H), 4.35 (s, 2H). MS (m/z) 465.2 (M-1). HPLC 96% at 16.38 min.
1-(4-Chlorophenyl)-N-{3-[2-cyano-3-(trifluoromethyl)phenoxy]phenyl}methanesulfonamide(13). The title compound was prepared by the general procedure A from 3A (42 mg, 0.15 mmol), 1-(4-chlorophenyl)methanesulfonyl chloride (41 mg, 1.2 equiv.), and pyridine (0.036 mL, 3 equiv.) to provide 45 mg (64%) of a white amorphous solid, mp 115–116 °C. Rf = 0.27 (30% ethyl acetate/hexanes; UV active). 1H NMR (300 MHz, CDCl3) δ 7.58–7.71 (m, 1H), 7.52 (d, J = 7.5 Hz, 1H), 7.35–7.43 (m, 1H), 7.31 (d, J = 8.2 Hz, 2H), 7.20 (d, J = 8.2 Hz, 2H), 7.12 (d, J = 8.5 Hz, 1H), 6.90–7.00 (m, 2H), 6.86 (d, J = 7.4 Hz, 1H), 6.69 (s, 1H), 4.26–4.44 (m, 2H). MS (m/z) 465.2 (M-1). HPLC 95% at 16.47 min.
N-{3-[2-Cyano-3-(trifluoromethyl)phenoxy]phenyl}-3-(trifluoromethoxy)benzene-1-sulfonamide(14). The title compound was prepared by the general procedure A from 3A (42 mg, 0.15 mmol), 1-(3-trifluoromethoxyphenyl)methanesulfonyl chloride (0.032 mL, 1.1 equiv.), and triethylamine (0.025 mL, 1.2 equiv.) to provide 55 mg (73%) of a white crystalline solid, mp 120–121 °C. Rf = 0.26 (30% ethyl acetate/hexanes; UV active). 1H NMR (300 MHz, CDCl3) δ 7.73 (d, J = 7.7 Hz, 1H), 7.39–7.68 (m, 5H), 7.28–7.38 (m, 1H), 6.83–7.06 (m, 4H), 6.77 (s, 1H). MS (m/z) 501.6 (M-1). HPLC > 99% at 16.74 min.
N-{3-[2-Cyano-3-(trifluoromethyl)phenoxy]phenyl}-4-(trifluoromethoxy)benzene-1-sulfonamide(15). The title compound was prepared by the general procedure A from 3A (42 mg, 0.15 mmol), 1-(4-trifluoromethoxyphenyl)methanesulfonyl chloride (0.032 mL, 1.1 equiv.), and triethylamine (0.025 mL, 1.2 equiv.) to provide 47 mg (62%) of a white crystalline solid, mp 164–165 °C. Rf = 0.28 (40% ethyl acetate/hexanes; UV active). 1H NMR (300 MHz, CDCl3) δ 7.85 (d, J = 8.5 Hz, 2H), 7.54–7.68 (m, 1H), 7.50 (d, J = 7.5 Hz, 1H), 7.28–7.41 (m, 3H), 7.00 (d, J = 7.7 Hz, 1H), 6.83–6.97 (m, 3H), 6.80 (s, 1H). MS (m/z) 501.4 (M-1). HPLC > 99% at 16.80 min.
N-{3-[2-Cyano-3-(trifluoromethyl)phenoxy]phenyl}hexanamide(16). The title compound was prepared by the general procedure A from 3A (42 mg, 0.15 mmol), n-pentanecarbonyl chloride (0.025 mL, 1.2 equiv.), and triethylamine (0.027 mL, 1.3 equiv.) to provide 56 mg (99%) of a white crystalline solid, mp 95–96 °C. Rf = 0.46 (40% ethyl acetate/hexanes; UV active). 1H NMR (300 MHz, CDCl3) δ 7.50–7.62 (m, 2H), 7.45 (d, J = 7.7 Hz, 1H), 7.32–7.40 (m, 1H), 7.22 (br s, 2H), 7.12 (d, J = 8.5 Hz, 1H), 6.85 (d, J = 6.6 Hz, 1H), 2.36 (t, J = 7.5 Hz, 2H), 1.67–1.76 (m, 2H), 1.31–1.40 (m, 4H), 0.91 (t, J = 7.5 Hz, 3H). MS (m/z) 377.3 (M+1), 375.3 (M-1). HPLC > 99% at 16.86 min.
Butyl N-{3-[2-Cyano-3-(trifluoromethyl)phenoxy]phenyl}carbamate(17). The title compound was prepared by the general procedure A from 3A (42 mg, 0.15 mmol), n-butyl chloroformate (0.021 mL, 1.2 equiv.), and triethylamine (0.027 mL, 1.3 equiv.) to provide 44 mg (78%) of a colorless oil. Rf = 0.24 (30% ethyl acetate/hexanes; UV active). 1H NMR (300 MHz, CDCl3) δ 7.57 (t, J = 7.7 Hz, 1H), 7.45 (d, J = 7.7 Hz, 1H), 7.30–7.41 (m, 2H), 7.05–7.19 (m, 2H), 6.80 (dd, J = 8.1, 1.5 Hz, 1H), 6.68 (s, 1H), 4.16 (t, J = 7.4 Hz, 2H), 1.59–1.73 (m, 2H), 1.33–1.47 (m, 2H), 0.95 (t, J = 7.4 Hz, 3H). MS (m/z) 379.2 (M+1), 377.3 (M-1). HPLC > 99% at 17.21 min.
3-Butyl-1-{3-[2-cyano-3-(trifluoromethyl)phenoxy]phenyl}urea(18). The title compound was prepared by the general procedure A from 3A (42 mg, 0.15 mmol), n-butyl isocyanate (0.021 mL, 1.2 equiv.), and triethylamine (0.025 mL, 1.2 equiv.) to provide 51 mg (90%) of a colorless oil. Rf = 0.23 (40% ethyl acetate/hexanes; UV active). 1H NMR (300 MHz, CDCl3) δ 7.56 (t, J = 7.7 Hz, 1H), 7.43 (d, J = 7.7 Hz, 1H), 7.27–7.36 (m, 2H), 7.02–7.19 (m, 2H), 6.79 (s, 1H), 6.73 (d, J = 7.9 Hz, 1H), 4.94 (br s, 1H), 3.16–3.32 (m, 2H), 1.43–1.55 (m, 2H), 1.27–1.42 (m, 2H), 0.92 (t, J = 7.2 Hz, 3H). MS (m/z) 378.3 (M+1), 376.2 (M-1). HPLC > 96% at 15.68 min.
3-Butyl-1-{3-[2-cyano-3-(trifluoromethyl)phenoxy]phenyl}-3-methylurea(19). To an ice-cold solution of 3A (42 mg, 0.15 mmol) in CH2Cl2 (1 mL) was added NaHCO3 (62 mg, 5 equiv.), followed by saturated NaHCO3 solution (0.3 mL). Triphosgene (45 mg, 1 equiv.) was added, and after 10 min, the ice bath was removed, and the mixture was stirred at rt for 1 h (gas evolution). Water (0.6 mL) was added and after 10 min, the aqueous layer was removed. The organic layer dried with sodium sulfate (20 min) and filtered. Toluene (0.5 mL) was added and most of the solvent evaporated. THF (1 mL) was added, and the mixture cooled in an ice bath. N-Methyl-n-butylamine (0.053 mL, 3 equiv.) was added, followed by triethylamine (0.042 mL, 2 equiv.). The mixture was stirred at rt for 20 h. Ethyl acetate (3 mL) was added, followed by saturated NaHCO3 solution (0.8 mL) and water (0.4 mL). After 10 min, the aqueous layer was removed. Celite (600 mg) was added to the organic layer and the solvent was evaporated. Flash chromatography using silica gel with an ethyl acetate/hexanes gradient provided 58 mg (99%) of a colorless oil. Rf = 0.22 (40% ethyl acetate/hexanes; UV active). 1H NMR (300 MHz, CDCl3) δ 7.55 (t, J = 7.7 Hz, 1H), 7.37–7.47 (m, 2H), 7.28–7.37 (m, 1H), 7.05–7.20 (m, 2H), 6.70–6.84 (m, 1H), 6.41 (s, 1H), 3.34 (t, J = 7.4 Hz, 2H), 3.01 (s, 3H), 1.48–1.69 (m, 2H), 1.28–1.45 (m, 2H), 0.95 (t, J = 7.4 Hz, 3H). MS (m/z) x (M+1). HPLC > 99% at 16.33 min.
N-{3-[2-Cyano-3-(trifluoromethyl)phenoxy]phenyl}-2-phenylacetamide(20). The title compound was prepared by the general procedure A from 3A (42 mg, 0.15 mmol), phenylacetyl chloride (0.024 mL, 1.2 equiv.), and triethylamine (0.027 mL, 1.3 equiv.) to provide 61 mg (100%) of a white crystalline solid, mp 136–137 °C. Rf = 0.34 (40% ethyl acetate/hexanes; UV active). 1H NMR (300 MHz, CDCl3) δ 7.55 (br s, 1H), 7.30–7.49 (m, 8H), 7.12–7.22 (m, 2H), 7.00–7.11 (m, 1H), 6.84 (br s, 1H), 3.74 (s, 2H). MS (m/z) 397.4 (M+1), 395.5 (M-1). HPLC > 99% at 16.07 min.
N-Cyclohexyl({3-[2-cyano-3-(trifluoromethyl)phenoxy]phenyl}m amino)sulfonamide(21). The title compound was prepared by the general procedure A from 3A (42 mg, 0.15 mmol), cyclohexylsulfamoyl chloride (45 mg, 1.5 equiv.), and triethylamine (0.027 mL, 1.3 equiv.) to provide 63 mg (96%) of a white amorphous solid, mp 199–200 °C. Rf = 0.46 (40% ethyl acetate/hexanes; UV active). 1H NMR (300 MHz, CDCl3) δ 7.53–7.70 (m, 1H), 7.47 (d, J = 7.5 Hz, 1H), 7.30–7.42 (m, 1H), 7.08 (d, J = 8.5 Hz, 1H), 7.00 (br s, 3H), 6.81 (d, J = 7.5 Hz, 1H), 4.66 (d, J = 6.8 Hz, 1H), 3.13–3.38 (m, 1H), 1.77–1.96 (m, 2H), 1.65 (br s, 2H), 1.47–1.59 (m, 1H), 1.06–1.37 (m, 5H). 13C NMR (100 MHz, CDCl3) δ 161.2, 155.3, 139.7, 134.7, 134.2, 131.2, 120.6, 120.2, 115.8, 115.3, 112.3, 110.6, 101.1, 53.5, 33.8, 25.2, 24.7. MS (m/z) 438.3 (M-1). HPLC > 99% at 16.17 min.
N-{4-[2-Cyano-3-(trifluoromethyl)phenoxy]phenyl}pentane-1-sulfonamide(22). The title compound was prepared by the general procedure A from 3B (42 mg, 0.15 mmol), n-pentane-1-sulfonyl chloride (0.026 mL, 1.2 equiv.), and triethylamine (0.027 mL, 1.3 equiv.) to provide 53 mg (86%) of a white crystalline solid, mp 94–95 °C. Rf = 0.49 (40% ethyl acetate/hexanes; UV active). 1H NMR (300 MHz, CDCl3) δ 7.59 (t, J = 8.1 Hz, 1H), 7.48 (d, J = 7.7 Hz, 1H), 7.29 (d, J = 8.9 Hz, 2H), 7.11 (d, J = 9.0 Hz, 2H), 7.07 (d, J = 8.7 Hz, 1H), 6.49 (s, 1 H), 2.96–3.21 (m, 2H), 1.77–1.94 (m, 2H), 1.24–1.48 (m, 4H), 0.90 (t, J = 7.4 Hz, 3H). MS (m/z) 413.1 (M+1), 411.3 (M-1). HPLC > 99% at 16.38 min.
N-{4-[2-Cyano-3-(trifluoromethyl)phenoxy]phenyl}-1-phenylmethanesulfonamide(23). The title compound was prepared by the general procedure A from 3B (42 mg, 0.15 mmol), 1-phenylmethanesulfonyl chloride (35 mg, 1.2 equiv.), and triethylamine (0.027 mL, 1.3 equiv.) to provide 28 mg (43%) of an off-white crystalline solid, mp 166–167 °C. Rf = 0.44 (40% ethyl acetate/hexanes; UV active). 1H NMR (300 MHz, CDCl3) δ 7.55–7.68 (m, 1H), 7.48 (d, J = 7.7 Hz, 1H), 7.27–7.42 (m, 6H), 7.15 (d, J = 7.9 Hz, 2H), 7.00–7.13 (m, 3H), 6.47 (s, 1H), 4.37 (s, 2H). MS (m/z) 431.2 (M-1). HPLC > 99% at 15.89 min.
N-{4-[2-Cyano-3-(trifluoromethyl)phenoxy]phenyl}-2-phenylacetamide(24). The title compound was prepared by the general procedure A from 3B (42 mg, 0.15 mmol), phenylacetyl chloride (0.024 mL, 1.2 equiv.), and triethylamine (0.027 mL, 1.3 equiv.) to provide 49 mg (82%) of a white crystalline solid, mp 200–201 °C. Rf = 0.31 (40% ethyl acetate/hexanes; UV active). 1H NMR (300 MHz, CDCl3) δ 7.46–7.60 (m, 3H), 7.27–7.45 (m, 6H), 7.19 (br s, 1H), 6.93–7.11 (m, 3H), 3.77 (s, 2H). MS (m/z) 397.3 (M+1), 395.4 (M-1). HPLC > 99% at 15.91 min.
4.1. cAMP Accumulation Assay
The cAMP assays were performed in Chinese Hamster Ovary (CHO) cells (Perkin Elmer, Cat # ES-110-C) stably expressing the human CB1 receptor (hCB1) cultured under standard cell culture conditions (37 °C, 5% carbon dioxide, DMEM media with 1% Penicillin/Streptomycin and 400 µg/mL G418) using the Lance™ assay kit and manufacturer’s instructions were closely followed (Perkin Elmer). In brief, stimulation buffer containing 1X Hank’s Balanced Salt Solution (HBSS), 5 mM HEPES, 0.1% BSA stabilizer, and 0.5 mM final IBMX was prepared and titrated to pH 7.4 at rt. Serial dilutions of the test compounds and 300 nM forskolin, both prepared at 4× the desired final concentration in stimulation buffer, were added to a 96-well white ½ area microplate (PerkinElmer). The CHO-hCB1 cells were lifted with a non-enzymatic solution (Cell-stripper, Mediatech Inc., Orlando, FL, USA), and 4000 cells were added to each well. After incubating for 30 min at room temperature, Eu-cAMP tracer and uLIGHT-anti-cAMP working solutions were added per the manufacturer’s instructions. After incubation for 1 h, the TR-FRET signal (ex 337 nm, em 620 and 650 nm) was read on a CLARIO star multimode plate reader (BMG Biotech, Cary, NC, USA). Data were analyzed using Prism software (GraphPad, La Jolla, CA, USA). Nonlinear regression analysis was performed to fit data and obtain maximum response (Emax), EC50, correlation coefficient (r2), and other parameters. All experiments were performed in duplicate 2–3 times to ensure reproducibility and data are reported as mean ± standard error of mean unless noted otherwise.
4.2. Radioligand Displacement Assay
Further characterization of
21 was performed using radioligand displacement of [
3H]CP55940 and equilibrium dissociation constant (Ki) value was determined as described previously [
27,
28]. Data reported are average values from 3 measurements with <30% standard error.
4.3. Pharmacokinetic Testing
Female C57BL/6 mice were bred in house and used at ~10 weeks of age for pharmacokinetic (PK) testing. Three animals were tested per time point. Doses were formulated in 2% NMP in canola oil, and all compounds were delivered at 3 mg/kg by intraperitoneal injection (IP). Tissues were taken at 0.5, 1, 2, and 4 h post dose. Animals were subjected to whole body perfusion using saline prior to tissue collection. Brain samples were homogenized with 50:50 ethanol:water (1:5, v/v). Forty µL of the homogenate, 10 µL of acetonitrile, and 150 µL of 100 ng/mL reserpine in acetonitrile containing 0.1% formic acid were vortexed and centrifuged. Plasma samples were diluted with 10 µL of acetonitrile, 150 µL of 100 ng/mL reserpine in acetonitrile containing 0.1% formic acid, vortexed and centrifuged. Samples were subjected to LC/MS/MS analysis. Standards were prepared in blank samples and used for calibration curves. Chromatography was performed using a Phenomenex Luna C18 column.
4.4. Computational Methods
Docking and Induced Fit. Two parallel and independent approaches with different scoring functions, Schrodinger’s XP/Induced Fit [
29] and Autodock VINA with an AMBER18 [
30] molecular mechanics generalized Born surface area MMGBSA rescoring, were used to provide predictions for ligand configurations allowing for both ligand and receptor binding site flexibility. Both methods include desolvation components in the scoring function. Following initial GLIDE-XP docking, we probed the importance of binding site flexibility within a 5 Å window of any atom in the best docking poses using Schrodinger’s Induced Fit [
29]. The docking box employed for VINA was ca 15 × 15 × 15 Å
3 in spatial extent, while the default box size employed in computing the GLIDE-XP docking grid was based on the ligand size. We employ Autodock VINA’s scoring function scores based on hydrophobic contacts, hydrogen bonds, lack of steric clash. This informatics-based scoring function serves primarily as a means of selecting the most plausible collection of poses from a large number of sampled docked configurations which we rescore using AMBER-MMGBSA. We collected 20 poses per ligand with a high “exhaustiveness” setting of 80. Similarly, the GLIDE-XP workflow and scoring function was used to obtain up to 20 poses per ligand for post-docking minimization but retaining the best Emodel scored 5 poses in the final analysis. The GLIDE-XP scoring function contains terms such as desolvation, lipophilic/hydrophobic contact and cavity costs, in addition to physics based coulombic and van der Waals and ligand strain terms.
For the initial docking/induced-fit and MMGBSA-rescoring phase we chose to use the model completed 5XR8 structure employing simulated annealing with topological/stereochemical constraints in Modeller, followed by SCWRL rotameric sidechain adjustments for loop/terminus modeled residues. We then used AMBER18 employing the AMBER14SB forcefield to prepare the full-length All-H model as described in prior publications [
31,
32]. The structure prepared with TLEAP was then energy minimized for 8000 steps of conjugate gradient following a 400 steepest-descents minimization to remove initial inferior contacts.
Docked poses above were re-scored using an AMBER 18 MMGBSA and Prime/MMGBSA approaches. This was performed because the top ranked docked poses are often not the crystallographically observed pose as observed by us and others in PDB-Bind assessments [
33], whereas frequently the lowest MMGBSA/MMPBSA scored pose has lowest RMSD to the known crystallographic solution and provides the best affinity correlations [
34,
35,
36].
MD simulation. An MD simulation was employed to track pose stability and ligand induced receptor binding site changes employing AMBER18 using ff14SB parameterization for protein residues with GAFF generalized forcefield parameters for internal forcefields of the ligand. ANTECHAMBER and PARMGEN were used to incorporate AM1-BCC charges into the ligand antechamber topology and parameter files. The LIPID14 forcefield was used for parameterization of 150 Å × 150 Å × 143 Å box of DOPC lipids (initially 323 lipids top and bottom). Elastic band pulls of the N− and C− terminal regions and simulated annealing were used to prepare low energy more compact conformations beyond initial extended constructed conformations after the MODELLER/SCWRL preparation steps. The box was used to immerse the CB1/8D0 complex and dynamics was used a method of ‘refining’ the N−/C− and loop conformations following production dynamics. The complex system was then energy minimized by a combination of 800 steepest descents followed by 8000 conjugate gradient steps. The system was then slowly heated to 300K over 200 ps followed by 5 ns of early equilibration under NPT ensemble simulation conditions followed by production dynamics under NVT conditions for 850 ns after establishing equilibration from the standpoint of RMSD variations from the starting structure and energetic and fluctuation criteria.
Native Contact Time Dependent Pharmacophore. Native contact analysis was conducted employing AMBER cpptraj, bash scripting and C++ code to determine the percentage of time of used defined regions in the 8D0/AM841 ligand contacted residues in the binding site. The percentage time of each region of the ligand with amino acids was computed from 850 ns of production dynamics, and a representation of those percentages was presented in a colored heatmap representation.