Macrophage-Targeted Punicalagin Nanoengineering to Alleviate Methotrexate-Induced Neutropenia: A Molecular Docking, DFT, and MD Simulation Analysis
Abstract
:1. Introduction
2. Methodology
2.1. Protein and Ligand Preparation
2.2. Molecular Docking Studies
2.3. QM-Based Density Functional Theory Analysis
2.4. Molecular Dynamics Simulations
3. Results
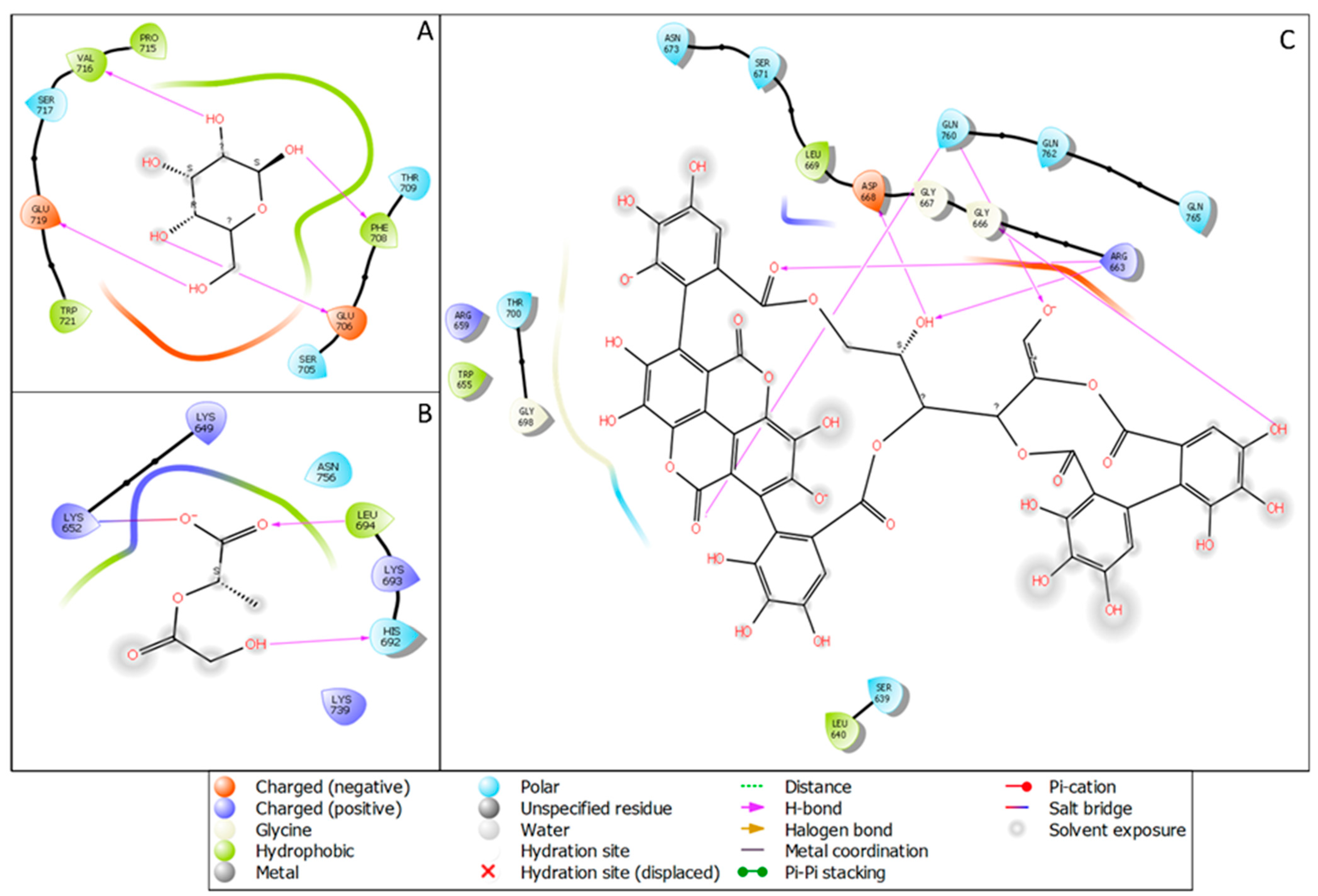
3.1. Interaction Analysis
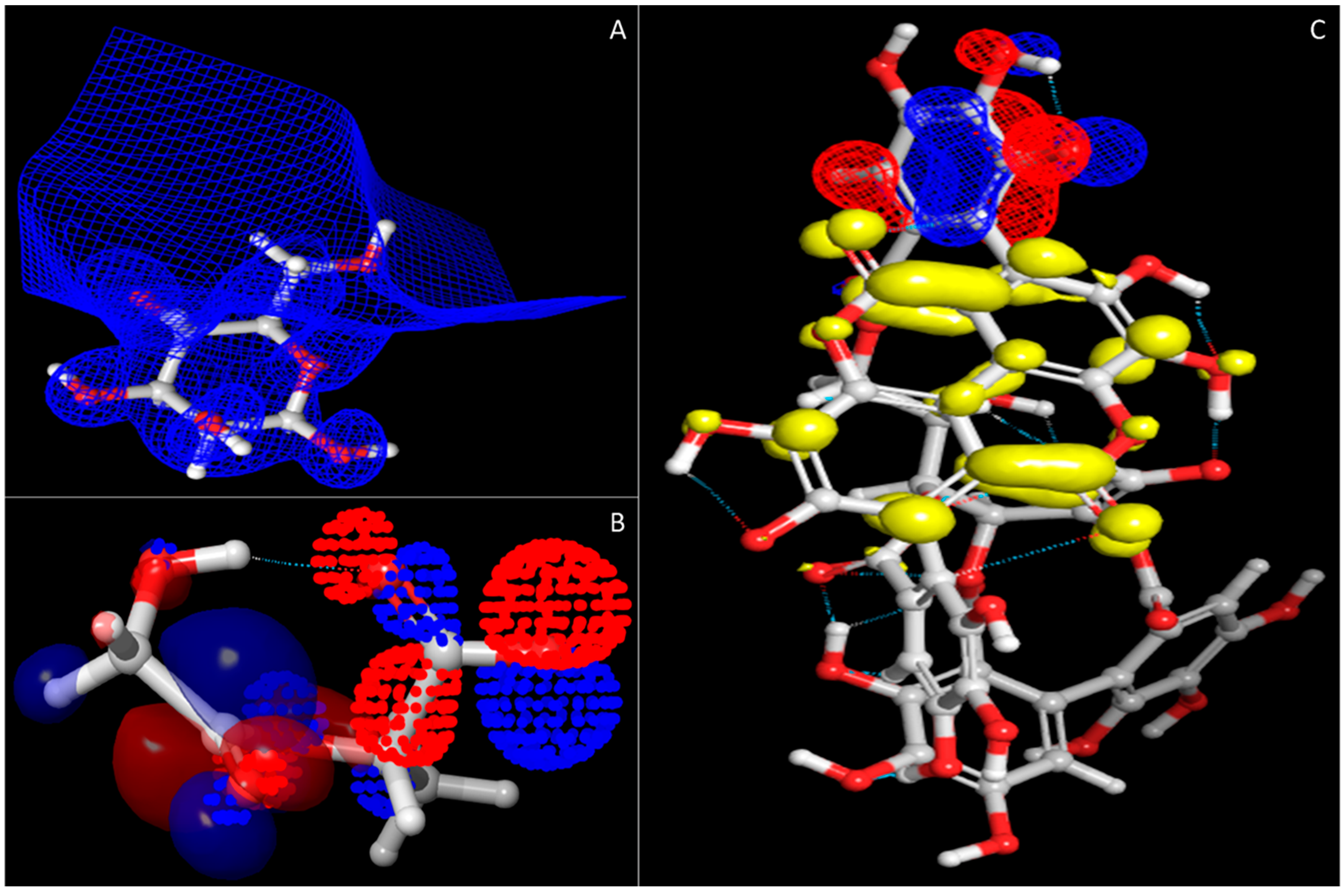
3.2. Density Functional Theory and HOMO–LUMO Analysis
3.3. Molecular Dynamics Simulation
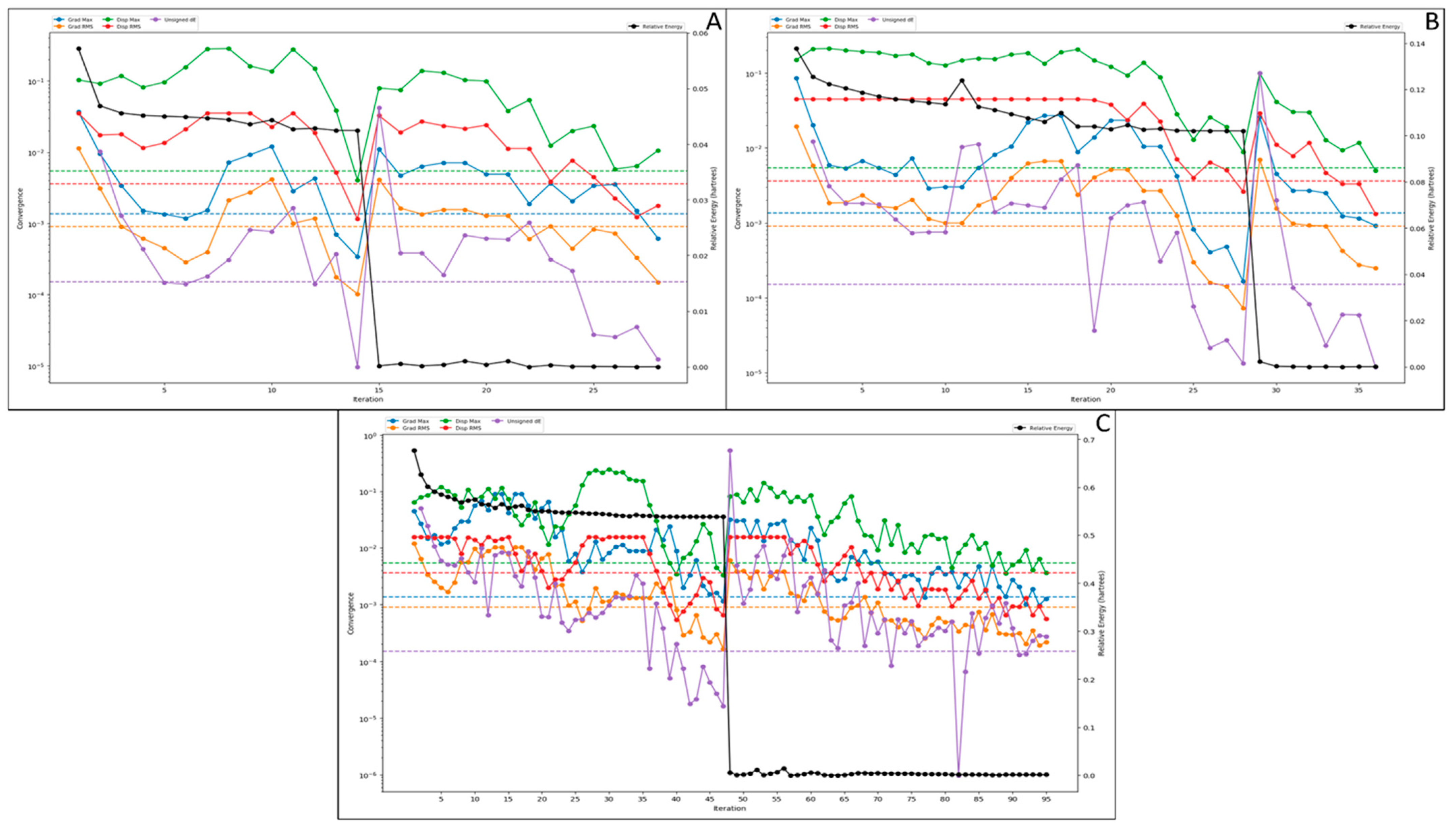
3.3.1. Root Mean Square Deviation
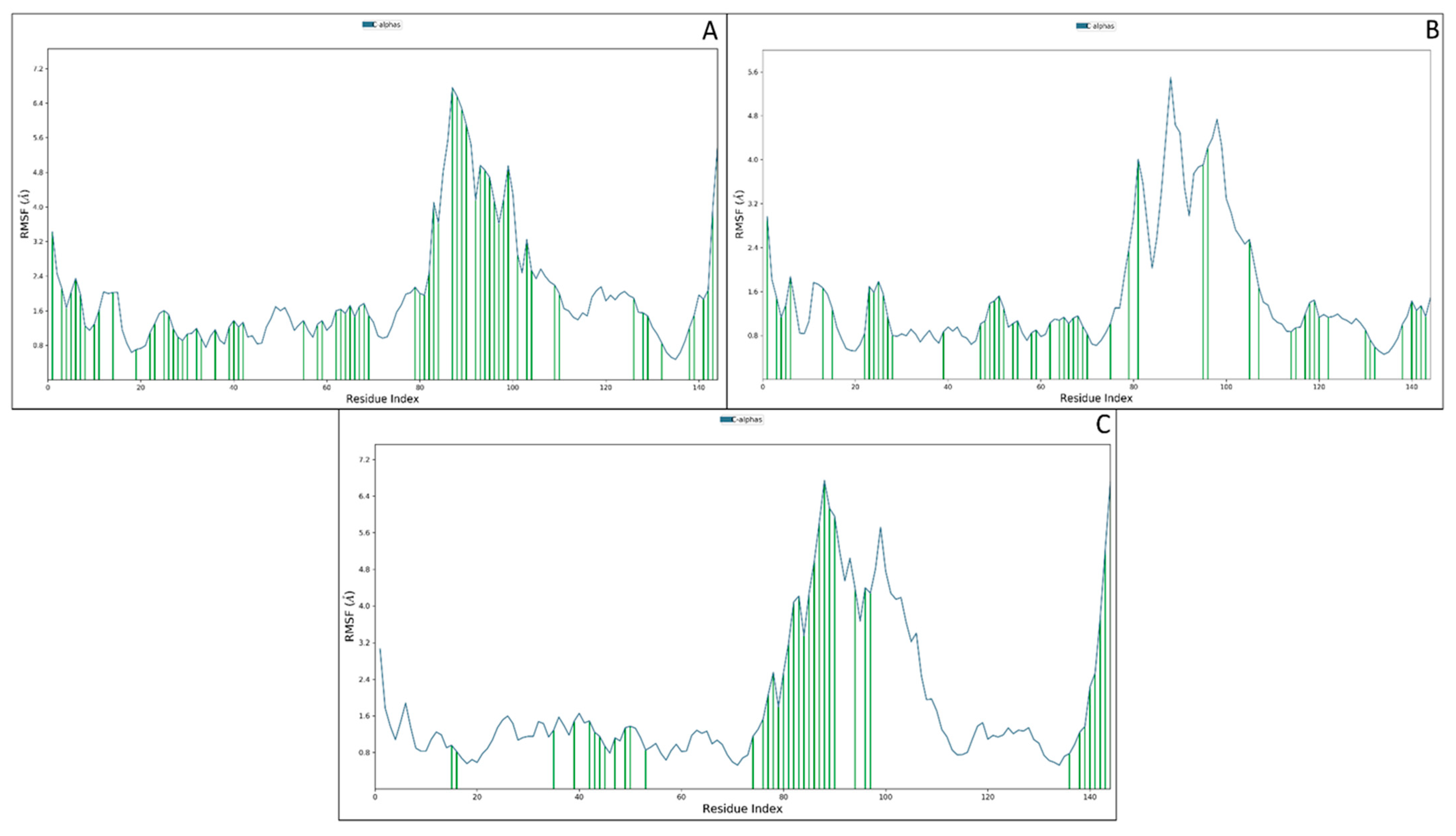
3.3.2. Root Mean Square Fluctuations
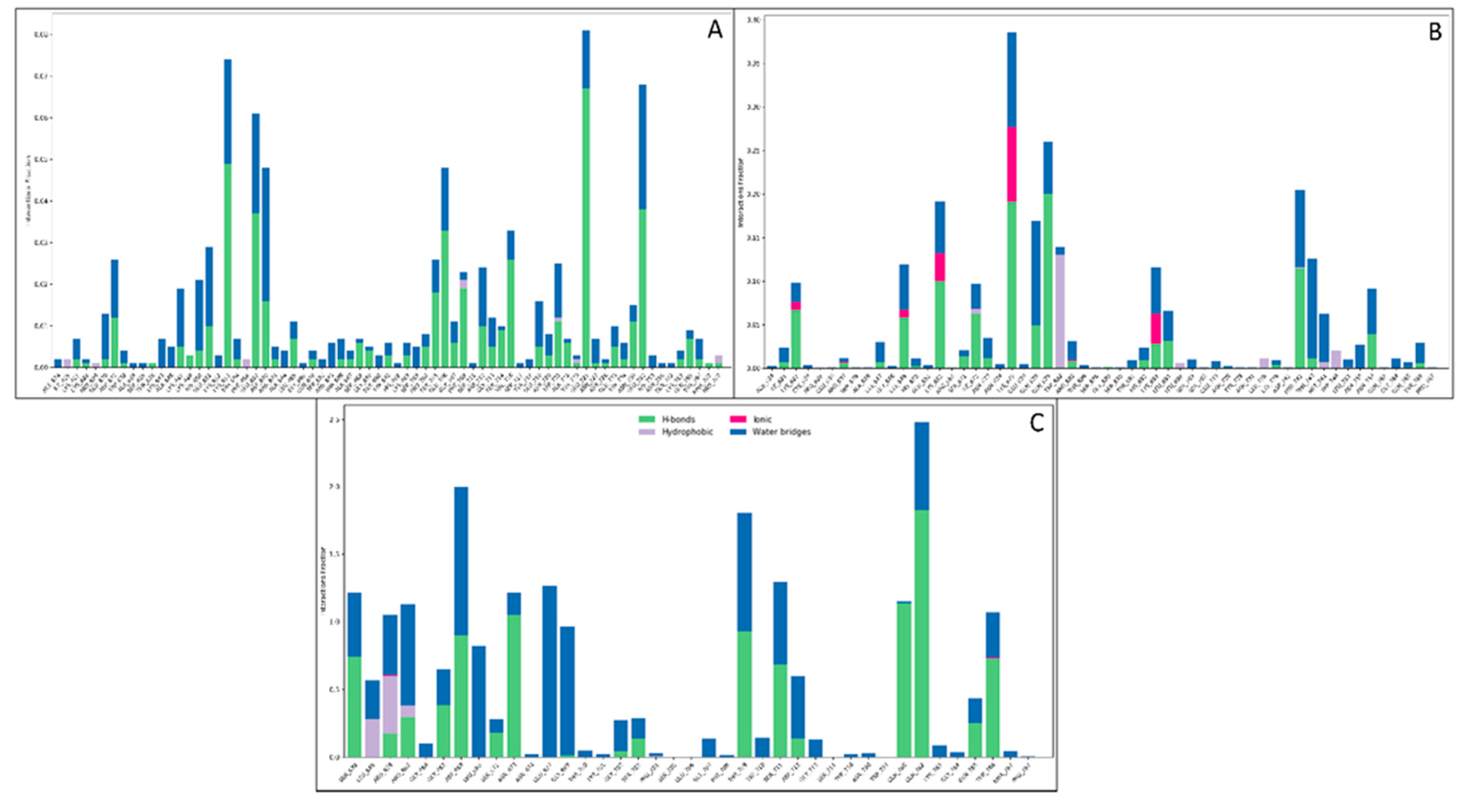
3.3.3. Simulative Interaction Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tran, S.; DeGiovanni, P.J.; Piel, B.; Rai, P. Cancer nanomedicine: A review of recent success in drug delivery. Clin. Transl. Med. 2017, 6, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Remon, B.; Houri, M.; El Achy, S.; Kamel, S.; Refaat, T. Cancer active targeting by nanoparticles: A comprehensive review of literature. J. Cancer Res. Clin. Oncol. 2016, 141, 769. [Google Scholar]
- Lookman, T.; Balachandran, P.V.; Xue, D.; Yuan, R. Active learning in materials science with emphasis on adaptive sampling using uncertainties for targeted design. NPJ Comput. Mater. 2019, 5, 1–17. [Google Scholar] [CrossRef]
- Wijeratne, P.A.; Vavourakis, V. A quantitative in silico platform for simulating cytotoxic and nanoparticle drug delivery to solid tumours. Interface Focus 2019, 9, 20180063. [Google Scholar] [CrossRef]
- Ramlal, A.; Ahmad, S.; Kumar, L.; Khan, F.N.; Chongtham, R. From molecules to patients: The clinical applications of biological databases and electronic health record. In Translational Bioinformatics in Healthcare and Medicine; Elsevier: Amsterdam, The Netherlands, 2021; pp. 107–125. [Google Scholar]
- Lim, A.; Gaffney, K.; Scott, D. Methotrexate-induced pancytopenia: Serious and under-reported? Our experience of 25 cases in 5 years. Rheumatology 2005, 44, 1051–1055. [Google Scholar] [CrossRef]
- Toth, P.; Bernd, R. Severe leukopenia in a rheumatoid arthritis patient treated with a methotrexate/leflunomide combination. Rev. Bras. Reumatol. Engl. Ed. 2014, 54, 152–154. [Google Scholar] [CrossRef]
- Advani, S.; Achreckar, S.; Thomas, D.; Krishnankutty, B. Granulocyte colony-stimulating factor (filgrastim) in chemotherapy-induced febrile neutropenia. Indian J. Med Paediatr. Oncol. 2010, 31, 79–82. [Google Scholar] [CrossRef]
- Atrahimovich, D.; Khatib, S.; Sela, S.; Vaya, J.; Samson, A.O. Punicalagin induces serum low-density lipoprotein influx to macrophages. Oxidative Med. Cell. Longev. 2016, 2016, 7124251. [Google Scholar] [CrossRef]
- Rosillo, M.A.; Alarcón-de-la-Lastra, C.; Sánchez-Hidalgo, M. An update on dietary phenolic compounds in the prevention and management of rheumatoid arthritis. Food Funct. 2016, 7, 2943–2969. [Google Scholar] [CrossRef]
- Karwasra, R.; Singh, S.; Sharma, D.; Sharma, S.; Sharma, N.; Khanna, K. Pomegranate supplementation attenuates inflammation, joint dysfunction via inhibition of NF-κB signaling pathway in experimental models of rheumatoid arthritis. J. Food Biochem. 2019, 43, e12959. [Google Scholar] [CrossRef]
- Karwasra, R.; Kalra, P.; Gupta, Y.K.; Saini, D.; Kumar, A.; Singh, S. Antioxidant and anti-inflammatory potential of pomegranate rind extract to ameliorate cisplatin-induced acute kidney injury. Food Funct. 2016, 7, 3091–3101. [Google Scholar] [CrossRef] [PubMed]
- Quirós-Fernández, R.; López-Plaza, B.; Bermejo, L.M.; Palma-Milla, S.; Gómez-Candela, C. Supplementation with hydroxytyrosol and punicalagin improves early atherosclerosis markers involved in the asymptomatic phase of atherosclerosis in the adult population: A randomised, placebo-controlled, crossover trial. Nutrients 2019, 11, 640. [Google Scholar] [CrossRef]
- Feinberg, H.; Feinberg, H.; Park-Snyder, S.; Kolatkar, A.R.; Heise, C.T.; Taylor, M.; Weis, W. structure of a C-type carbohydrate recognition domain from the macrophage mannose receptor. J. Biol. Chem. 2000, 275, 21539–21548. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Release, S. 1: Maestro Desmond Interoperability Tools; Schrödinger: New York, NY, USA, 2020. [Google Scholar]
- Schrodinger, L. The PyMOL Molecular Graphics System, Version 1.3r1; DeLano Scientific LLC: San Francisco, CA, USA, 2010.
- Bochevarov, A.D.; Harder, E.; Hughes, T.F.; Greenwood, J.R.; Braden, D.A.; Philipp, D.M.; Rinaldo, D.; Halls, M.D.; Zhang, J.; Friesner, R.A. Jaguar: A high-performance quantum chemistry software program with strengths in life and materials sciences. Int. J. Quantum Chem. 2013, 113, 2110–2142. [Google Scholar] [CrossRef]
- Ganji, M.D.; Hosseini-Khah, S.; Amini-Tabar, Z. Theoretical insight into hydrogen adsorption onto graphene: A first-principles B3LYP-D3 study. Phys. Chem. Chem. Phys. 2015, 17, 2504–2511. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Tirado-Rives, J. The OPLS [optimised potentials for liquid simulations] potential functions for proteins, energy minimisations for crystals of cyclic peptides and crambin. J. Am. Chem. Soc. 1988, 110, 1657–1666. [Google Scholar] [CrossRef]
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; Von Bargen, C.D.; et al. OPLS4: Improving force field accuracy on challenging regimes of chemical space. J. Chem. Theory Comput. 2021, 17, 4291–4300. [Google Scholar] [CrossRef]
- Mark, P.; Nilsson, L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- McDonald, I. NpT-ensemble Monte Carlo calculations for binary liquid mixtures. Mol. Phys. 1972, 23, 41–58. [Google Scholar] [CrossRef]
- Bowers, K.J.; Chow, D.E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL, USA, 11–17 November 2006; IEEE: New York, NY, USA, 2006. [Google Scholar]
- Kaul, T.; Eswaran, M.; Ahmad, S.; Thangaraj, A.; Jain, R.; Kaul, R.; Raman, N.M.; Bharti, J. Probing the effect of a plus 1bp frameshift mutation in protein-DNA interface of domestication gene, NAMB1, in wheat. J. Biomol. Struct. Dyn. 2020, 38, 3633–3647. [Google Scholar] [CrossRef] [PubMed]
- Deisboeck, T.S.; Zhang, L.; Yoon, J.; Costa, J. In silico cancer modeling: Is it ready for prime time? Nat. Clin. Pract. Oncol. 2009, 6, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Sedykh, A.; Wang, W.; Zhao, X.; Yan, B.; Zhu, H. In silico profiling nanoparticles: Predictive nanomodeling using universal nanodescriptors and various machine learning approaches. Nanoscale 2019, 11, 8352–8362. [Google Scholar] [CrossRef] [PubMed]
- Lunnoo, T.; Assawakhajornsak, J.; Puangmali, T. In silico study of gold nanoparticle uptake into a mammalian cell: Interplay of size, shape, surface charge, and aggregation. J. Phys. Chem. C 2019, 123, 3801–3810. [Google Scholar] [CrossRef]
- Weiss, C.; Carriere, M.; Fusco, L.; Capua, I.; Regla-Nava, J.A.; Pasquali, M.; Scott, J.A.; Vitale, F.; Unal, M.A.; Mattevi, C. Toward nanotechnology-enabled approaches against the COVID-19 pandemic. ACS Nano 2020, 14, 6383–6406. [Google Scholar] [CrossRef]
- Ahmad, S.; Chitkara, P.; Khan, F.N.; Kishan, A.; Alok, V.; Ramlal, A.; Mehta, S. Mobile technology solution for COVID-19: Surveillance and prevention. In Computational Intelligence Methods in COVID-19: Surveillance, Prevention, Prediction and Diagnosis; Springer: Berlin/Heidelberg, Germany, 2021; pp. 79–108. [Google Scholar]
- Khan, F.N.; Khanam, A.A.; Ramlal, A.; Ahmad, S. A Review on Predictive Systems and Data Models for COVID-19. In Computational Intelligence Methods in COVID-19: Surveillance, Prevention, Prediction and Diagnosis; Springer: Berlin/Heidelberg, Germany, 2021; pp. 123–164. [Google Scholar]
- Ou, L.; Corradi, V.; Tieleman, D.P.; Liang, Q. Atomistic simulations on interactions between amphiphilic Janus nanoparticles and lipid bilayers: Effects of lipid ordering and leaflet asymmetry. J. Phys. Chem. B 2020, 124, 4466–4475. [Google Scholar] [CrossRef]
- Casalini, T.; Limongelli, V.; Schmutz, M.; Som, C.; Jordan, O.; Wick, P.; Borchard, G.; Perale, G. Molecular modeling for nanomaterial–biology interactions: Opportunities, challenges, and perspectives. Front. Bioeng. Biotechnol. 2019, 7, 268. [Google Scholar] [CrossRef] [Green Version]


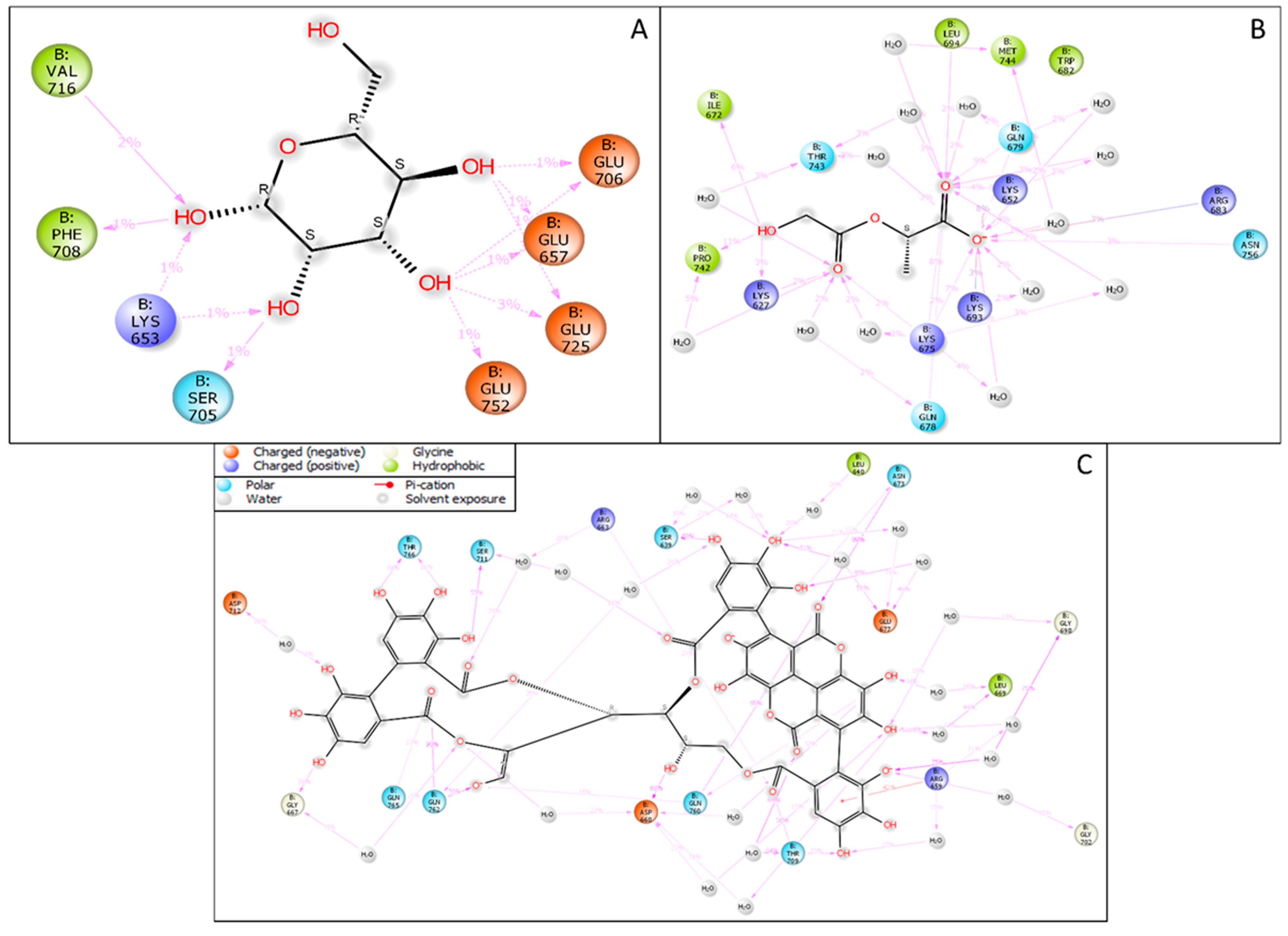
| PDB ID | Ligand Name | PubChem CID | Docking Score | MM\GBSA dG Bind | Ligand Efficiency Sa | Ligand Efficiency ln | Glide Evdw | Glide Ecoul | Prime Hbond | H-Bond Count |
|---|---|---|---|---|---|---|---|---|---|---|
| 1EGI | Mannose | 18950 | −5.811 | −19.28 | −1.109 | −1.667 | −6.965 | −20.241 | −68.76 | 4 |
| 1EGI | PLGA | 23111554 | −4.334 | −18.38 | −0.934 | −1.312 | −6.634 | −17.529 | −68 | 2 |
| 1EGI | Punicalagin | 44584733 | −2.931 | −14.1 | −0.161 | −0.547 | −31.984 | −13.778 | −70.84 | 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karwasra, R.; Ahmad, S.; Bano, N.; Qazi, S.; Raza, K.; Singh, S.; Varma, S. Macrophage-Targeted Punicalagin Nanoengineering to Alleviate Methotrexate-Induced Neutropenia: A Molecular Docking, DFT, and MD Simulation Analysis. Molecules 2022, 27, 6034. https://doi.org/10.3390/molecules27186034
Karwasra R, Ahmad S, Bano N, Qazi S, Raza K, Singh S, Varma S. Macrophage-Targeted Punicalagin Nanoengineering to Alleviate Methotrexate-Induced Neutropenia: A Molecular Docking, DFT, and MD Simulation Analysis. Molecules. 2022; 27(18):6034. https://doi.org/10.3390/molecules27186034
Chicago/Turabian StyleKarwasra, Ritu, Shaban Ahmad, Nagmi Bano, Sahar Qazi, Khalid Raza, Surender Singh, and Saurabh Varma. 2022. "Macrophage-Targeted Punicalagin Nanoengineering to Alleviate Methotrexate-Induced Neutropenia: A Molecular Docking, DFT, and MD Simulation Analysis" Molecules 27, no. 18: 6034. https://doi.org/10.3390/molecules27186034
APA StyleKarwasra, R., Ahmad, S., Bano, N., Qazi, S., Raza, K., Singh, S., & Varma, S. (2022). Macrophage-Targeted Punicalagin Nanoengineering to Alleviate Methotrexate-Induced Neutropenia: A Molecular Docking, DFT, and MD Simulation Analysis. Molecules, 27(18), 6034. https://doi.org/10.3390/molecules27186034







