A Label-Free Fluorescence Aptasensor Based on G-Quadruplex/Thioflavin T Complex for the Detection of Trypsin
Abstract
1. Introduction
2. Results and Discussion
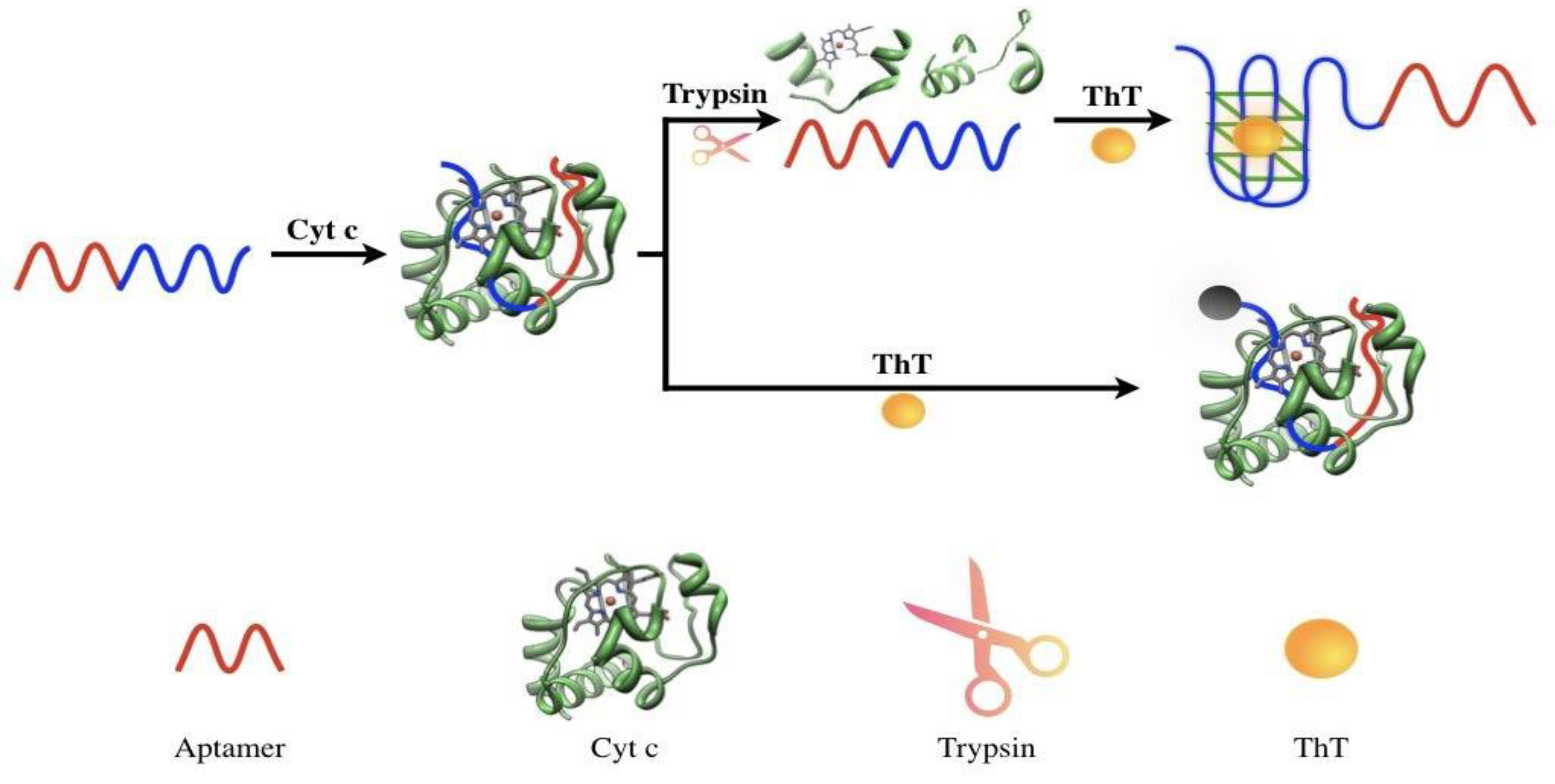
2.1. Principle of Trypsin Detection
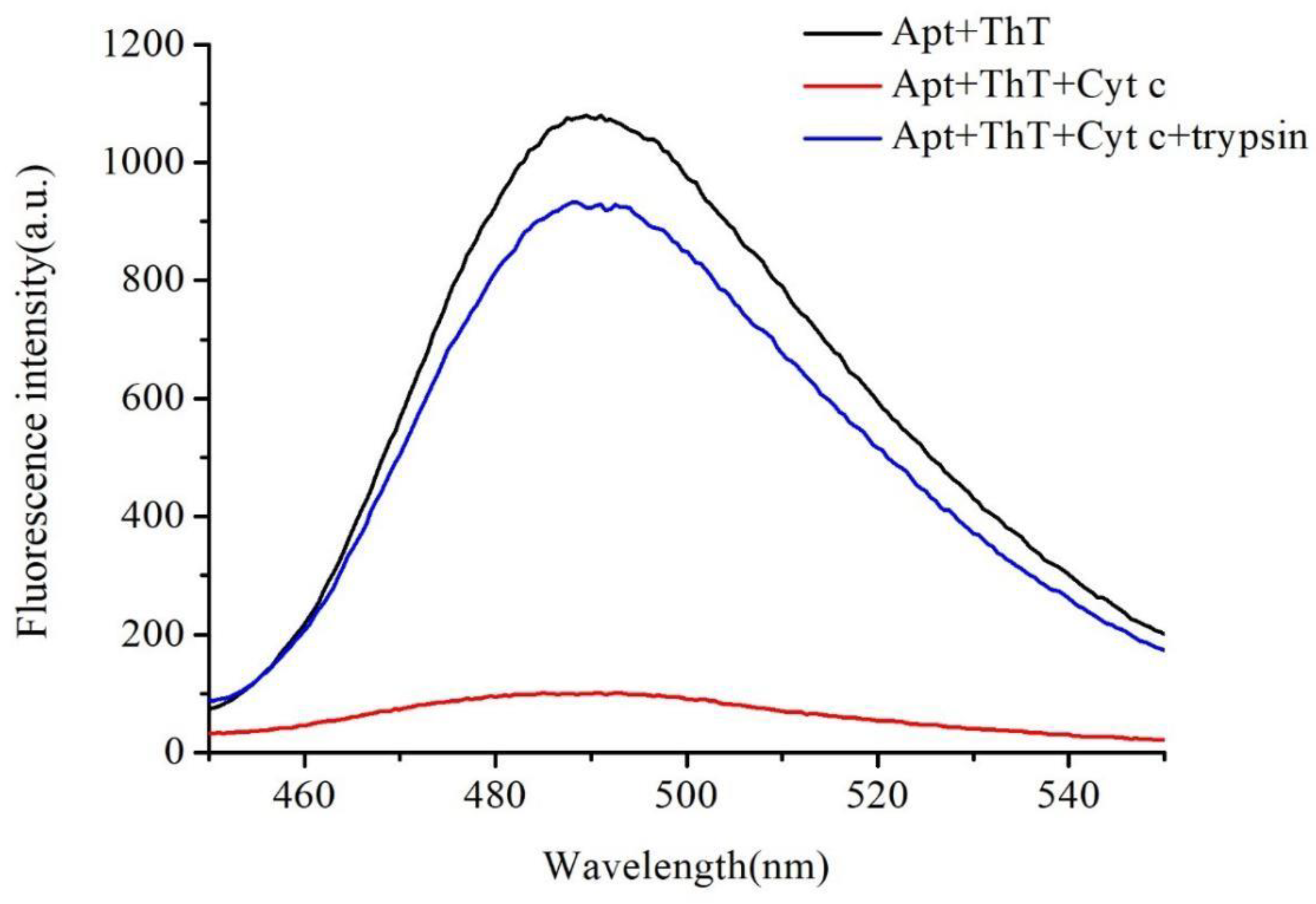
2.2. Verification of the Feasibility of Trypsin Detection
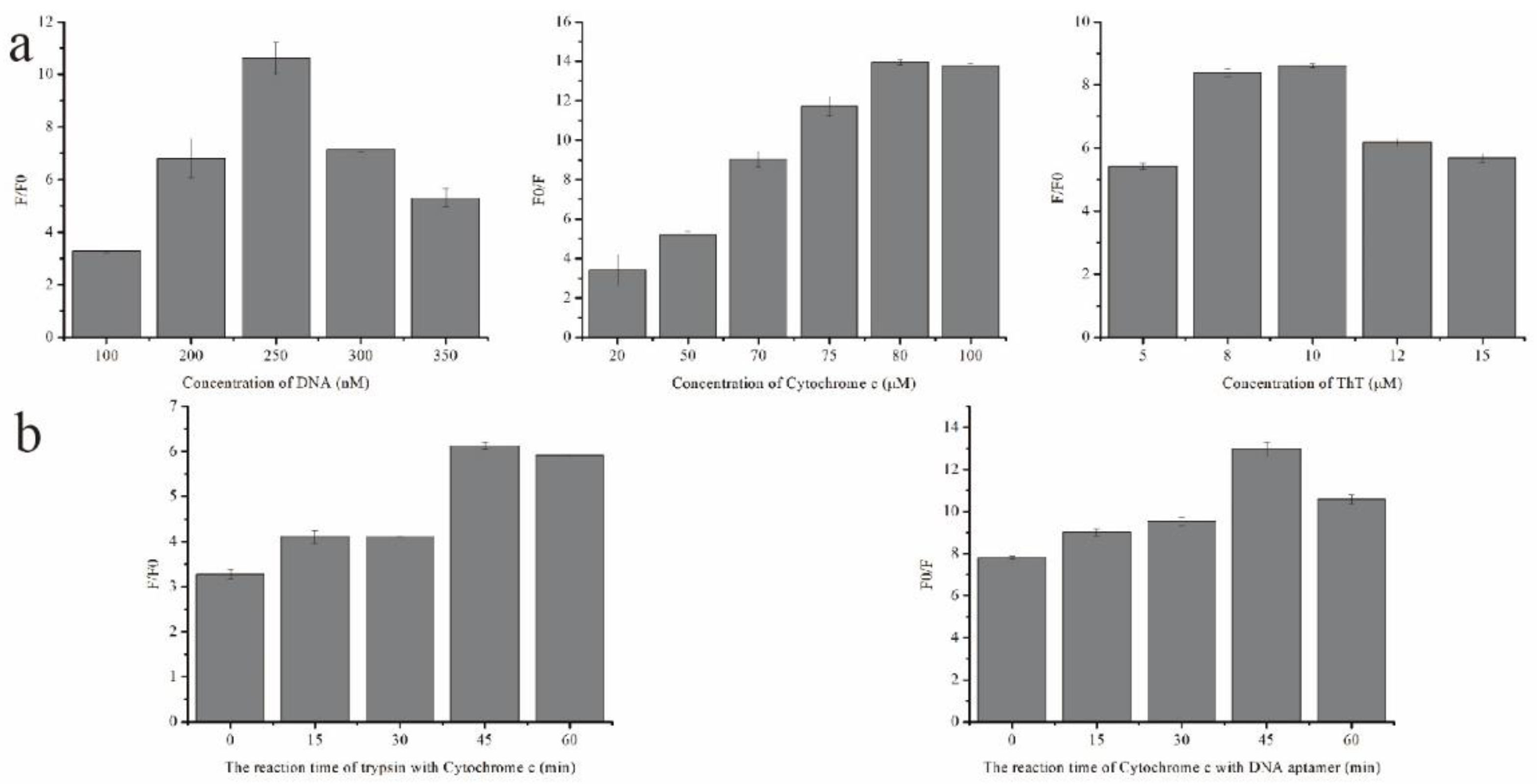
2.3. Optimization of Experimental Conditions
2.4. Quantitative Measurement of Trypsin
2.5. Study of Interferences
2.6. Trypsin Detection Assay in Real Samples
3. Materials and Methods
3.1. Materials and Reagents
3.2. Apparatus
3.3. Trypsin Detection Assays
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Atkinson, M.A.; Campbell-Thompson, M.; Kusmartseva, I.; Kaestner, K.H. Organisation of the human pancreas in health and in diabetes. Diabetologia 2020, 63, 1966–1973. [Google Scholar] [CrossRef] [PubMed]
- Sensoy, I. A review on the food digestion in the digestive tract and the used in vitro models. Curr. Res. Food Sci. 2021, 4, 308–319. [Google Scholar] [CrossRef]
- Hegyi, E.; Tóth, A.Z.; Vincze, Á.; Szentesi, A.; Hegyi, P.; Sahin-Tóth, M. Alcohol-dependent effect of PRSS1-PRSS2 haplotype in chronic pancreatitis. Gut 2020, 69, 1713–1715. [Google Scholar] [CrossRef] [PubMed]
- Hirota, M.; Ohmuraya, M.; Hashimoto, D.; Suyama, K.; Sugita, H.; Ogawa, M. Roles of Autophagy and Pancreatic Secretory Trypsin Inhibitor in Trypsinogen Activation in Acute Pancreatitis. Pancreas 2020, 49, 493–497. [Google Scholar] [CrossRef] [PubMed]
- Lasher, D.; Szabó, A.; Masamune, A.; Chen, J.M.; Xiao, X.; Whitcomb, D.C.; Barmada, M.M.; Ewers, M.; Ruffert, C.; Paliwal, S.; et al. Protease-Sensitive Pancreatic Lipase Variants Are Associated With Early Onset Chronic Pancreatitis. Am. J. Gastroenterol. 2019, 114, 974–983. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Liu, J.; Zou, B.; Li, C.; Zeh, H.J.; Kang, R.; Kroemer, G.; Huang, J.; Tang, D. Trypsin-Mediated Sensitization to Ferroptosis Increases the Severity of Pancreatitis in Mice. Cell. Mol. Gastroenterol. Hepatol. 2022, 13, 483–500. [Google Scholar] [CrossRef]
- Malla, S.R.; Krueger, B.; Wartmann, T.; Sendler, M.; Mahajan, U.M.; Weiss, F.U.; Thiel, F.G.; De Boni, C.; Gorelick, F.S.; Halangk, W.; et al. Early trypsin activation develops independently of autophagy in caerulein-induced pancreatitis in mice. Cell. Mol. Life Sci. 2020, 77, 1811–1825. [Google Scholar] [CrossRef]
- Kim, K.K.; Turner, R.; Khazan, N.; Kodza, A.; Jones, A.; Singh, R.K.; Moore, R.G. Role of trypsin and protease-activated receptor-2 in ovarian cancer. PLoS ONE 2020, 15, e0232253. [Google Scholar] [CrossRef]
- Søreide, K.; Roalsø, M.; Aunan, J.R. Is There a Trojan Horse to Aggressive Pancreatic Cancer Biology? A Review of the Trypsin-PAR2 Axis to Proliferation, Early Invasion, and Metastasis. J. Pancreat. Cancer 2020, 6, 12–20. [Google Scholar] [CrossRef]
- Thomas, R.G.; Surendran, S.P.; Jeong, Y.Y. Tumor Microenvironment-Stimuli Responsive Nanoparticles for Anticancer Therapy. Front. Mol. Biosci. 2020, 7, 610533. [Google Scholar] [CrossRef]
- Dutta, B.; Arya, R.K.; Goswami, R.; Alharbi, M.O.; Sharma, S.; Rahaman, S.O. Role of macrophage TRPV4 in inflammation. Lab. Investig. 2020, 100, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.C.; Zhang, B. Effect of Early Continuous Veno-Venous Haemofiltration in Severe Acute Pancreatitis for the Prevention of Local Pancreatic Complications. Gastroenterol. Res. Pract. 2022, 2022, 7575231. [Google Scholar] [CrossRef] [PubMed]
- Meyer-Hoffert, U.; Rogalski, C.; Seifert, S.; Schmeling, G.; Wingertszahn, J.; Proksch, E.; Wiedow, O. Trypsin induces epidermal proliferation and inflammation in murine skin. Exp. Dermatol. 2004, 13, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Dan, Z.Q.Z.; Liao, X.L.; Hou, C.S.; Xu, B.B.; Yang, J.; Kang, Y. The expression of trypsin in serum and vital organs of septic rats. Zhonghua Nei Ke Za Zhi 2018, 57, 505–510. [Google Scholar] [CrossRef]
- Zhou, S.; Wang, L.; Chen, X.; Guan, X. Label-free nanopore single-molecule measurement of trypsin activity. ACS Sens. 2016, 1, 607–613. [Google Scholar] [CrossRef]
- Pezzilli, R.; Caccialanza, R.; Capurso, G.; Brunetti, O.; Milella, M.; Falconi, M. Pancreatic Enzyme Replacement Therapy in Pancreatic Cancer. Cancers 2020, 12, 275. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, G.; Zhou, B.; Tang, Y.; Liu, X.; Wu, Y.; Wang, Y.; Kong, J.; Xu, T.; He, C.; et al. Effects of acids, pepsin, bile acids, and trypsin on laryngopharyngeal reflux diseases: Physiopathology and therapeutic targets. Eur. Arch. Otorhinolaryngol. 2022, 279, 2743–2752. [Google Scholar] [CrossRef]
- Lohman, R.J.; O’Brien, T.J.; Cocks, T.M. Protease-activated receptor-2 regulates trypsin expression in the brain and protects against seizures and epileptogenesis. Neurobiol. Dis. 2008, 30, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Liu, L.; Li, X.; Chen, H.; Wang, Z.; Yang, W.; Zhang, H.; Zhang, H. Peptide modified manganese-doped iron oxide nanoparticles as a sensitive fluorescence nanosensor for non-invasive detection of trypsin activity in vitro and in vivo. RSC Adv. 2021, 11, 2213–2220. [Google Scholar] [CrossRef]
- Rainio, M.; Lindström, O.; Penttilä, A.; Itkonen, O.; Kemppainen, E.; Stenman, U.H.; Kylänpää, L. Serum Serine Peptidase Inhibitor Kazal-Type 1, Trypsinogens 1 to 3, and Complex of Trypsin 2 and α1-Antitrypsin in the Diagnosis of Severe Acute Pancreatitis. Pancreas 2019, 48, 374–380. [Google Scholar] [CrossRef]
- Anuracpreeda, P.; Chawengkirtikul, R.; Tinikul, Y.; Poljaroen, J.; Chotwiwatthanakun, C.; Sobhon, P. Diagnosis of Fasciola gigantica infection using a monoclonal antibody-based sandwich ELISA for detection of circulating cathepsin B3 protease. Acta Trop. 2013, 127, 38–45. [Google Scholar] [CrossRef] [PubMed]
- See, W.A.; Smith, J.L. Urinary levels of activated trypsin in whole-organ pancreas transplant patients with duodenocystostomies. Transplantation 1991, 52, 630–633. [Google Scholar] [CrossRef] [PubMed]
- Schebb, N.H.; Vielhaber, T.; Jousset, A.; Karst, U. Development of a liquid chromatography-based screening methodology for proteolytic enzyme activity. J. Chromatogr. A 2009, 1216, 4407–4415. [Google Scholar] [CrossRef] [PubMed]
- Stoytcheva, M.; Zlatev, R.; Cosnier, S.; Arredondo, M. Square wave voltammetric determination of trypsin activity. Electrochim. Acta 2012, 76, 43–47. [Google Scholar] [CrossRef]
- Lou, X.; Zhang, L.; Qin, J.; Li, Z. Colorimetric sensing of alpha-amino acids and its application for the “label-free” detection of protease. Langmuir 2010, 26, 1566–1569. [Google Scholar] [CrossRef]
- Wang, G.L.; Jin, L.Y.; Dong, Y.M.; Wu, X.M.; Li, Z.J. Intrinsic enzyme mimicking activity of gold nanoclusters upon visible light triggering and its application for colorimetric trypsin detection. Biosens. Bioelectron. 2015, 64, 523–529. [Google Scholar] [CrossRef]
- Zhang, L.; Du, J. A sensitive and label-free trypsin colorimetric sensor with cytochrome c as a substrate. Biosens. Bioelectron. 2016, 79, 347–352. [Google Scholar] [CrossRef]
- Stoytcheva, M.; Zlatev, R.; Cosnier, S.; Arredondo, M.; Valdez, B. High sensitive trypsin activity evaluation applying a nanostructured QCM-sensor. Biosens. Bioelectron. 2013, 41, 862–866. [Google Scholar] [CrossRef]
- Wang, M.; Su, D.; Wang, G.; Su, X. A fluorometric sensing method for sensitive detection of trypsin and its inhibitor based on gold nanoclusters and gold nanoparticles. Anal. Bioanal. Chem. 2018, 410, 6891–6900. [Google Scholar] [CrossRef]
- Zhuo, C.X.; Wang, L.H.; Feng, J.J.; Zhang, Y.D. Label-Free Fluorescent Detection of Trypsin Activity Based on DNA-Stabilized Silver Nanocluster-Peptide Conjugates. Sensors 2016, 16, 1477. [Google Scholar] [CrossRef]
- Huang, S.; Li, F.; Liao, C.; Zheng, B.; Du, J.; Xiao, D. A selective and sensitive fluorescent probe for the determination of HSA and trypsin. Talanta 2017, 170, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Amouzadeh Tabrizi, M.; Ferré-Borrull, J.; Marsal, L.F. Remote biosensor for the determination of trypsin by using nanoporous anodic alumina as a three-dimensional nanostructured material. Sci. Rep. 2020, 10, 2356. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Shen, R.; Liu, N.; Yi, H.; Dai, H.; Lin, J. A highly sensitive peptide-based biosensor using NiCo(2)O(4) nanosheets and g-C(3)N(4) nanocomposite to construct amplified strategy for trypsin detection. Anal. Chim. Acta 2018, 1035, 175–183. [Google Scholar] [CrossRef]
- Hu, Y.; He, Y.; Han, Y.; Ge, Y.; Song, G.; Zhou, J. Poly(styrene-4-sulfonate)-protected copper nanoclusters as a fluorometric probe for sequential detection of cytochrome c and trypsin. Mikrochim. Acta 2018, 185, 383. [Google Scholar] [CrossRef]
- Zhao, D.; Chen, C.; Zhao, J.; Sun, J.; Yang, X. Label-free fluorescence turn-on strategy for trypsin activity based on thiolate-protected gold nanoclusters with bovine serum albumin as the substrate. Sens. Actuators B Chem. 2017, 247, 392–399. [Google Scholar] [CrossRef]
- Wu, P.; Zhao, T.; Zhang, J.; Wu, L.; Hou, X. Analyte-activable probe for protease based on cytochrome C-capped Mn: ZnS quantum dots. Anal. Chem. 2014, 86, 10078–10083. [Google Scholar] [CrossRef] [PubMed]
- Eissa, S.; Siddiqua, A.; Chinnappan, R.; Zourob, M. Electrochemical SELEX Technique for the Selection of DNA Aptamers against the Small Molecule 11-Deoxycortisol. ACS Appl. Bio Mater. 2019, 2, 2624–2632. [Google Scholar] [CrossRef]
- Dai, S.; Wu, S.; Duan, N.; Chen, J.; Zheng, Z.; Wang, Z. An ultrasensitive aptasensor for Ochratoxin A using hexagonal core/shell upconversion nanoparticles as luminophores. Biosens. Bioelectron. 2017, 91, 538–544. [Google Scholar] [CrossRef]
- Abnous, K.; Danesh, N.M.; Alibolandi, M.; Ramezani, M.; Taghdisi, S.M. Amperometric aptasensor for ochratoxin A based on the use of a gold electrode modified with aptamer, complementary DNA, SWCNTs and the redox marker Methylene Blue. Microchim. Acta 2017, 184, 1151–1159. [Google Scholar] [CrossRef]
- Cetin, A.E.; Coskun, A.F.; Galarreta, B.C.; Huang, M.; Herman, D.; Ozcan, A.; Altug, H. Handheld high-throughput plasmonic biosensor using computational on-chip imaging. Light Sci. Appl. 2014, 3, e122. [Google Scholar] [CrossRef]
- Mascini, M.; Palchetti, I.; Tombelli, S. Nucleic acid and peptide aptamers: Fundamentals and bioanalytical aspects. Angew. Chem. Int. Ed. 2012, 51, 1316–1332. [Google Scholar] [CrossRef] [PubMed]
- Taghdisi, S.M.; Danesh, N.M.; Nameghi, M.A.; Ramezani, M.; Abnous, K. A label-free fluorescent aptasensor for selective and sensitive detection of streptomycin in milk and blood serum. Food Chem. 2016, 203, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Roushani, M.; Shahdost-fard, F. A novel ultrasensitive aptasensor based on silver nanoparticles measured via enhanced voltammetric response of electrochemical reduction of riboflavin as redox probe for cocaine detection. Sens. Actuators B Chem. 2015, 207, 764–771. [Google Scholar] [CrossRef]
- Zheng, H.; Lang, Y.; Yu, J.; Han, Z.; Chen, B.; Wang, Y. Affinity binding of aptamers to agarose with DNA tetrahedron for removal of hepatitis B virus surface antigen. Colloids Surf. B Biointerfaces 2019, 178, 80–86. [Google Scholar] [CrossRef]
- Zhao, H.; Xiang, X.; Chen, M.; Ma, C. Aptamer-Based Fluorometric Ochratoxin A Assay Based on Photoinduced Electron Transfer. Toxins 2019, 11, 65. [Google Scholar] [CrossRef]
- Tang, X.; Wu, K.; Zhao, H.; Chen, M.; Ma, C. A Label-Free Fluorescent Assay for the Rapid and Sensitive Detection of Adenosine Deaminase Activity and Inhibition. Sensors 2018, 18, 2441. [Google Scholar] [CrossRef]
- Wang, J.; Liu, H.; Ma, C.; Wang, J.; Zhong, L.; Wu, K. Label-free monitoring of DNA polymerase activity based on a thrombin-binding aptamer G-quadruplex. Mol. Cell. Probes 2017, 32, 13–17. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Zaidi, S.; Hassan, M.I.; Islam, A.; Ahmad, F. The role of key residues in structure, function, and stability of cytochrome-c. Cell. Mol. Life Sci. 2014, 71, 229–255. [Google Scholar] [CrossRef]
- Ge, J.; Li, X.P.; Jiang, J.H.; Yu, R.Q. A highly sensitive label-free sensor for Mercury ion (Hg2+) by inhibiting thioflavin T as DNA G-quadruplexes fluorescent inducer. Talanta 2014, 122, 85–90. [Google Scholar] [CrossRef]
- Wu, K.; Ma, C.; Zhao, H.; He, H.; Chen, H. Label-Free G-Quadruplex Aptamer Fluorescence Assay for Ochratoxin A Using a Thioflavin T Probe. Toxins 2018, 10, 198. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Ma, C.; Chen, M. A novel fluorometric method for inorganic pyrophosphatase detection based on G-quadruplex-thioflavin T. Mol. Cell. Probes 2019, 43, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hua, X.; Fan, Q.; Chao, J.; Su, S.; Huang, Y.Q.; Wang, L.; Huang, W. Thioflavin T as an Efficient G-Quadruplex Inducer for the Highly Sensitive Detection of Thrombin Using a New Föster Resonance Energy Transfer System. ACS Appl. Mater. Interfaces 2015, 7, 16458–16465. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Yang, Y.; Zou, S.; Ouyang, P.; Qing, Y.; Han, J.; Li, H.; Wang, Z.; Du, J. Signal-On Fluorescence Biosensor for Highly Sensitive Detection of miRNA-21 Based on DNAzyme Assisted Double-Hairpin Molecular Beacon. Biosensors 2022, 12, 276. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Liu, Q.; Liu, W.; Jin, Y.; Li, B. Comparative evaluation and design of a G-triplex/thioflavin T-based molecular beacon. Analyst 2021, 146, 2567–2573. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Guo, Q.; Zhang, C.; Sun, Z.; Weng, X. Microfluidic origami nano-aptasensor for peanut allergen Ara h1 detection. Food Chem. 2021, 365, 130511. [Google Scholar] [CrossRef]
- Jiang, J.; Cai, Q.; Deng, M. Construction of Electrochemical Aptamer Sensor Based on Pt-Coordinated Titanium-Based Porphyrin MOF for Thrombin Detection. Front Chem. 2021, 9, 812983. [Google Scholar] [CrossRef]
- Hong, M.-L.; Li, L.-J.; Han, H.-X.; Chu, X. A Label-free Fluorescence Assay for Trypsin Based on the Electron Transfer between Oligonucleotide-stabilized Ag Nanoclusters and Cytochrome c. Anal. Sci. 2014, 30, 811–815. [Google Scholar] [CrossRef]
- Li, H.; Yang, M.; Kong, D.; Jin, R.; Zhao, X.; Liu, F.; Yan, X.; Lin, Y.; Lu, G. Sensitive fluorescence sensor for point-of-care detection of trypsin using glutathione-stabilized gold nanoclusters. Sens. Actuators B Chem. 2019, 282, 366–372. [Google Scholar] [CrossRef]
- Zhu, Q.; Zhan, R.; Liu, B. Homogeneous Detection of Trypsin in Protein Mixtures Based on Fluorescence Resonance Energy Transfer between Anionic Conjugated Polymer and Fluorescent Probe. Macromol. Rapid Commun. 2010, 31, 1060–1064. [Google Scholar] [CrossRef]
- Poon, C.-Y.; Li, Q.; Zhang, J.; Li, Z.; Dong, C.; Lee, A.W.-M.; Chan, W.-H.; Li, H.-W. FRET-based modified graphene quantum dots for direct trypsin quantification in urine. Anal. Chim. Acta 2016, 917, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Tian, X.; Liu, C.; Ge, J.; Chu, X.; Li, Y.-F. Fluorescence activation imaging of cytochrome c released from mitochondria using aptameric nanosensor. J. Am. Chem. Soc. 2015, 137, 982–989. [Google Scholar] [CrossRef] [PubMed]


| Method | Material | LOD 1 (μg/mL) | Linear Range (μg/mL) | Reference |
|---|---|---|---|---|
| Colorimetric | Copper ion chemosensor | 1.0 | 1.0–5.0 | [25] |
| Colorimetric | Gold nanoclusters | 0.6 | 0.9–1000 | [26] |
| Fluorescent | Ag nanoclusters | 0.06 | 0.7–4.0 | [58] |
| Fluorescent | Gold nanoclusters | 0.08 | 0.2–100 | [59] |
| Fluorescent | Conjugated polyelectrolyte | 0.2 | 0–2.5 | [60] |
| Fluorescent | Graphene quantum dots | 0.7 | 0–6.0 | [61] |
| Fluorescent | DNA aptamer | 0.2 | 0.2–60 | This work |
| Sample | Added (μg/mL) | Found (μg/mL) | Recovery (%) | R.S.D (%, n = 5) |
|---|---|---|---|---|
| 1 | 15 | 15.59 | 103.9 | 5.8 |
| 2 | 40 | 36.72 | 91.8 | 4.1 |
| 3 | 60 | 61.17 | 101.9 | 2.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, P.; Lu, Y.; Li, S.; Ma, C. A Label-Free Fluorescence Aptasensor Based on G-Quadruplex/Thioflavin T Complex for the Detection of Trypsin. Molecules 2022, 27, 6093. https://doi.org/10.3390/molecules27186093
Gu P, Lu Y, Li S, Ma C. A Label-Free Fluorescence Aptasensor Based on G-Quadruplex/Thioflavin T Complex for the Detection of Trypsin. Molecules. 2022; 27(18):6093. https://doi.org/10.3390/molecules27186093
Chicago/Turabian StyleGu, Pan, Yangfan Lu, Shanni Li, and Changbei Ma. 2022. "A Label-Free Fluorescence Aptasensor Based on G-Quadruplex/Thioflavin T Complex for the Detection of Trypsin" Molecules 27, no. 18: 6093. https://doi.org/10.3390/molecules27186093
APA StyleGu, P., Lu, Y., Li, S., & Ma, C. (2022). A Label-Free Fluorescence Aptasensor Based on G-Quadruplex/Thioflavin T Complex for the Detection of Trypsin. Molecules, 27(18), 6093. https://doi.org/10.3390/molecules27186093







