Phosphatidylcholine Cation—Tyrosine π Complexes: Motifs for Membrane Binding by a Bacterial Phospholipase C
Abstract
1. Introduction
2. Experimental Results—Characterization of Specific PC Binding Site(s) on BtPI-PLC
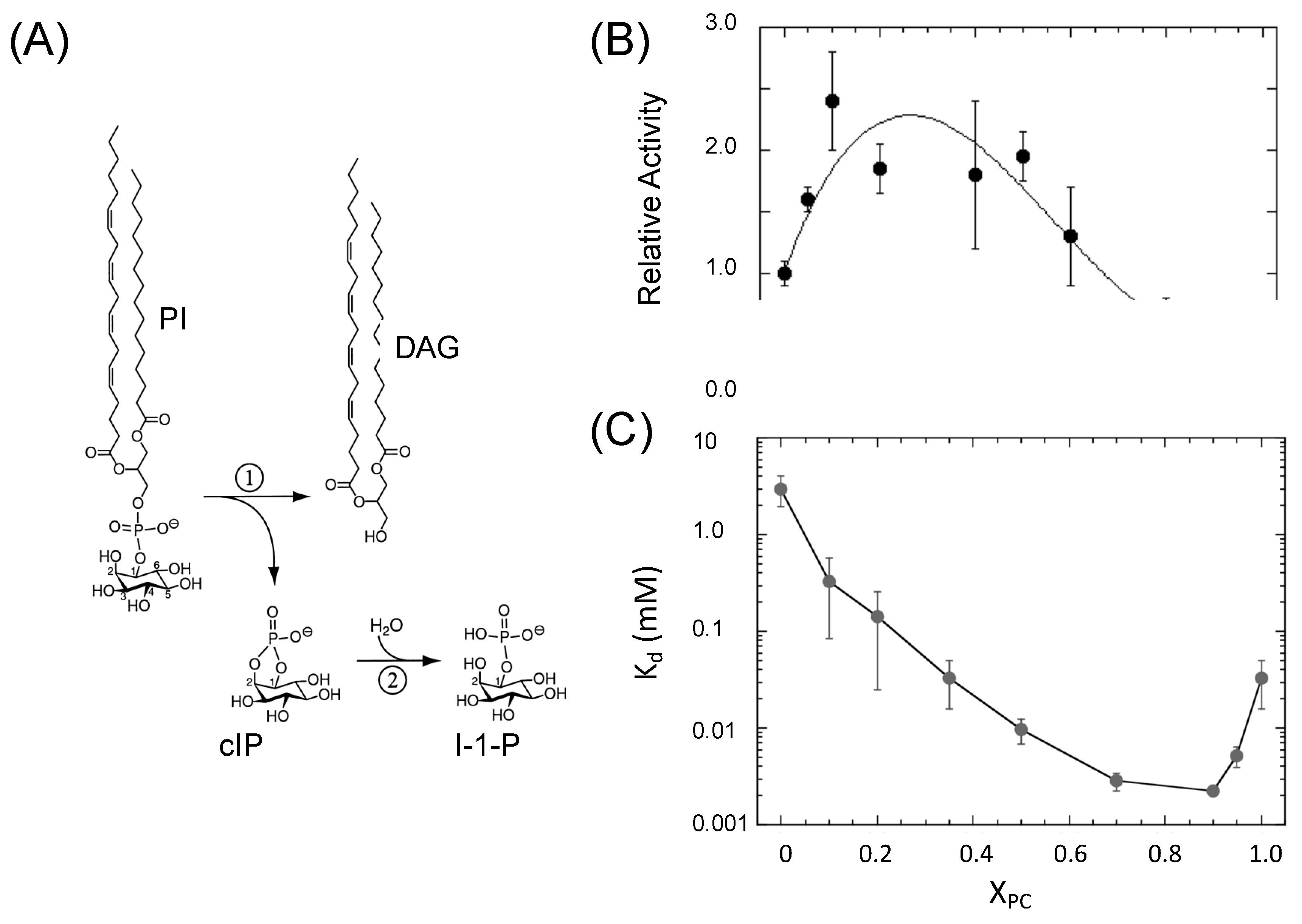
2.1. B. thuringiensis PI-PLC, a Member of the TIM Barrel Superfamily
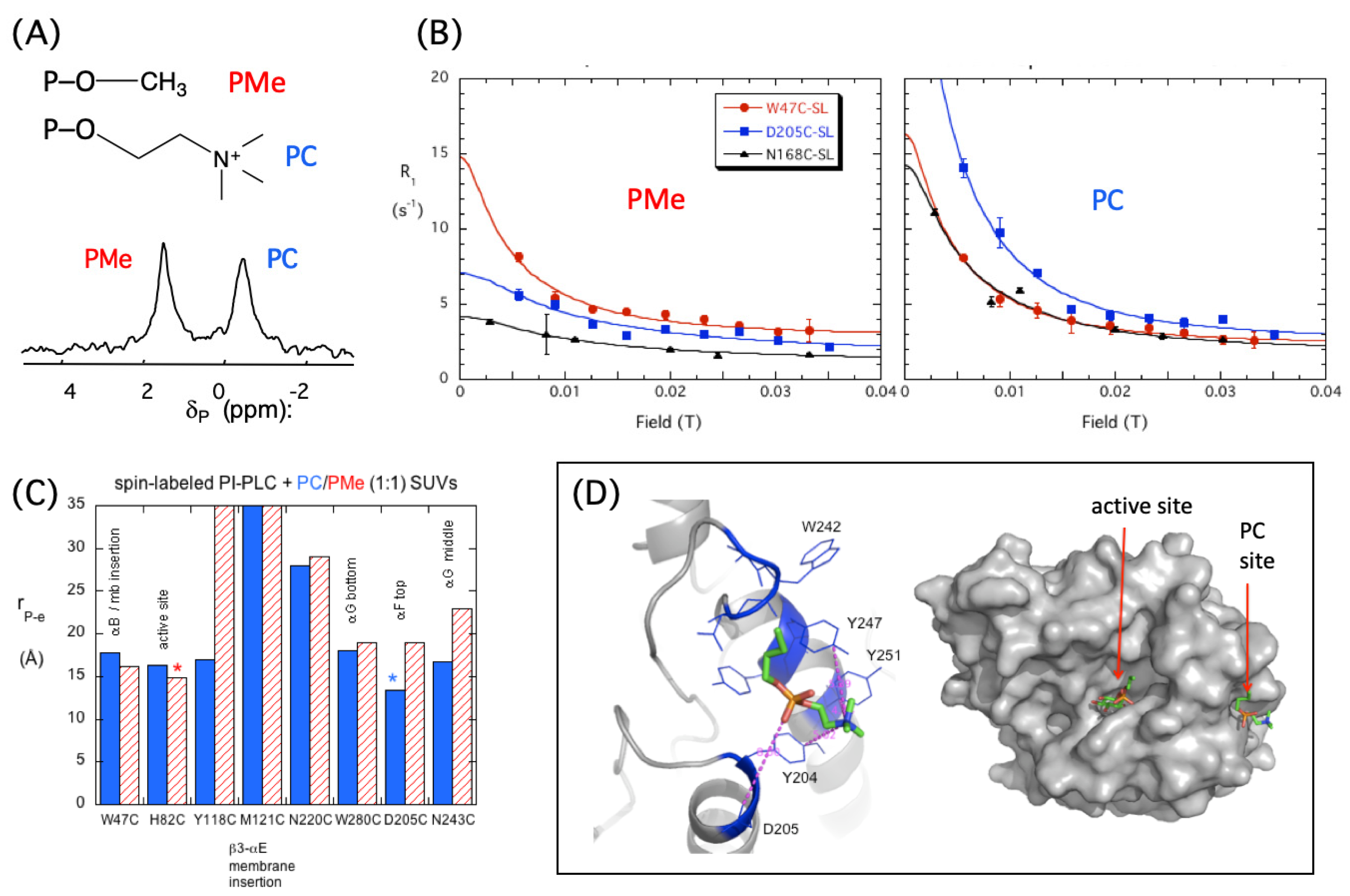
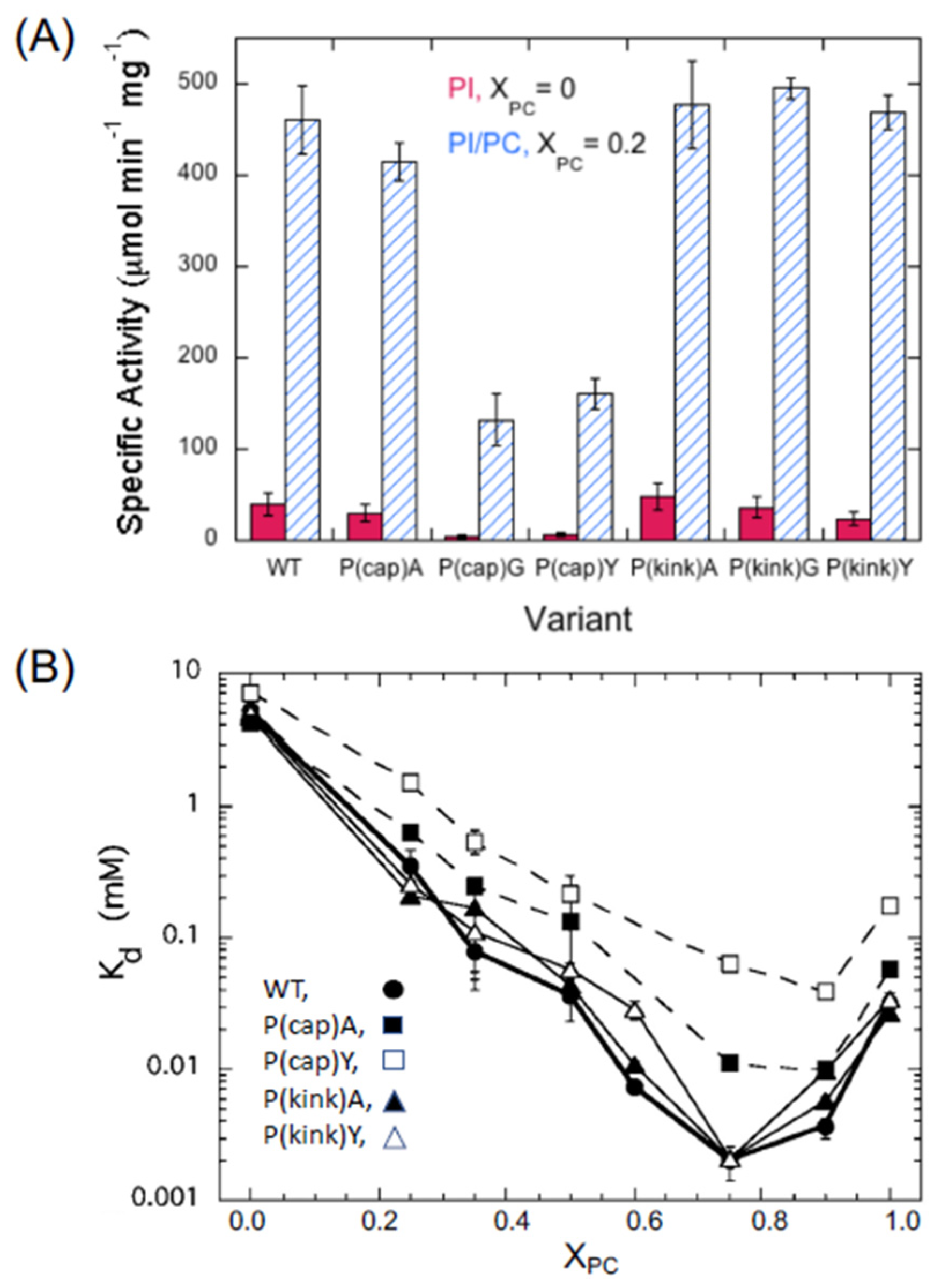
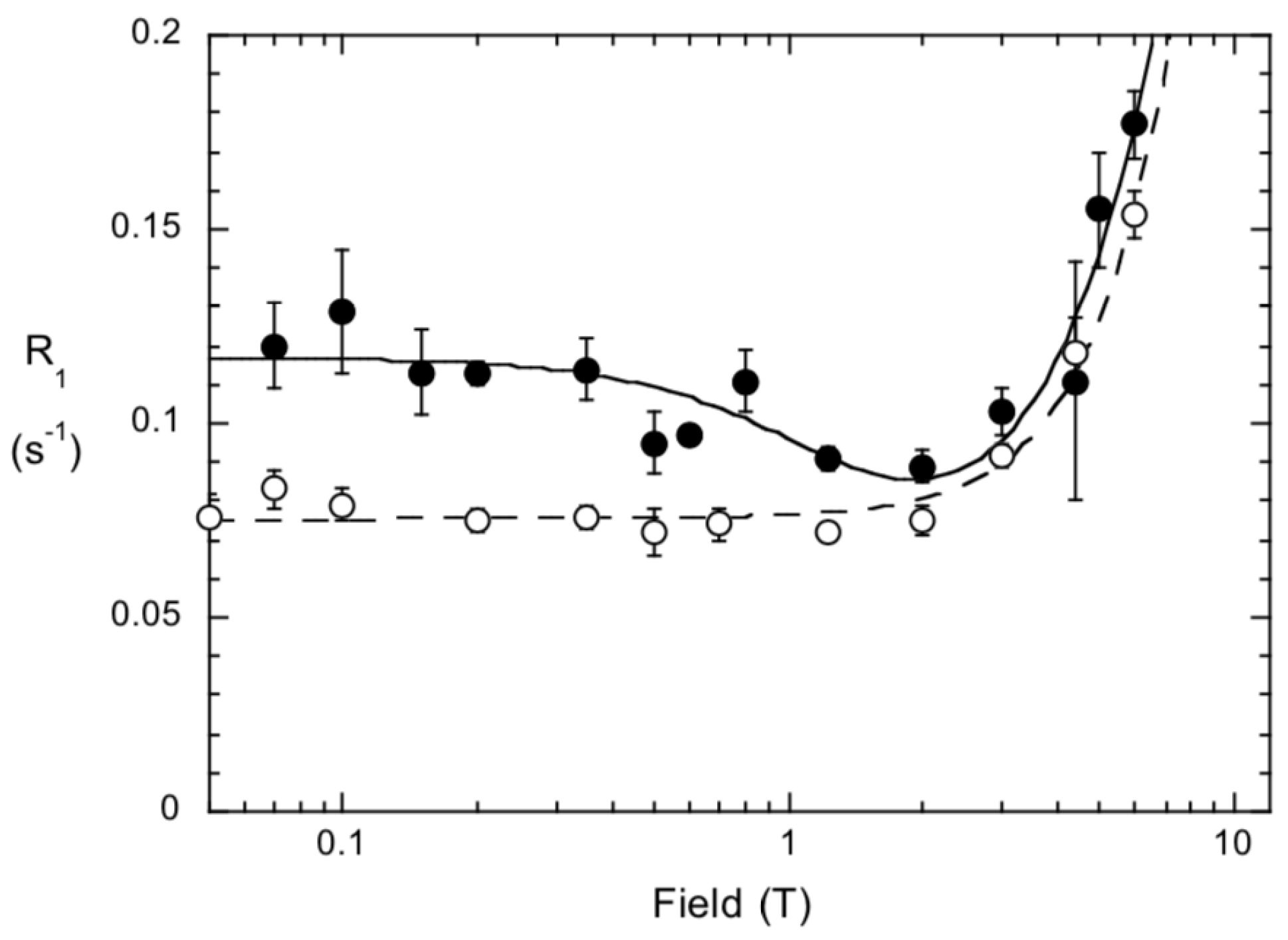
2.2. Experimental Evidence for a Specific PC Binding Site on B. thuringiensis PI-PLC
3. Computational Results—Identification of PC Binding Sites on BtPI-PLC
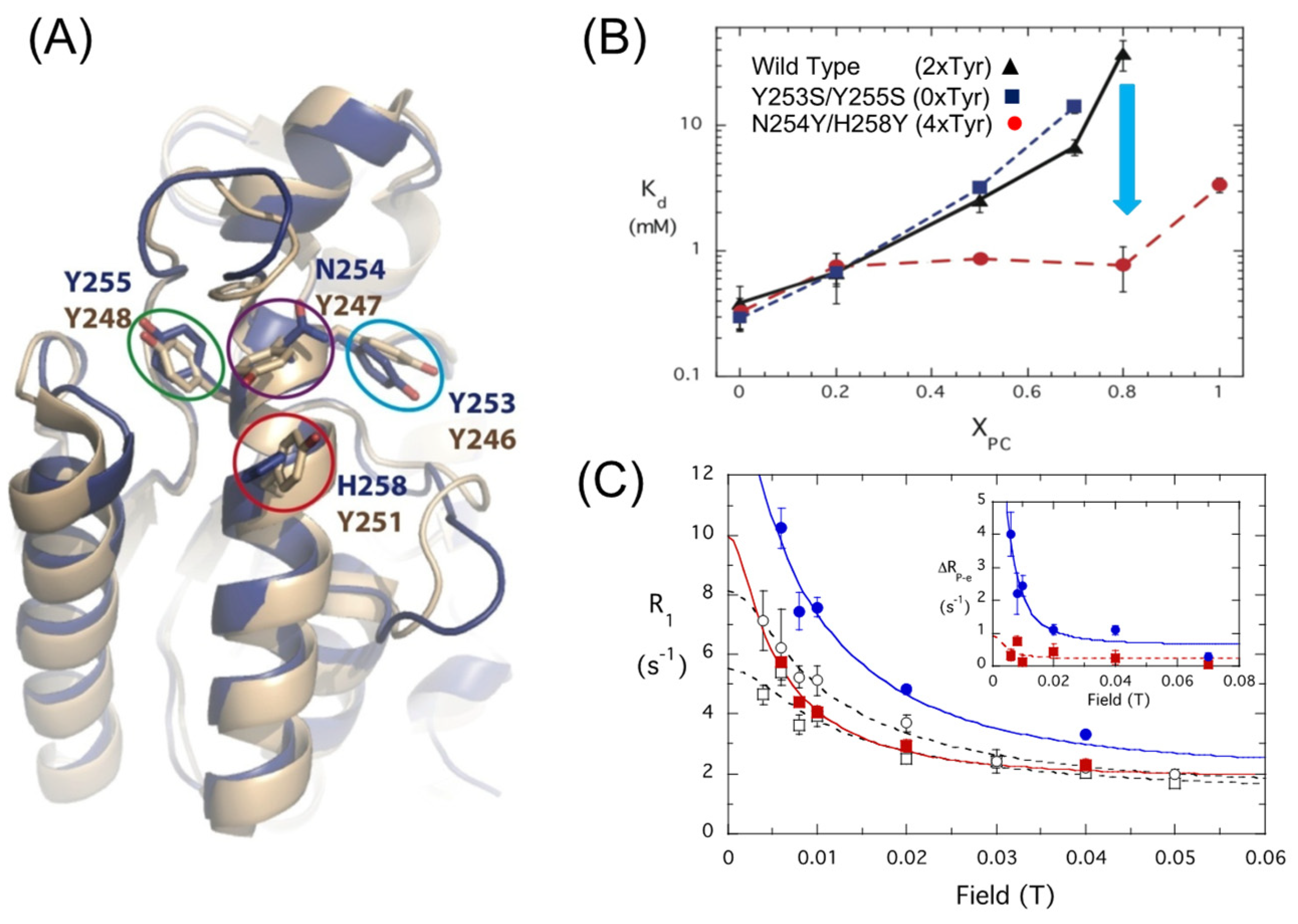
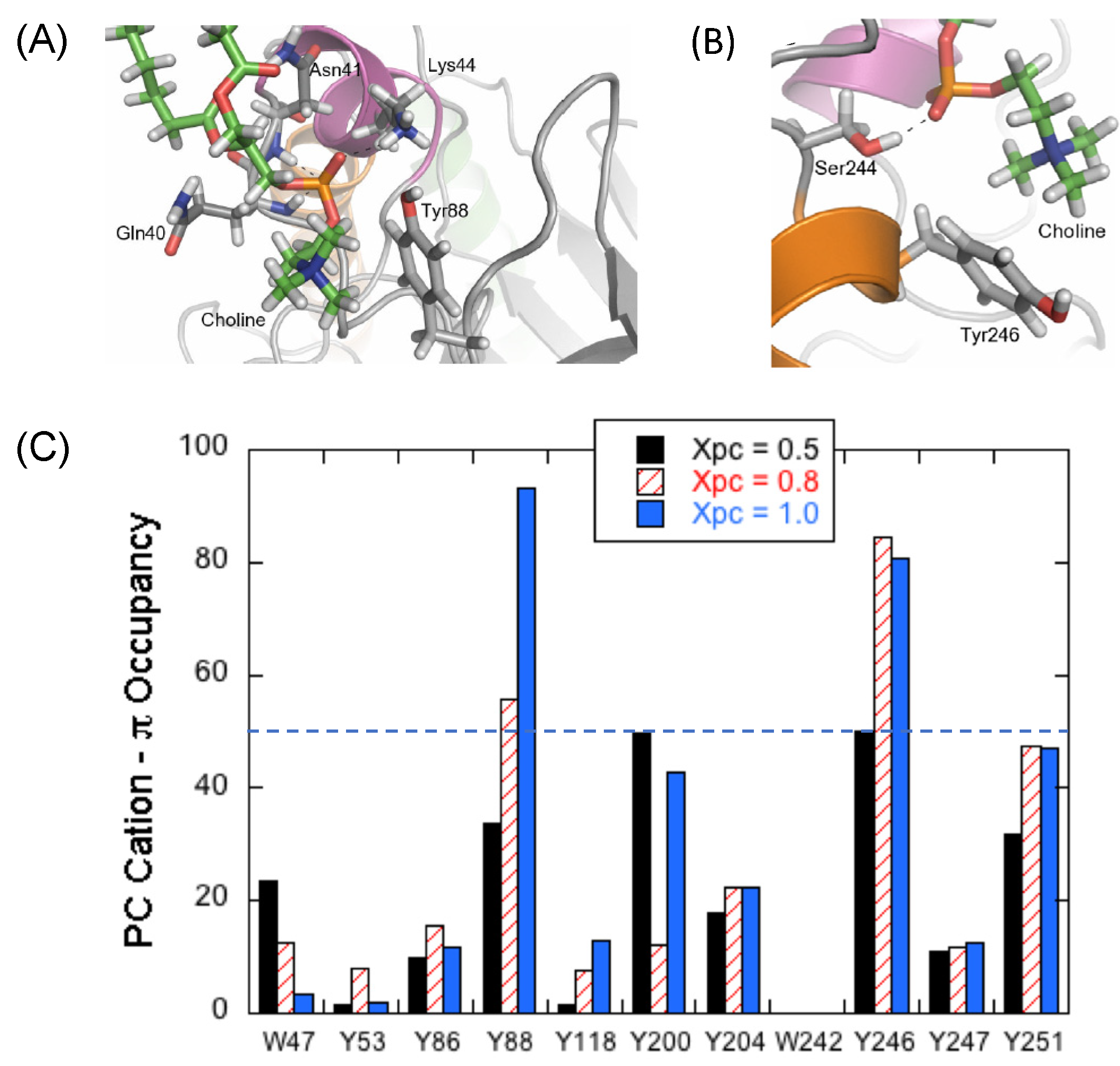
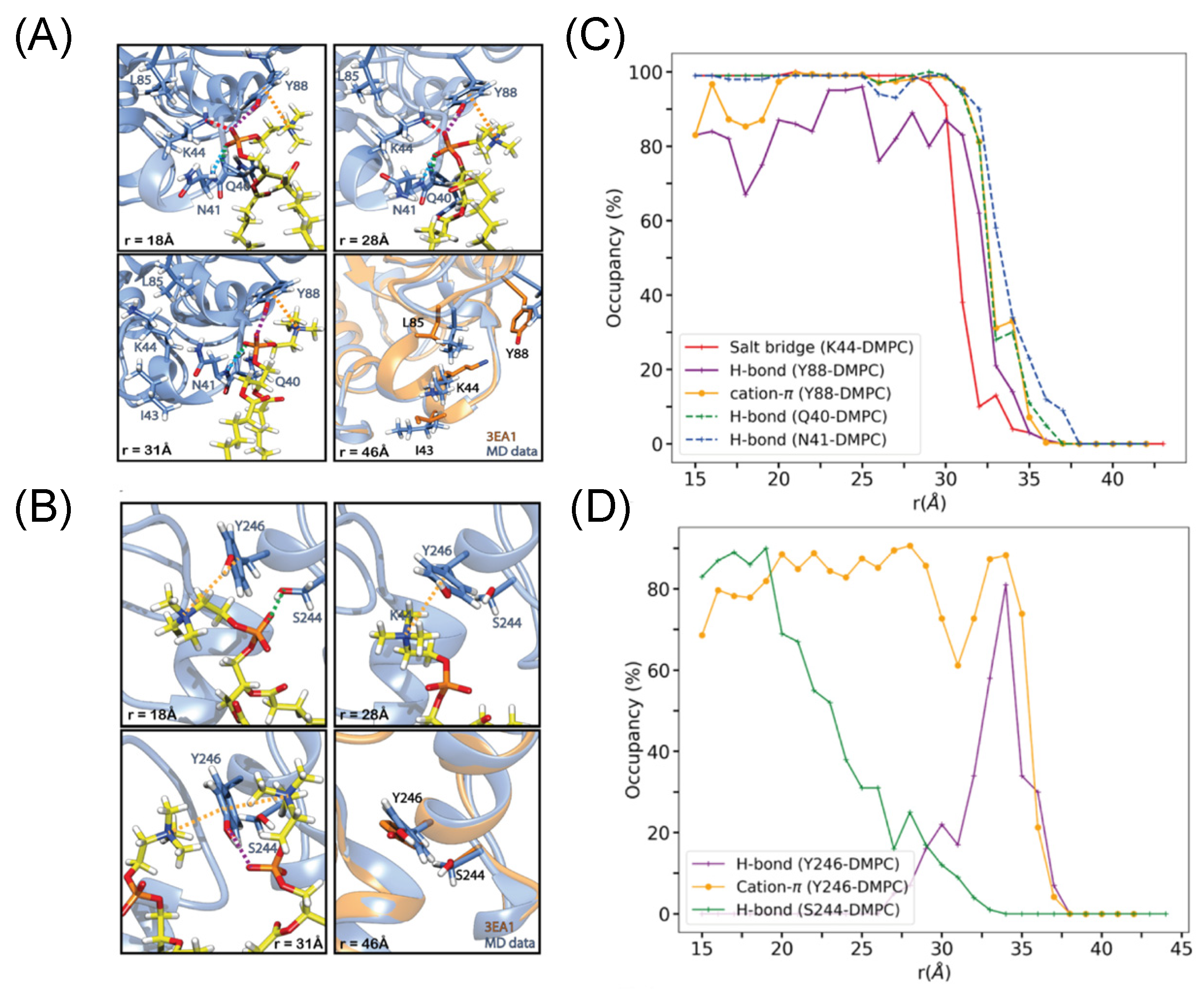
3.1. Transient Opportunistic and Very Specific PC Cation–PI-PLC Tyr-π Interactions
3.2. A Computational Approach to Estimating ΔG0 bind Provides a Membrane Desorption Pathway for BtPI-PLC
4. What Is the Allosteric Mechanism for PC Altering BtPI-PLC Enzymatic Activity?
5. Conclusions
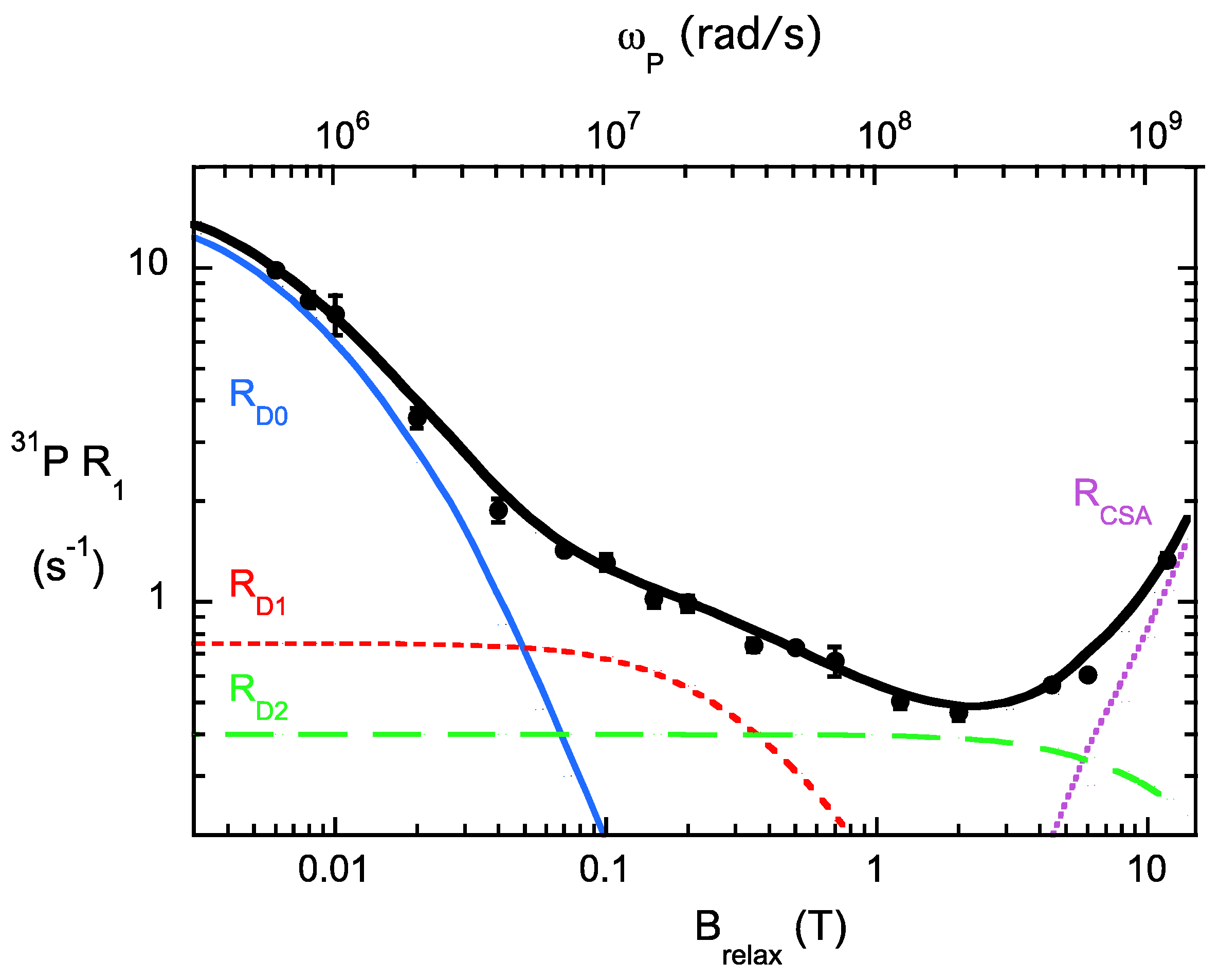
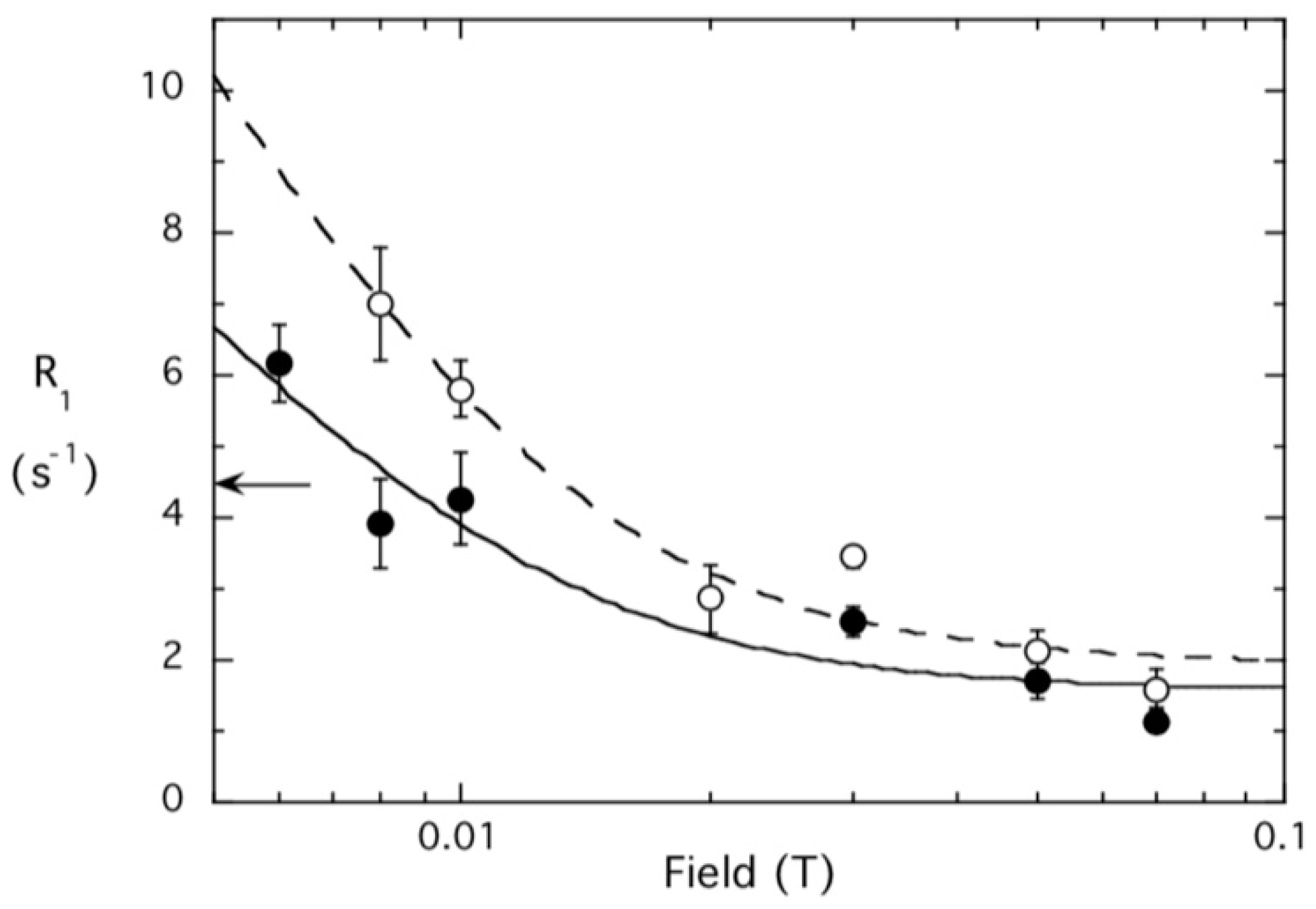
6. Appendix: Details on Analysis of 31P NMRDs and Examples
Author Contributions
Funding
Author Information
Acknowledgments
Notes
Conflicts of Interest
References
- Zenewicz, L.A.; Wei, Z.; Goldfine, H.; Shen, H. Phosphatidylinositol-specific phospholipase C of Bacillus anthracis down-modulates the immune response. J. Immunol. 2005, 174, 8011–8016. [Google Scholar] [CrossRef] [PubMed]
- Flores-Díaz, M.; Monturiol-Gross, L.; Naylor, C.; Alape-Girón, A.; Flieger, A. Bacterial sphingomyelinases and phospholipases as virulence factors. Microbiol. Mol. Biol Rev. 2016, 80, 597–628. [Google Scholar] [CrossRef] [PubMed]
- Lehto, M.T.; Sharom, F.J. Release of the glycosylphosphatidylinositol-anchored enzyme ecto-5’-nucleotidase by phospholipase C: Catalytic activation and modulation by the lipid bilayer. Biochem. J. 1998, 332, 101–109. [Google Scholar] [CrossRef]
- Lehto, M.T.; Sharom, F.J. PI-specific phospholipase C cleavage of a reconstituted GPI-anchored protein: Modulation by the lipid bilayer. Biochemistry 2002, 41, 1398–1408. [Google Scholar] [CrossRef] [PubMed]
- Sharom, F.J.; Lehto, M.T. Glycosylphosphatidylinositol-anchored proteins: Structure, function, and cleavage by phosphatidylinositol-specific phospholipase C. Biochem. Cell. Biol. 2002, 80, 535–549. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Wu, Y.; Roberts, M.F. Activation of phosphatidylinositol-specific phospholipase C towards inositol 1,2-(cyclic)-phosphate. Biochemistry 1997, 36, 347–355. [Google Scholar] [CrossRef]
- Qian, X.; Zhou, C.; Roberts, M.F. Phosphatidylcholine activation of bacterial phosphatidylinositol-specific phospholipase C towards PI vesicles. Biochemistry 1998, 37, 6513–6522. [Google Scholar] [CrossRef]
- Pu, M.; Roberts, M.F.; Gershenson, A. Fluorescence correlation spectroscopy of phosphatidylinositol-specific phospholipase C monitors the interplay of substrate and activator lipid binding. Biochemistry 2009, 48, 6835–6845. [Google Scholar] [CrossRef][Green Version]
- Yang, B.; Pu, M.; Khan, H.; Friedman, L.; Reuter, N.; Roberts, M.F.; Gershenson, A. Quantifying transient interactions between Bacillus phosphatidylinositol-specific phospholipase C and phosphatidylcholine-rich vesicles. J. Am. Chem. Soc. 2015, 137, 14–17. [Google Scholar] [CrossRef]
- Roberts, M.F.; Khan, H.M.; Goldstein, R.; Reuter, N.; Gershenson, A. Search and subvert: Minimalist bacterial phosphatidylinositol-specific phospholipase C (PI-PLC) enzymes. Chem. Rev. 2018, 118, 8435–8473. [Google Scholar] [CrossRef]
- Heinz, D.W.; Ryan, M.; Bullock, T.L.; Griffith, O.H. Crystal structure of the phosphatidylinositol-specific phospholipase C from Bacillus cereus in complex with myo-inositol. EMBO J. 1995, 14, 3855–3863. [Google Scholar] [CrossRef] [PubMed]
- Heinz, D.W.; Ryan, M.; Smith, M.P.; Weaver, L.H.; Keana, J.F.; Griffith, O.H. Crystal structure of phosphatidylinositol-specific phospholipase C from Bacillus cereus in complex with glucosaminyl(1→6)-D-myo-inositol, an essential fragment of GPI anchors. Biochemistry 1996, 35, 9496–9504. [Google Scholar] [CrossRef]
- Grauffel, C.; Yang, B.; He, T.; Roberts, M.F.; Gershenson, A.; Reuter, N. Cation-π interactions as lipid-specific anchors for phosphatidylinositol-specific phospholipase C. J. Am. Chem. Soc. 2013, 135, 5740–5750. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wehbi, H.; Roberts, M.F. Crosslinking phosphatidylinositol-specific phospholipase C traps two activating phosphatidylcholine molecules on the enzyme. J. Biol. Chem. 2004, 279, 20490–20500. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Wehbi, H.; Roberts, M.F. Role of tryptophan residues in interfacial binding of phosphatidylinositol-specific phospholipase C. J. Biol. Chem. 2002, 277, 19867–19875. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Bradley, W.; Roberts, M.F. Optimizing the interfacial activity of a bacterial phosphatidylinositol-specific phospholipase C. J. Biol. Chem. 2003, 278, 24651–24657. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Qian, X.; Roberts, M.F. Allosteric activation of phosphatidylinositol-specific phospholipase C: Phospholipid binding anchors the enzyme to the interface. Biochemistry 1997, 36, 10089–10097. [Google Scholar] [CrossRef]
- Roberts, M.F.; Redfield, A.G. High resolution 31P field cycling as a probe of phospholipid dynamics. J. Am. Chem. Soc. 2004, 126, 13765–13777. [Google Scholar] [CrossRef]
- Roberts, M.F.; Redfield, A.G. Phospholipid bilayer surface configuration probed quantitatively by 31P field-cycling NMR. Proc. Natl. Acad. Sci. USA 2004, 101, 17066–17071. [Google Scholar] [CrossRef]
- Gupta, S.; De Mel, J.U.; Schneider, G.J. Dynamics of liposomes in the fluid phase. Curr. Opin. Colloid Interface Sci. 2019, 42, 121–136. [Google Scholar] [CrossRef]
- Roberts, M.F.; Cai, J.; Natarajan, S.V.; Khan, H.M.; Reuter, N.; Gershenson, A.; Redfield, A.G. Phospholipids in motion: High-resolution 31P NMR field cycling studies. J. Phys. Chem. B 2021, 125, 8827–8838. [Google Scholar] [CrossRef] [PubMed]
- Pu, M.; Orr, A.; Redfield, A.G.; Roberts, M.F. Defining specific lipid binding sites for a membrane protein in situ using subtesla field-cycling. J. Biol. Chem. 2010, 285, 26916–26922. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Goldstein, R.; Gershenson, A.; Stec, B.; Roberts, M.F. The cation-π box is a specific phosphatidylcholine membrane targeting motif. J. Biol. Chem. 2013, 288, 14863–14873. [Google Scholar] [CrossRef]
- He, T.; Gershenson, A.; Eyles, S.J.; Lee, Y.-J.; Liu, W.R.; Wang, J.; Gao, J.; Roberts, M.F. Fluorinated aromatic amino acids distinguish cation-π interactions from membrane insertion. J. Biol. Chem. 2015, 290, 19334–19342. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.M.; He, T.; Fuglebakk, E.; Grauffel, C.; Yang, B.; Roberts, M.F.; Gershenson, A.; Reuter, N. A role for weak electrostatic interactions in peripheral membrane protein binding. Biophys. J. 2016, 110, 1367–1378. [Google Scholar] [CrossRef][Green Version]
- Moutoussamy, E.E.; Khan, H.M.; Roberts, M.F.; Gershenson, A.; Chipot, C.; Reuter, A. Standard binding free energy and membrane desorption mechanism for a phospholipase C. J. Chem. Inf. Model. 2022; online ahead of print. [Google Scholar]
- Cheng, J.; Karri, S.; Grauffel, C.; Reuter, N.; Roberts, M.F.; Wintrode, P.L.; Gershenson, A. Does changing the predicted dynamics of a phospholipase C alter activity and membrane binding? Biophys. J. 2013, 104, 185–195. [Google Scholar] [CrossRef][Green Version]
- Nagano, N.; Orengo, C.A.; Thornton, J.M. One fold with many functions: The evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol. 2002, 321, 741–765. [Google Scholar] [CrossRef]
- Wu, Y.; Zhou, C.; Roberts, M.F. Stereocontrolled syntheses of water soluble inhibitors of phosphatidylinositol-specific phospholipase C: Inhibition enhanced by an interface. Biochemistry 1997, 36, 356–363. [Google Scholar] [CrossRef]
- Mouchlis, V.D.; Bucher, D.; McCammon, J.A.; Dennis, E.A. Membranes serve as allosteric activators of phospholipase A2, enabling it to extract, bind, and hydrolyze phospholipid substrates. Proc. Natl. Acad. Sci. USA 2015, 112, E516–E525. [Google Scholar] [CrossRef]
- Mouchlis, V.D.; Chen, Y.; McCammon, J.A.; Dennis, E.A. Membrane allostery and unique hydrophobic sites promote enzyme substrate specificity. J. Am. Chem. Soc. 2018, 140, 3285–3291. [Google Scholar] [CrossRef]
- Mouchlis, V.D.; Hayashi, D.; Vasquez, A.M.; Cao, J.; McCammon, J.A.; Dennis, E.A. Lipoprotein-associated phospholipase A2: A paradigm for allosteric regulation by membranes. Proc. Natl. Acad. Sci. USA 2022, 119, e2102953118. [Google Scholar] [CrossRef] [PubMed]
- Hirano, Y.; Gao, Y.G.; Stephenson, D.J.; Vu, N.T.; Malinina, L.; Simanshu, D.K.; Chalfant, C.E.; Patel, D.J.; Brown, R.E. Structural basis of phosphatidylcholine recognition by the C2-domain of cytosolic phospholipase A2α. sLife 2019, 8, e44769. [Google Scholar] [CrossRef] [PubMed]
- Stahelin, R.V.; Cho, W. Differential roles of ionic, aliphatic, and aromatic residues in membrane-protein interactions: A surface plasmon resonance study on phospholipases A2. Biochemistry 2001, 40, 4672–4678. [Google Scholar] [CrossRef]
- Moutoussamy, E.E.; Waheed, Q.; Binford, G.J.; Khan, H.M.; Moran, S.M.; Eitel, A.R.; Cordes, M.H.J.; Reuter, N. Specificity of Loxosceles α clade phospholipase D enzymes for choline-containing lipids: Role of a conserved aromatic cage. PLoS Comput. Biol. 2022, 18, e1009871. [Google Scholar] [CrossRef] [PubMed]
- Broemstrup, T.; Reuter, N. How does proteinase 3 interact with lipid bilayers? Phys. Chem. Chem. Phys. 2010, 12, 7487–7796. [Google Scholar] [CrossRef]
- Goh, B.C.; Wu, H.; Rynkiewicz, M.J.; Schulten, K.; Seaton, B.A.; McCormack, F.X. Elucidation of lipid binding sites on lung surfactant protein A using x-ray crystallography, mutagenesis, and molecular dynamics simulations. Biochemistry 2016, 55, 3692–3701. [Google Scholar] [CrossRef]
- Weber, D.K.; Yao, S.; Rojko, N.; Anderluh, G.; Lybrand, T.P.; Downton, M.T.; Wagner, J.; Separovic, F. Characterization of the lipid-binding site of equinatoxin II by NMR and molecular dynamics simulation. Biophys. J. 2015, 108, 1987–1996. [Google Scholar] [CrossRef]
- Waheed, W.; Khan, H.M.; He, T.; Roberts, M.F.; Gershenson, A.; Reuter, N. Interfacial aromatics mediating cation—π interactions with choline containing lipids can contribute as much to peripheral protein affinity for membranes as aromatics inserted below the phosphates. J. Phys. Chem. Lett. 2019, 10, 3972–3977. [Google Scholar] [CrossRef]
- Klauda, J.B.; Roberts, M.F.; Redfield, A.G.; Brooks, B.R.; Pastor, R.W. Rotation of lipids in membranes: MD simulation, 31P spin-lattice relaxation, and rigid-body dynamics. Biophys. J. 2008, 94, 3074–3083. [Google Scholar] [CrossRef]
- Pu, M.; Feng, J.; Redfield, A.G.; Roberts, M.F. Enzymology with a spin-labeled phospholipase C: Soluble substrate binding by 31P NMR from 0.005 to 11.7 Tesla. Biochemistry 2009, 48, 8282–8284. [Google Scholar] [CrossRef][Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roberts, M.F.; Gershenson, A.; Reuter, N. Phosphatidylcholine Cation—Tyrosine π Complexes: Motifs for Membrane Binding by a Bacterial Phospholipase C. Molecules 2022, 27, 6184. https://doi.org/10.3390/molecules27196184
Roberts MF, Gershenson A, Reuter N. Phosphatidylcholine Cation—Tyrosine π Complexes: Motifs for Membrane Binding by a Bacterial Phospholipase C. Molecules. 2022; 27(19):6184. https://doi.org/10.3390/molecules27196184
Chicago/Turabian StyleRoberts, Mary F., Anne Gershenson, and Nathalie Reuter. 2022. "Phosphatidylcholine Cation—Tyrosine π Complexes: Motifs for Membrane Binding by a Bacterial Phospholipase C" Molecules 27, no. 19: 6184. https://doi.org/10.3390/molecules27196184
APA StyleRoberts, M. F., Gershenson, A., & Reuter, N. (2022). Phosphatidylcholine Cation—Tyrosine π Complexes: Motifs for Membrane Binding by a Bacterial Phospholipase C. Molecules, 27(19), 6184. https://doi.org/10.3390/molecules27196184






