Effect of Nanoparticles of DOX and miR-125b on DNA Damage Repair in Glioma U251 Cells and Underlying Mechanisms
Abstract
:1. Introduction
2. Results
2.1. Construction of Nanoparticles
2.2. miR-125b Targets the 3′UTR of YES1 mRNA
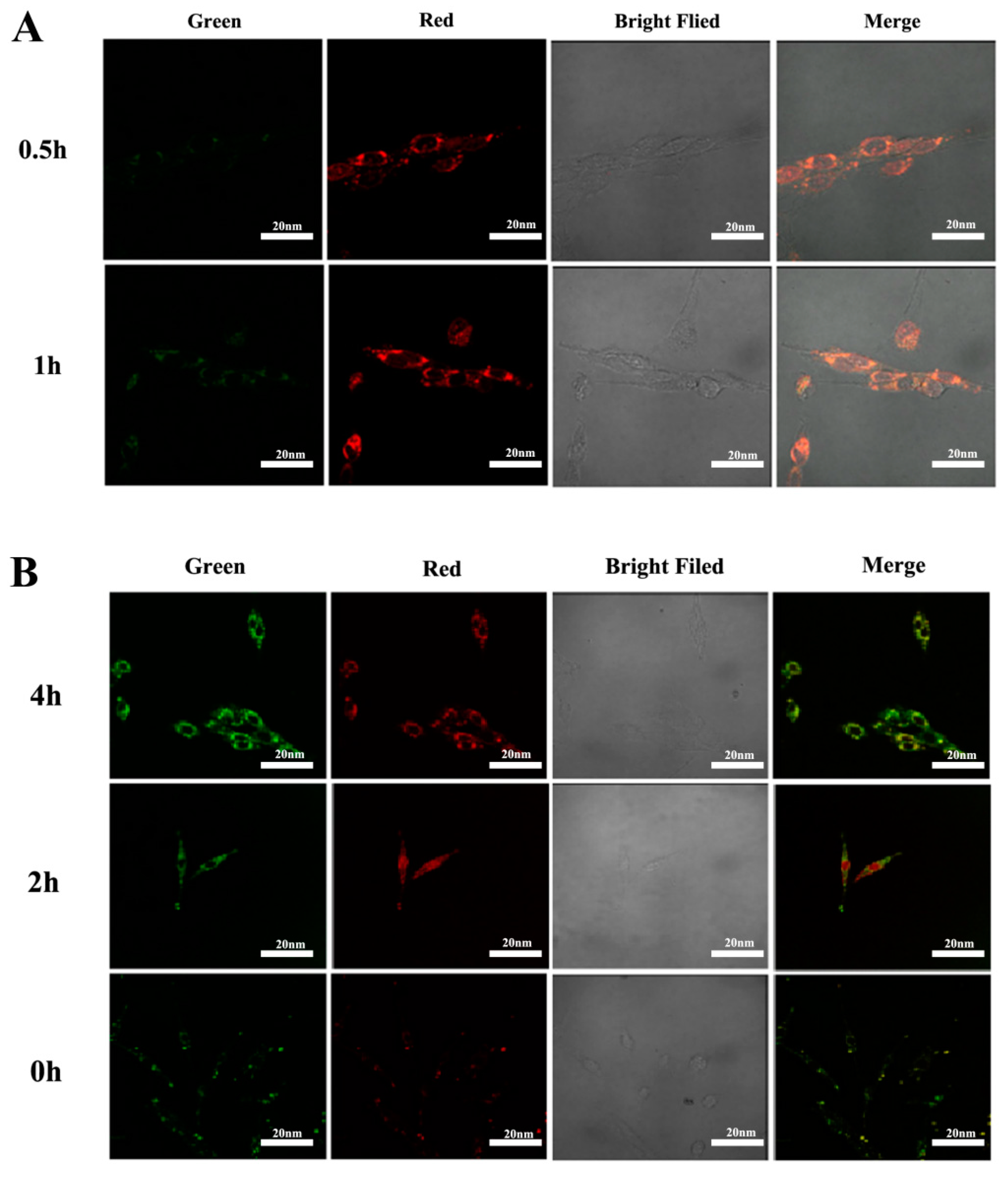
2.3. The Results of Cell Viability and Lysosome Localization
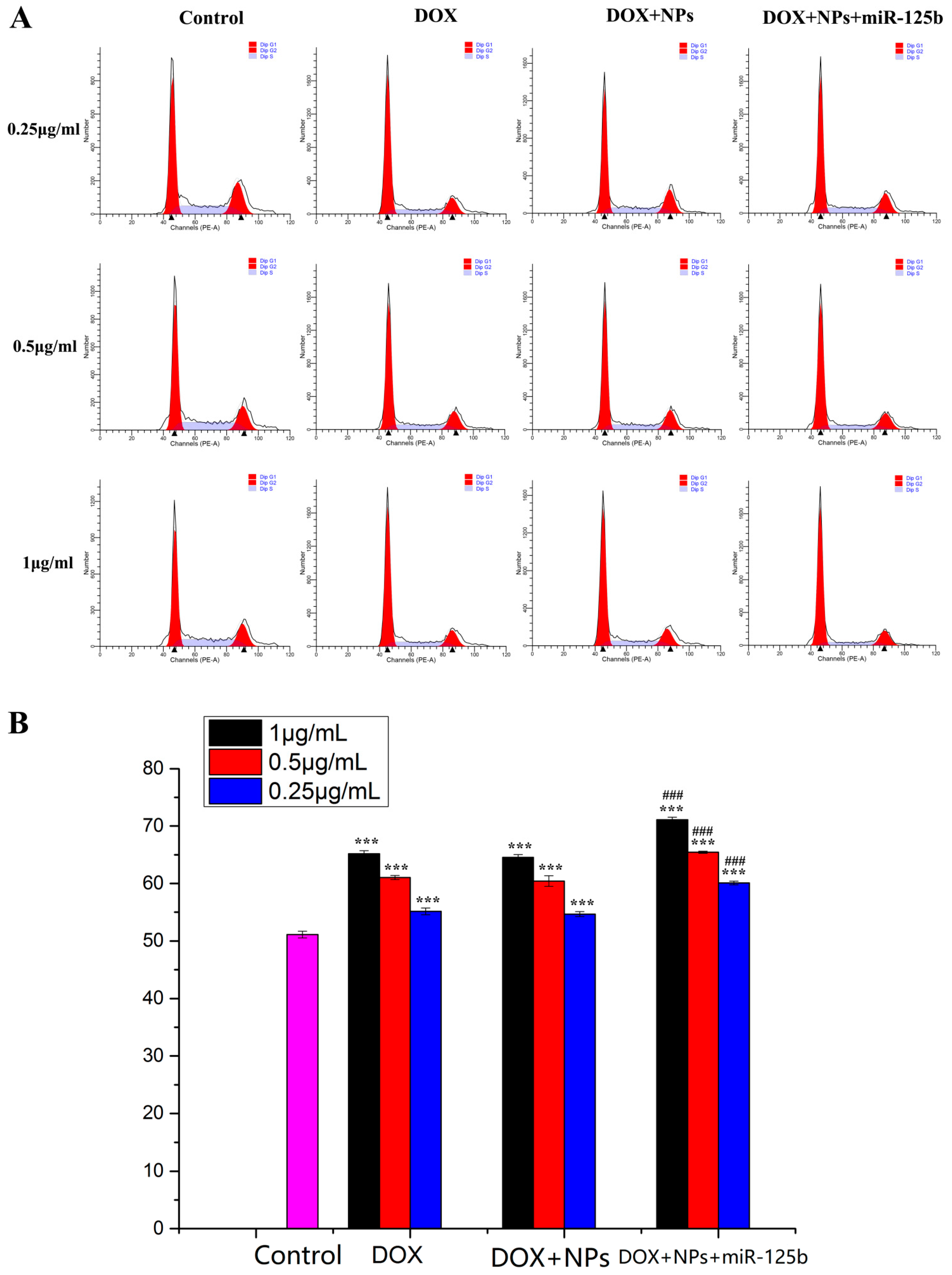
2.4. Arresting Effect of DOX+NPs+miR-125b on U251 Cell Cycle
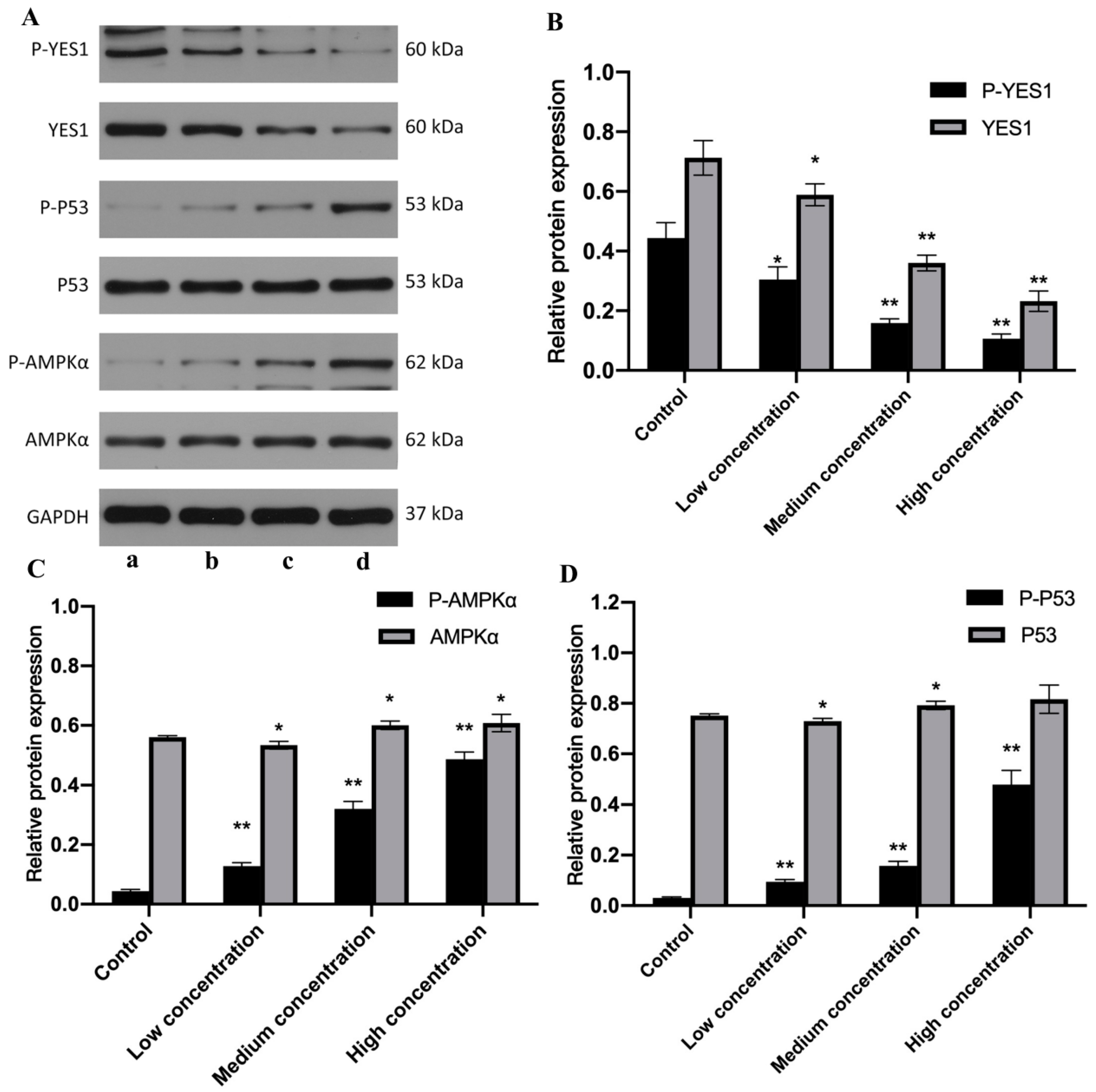
2.5. miR-125b and DOX+NPs+miR-125b Altered the Expression of YES1, AMPK, p53 and Corresponding Phosphorylated Proteins
3. Discussion
4. Materials and Methods
4.1. Drugs and Reagents
4.2. Construction of NPs
4.3. Cell Lines and Cell Culture
4.4. Cell Viability Assay
4.5. Dual-Luciferase Detection Experiment
4.6. Real Time-Polymerase Chain Reaction
4.7. Western Blot
4.8. Flow Cytometry
4.9. Laser Confocal Microscopy
4.9.1. Drug Uptake and Lysosomal Colocalization in U251 Cells
4.9.2. Immunofluorescence
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Puranik, A.D.; Rangarajan, V.; Dev, I.D.; Jain, Y.; Purandare, N.C.; Sahu, A.; Choudhary, A.; Gupta, T.; Chatterjee, A.; Moiyadi, A.; et al. Brain FET PET tumor-to-white mater ratio to differentiate recurrence from post-treatment changes in high-grade gliomas. J. Neuroimaging 2021, 31, 1211–1218. [Google Scholar] [CrossRef]
- Shen, J.; Xiong, J.; Shao, X.; Cheng, H.; Fang, X.; Sun, Y.; Di, G.; Mao, J.; Jiang, X. Knockdown of the long noncoding RNA XIST suppresses glioma progression by up-regulating miR-204-5p. J. Cancer 2020, 11, 4550–4559. [Google Scholar] [CrossRef]
- Liu, L.; Li, X.; Wu, H.; Tang, Y.; Li, X.; Shi, Y. The COX10-AS1/miR-641/E2F6 Feedback Loop Is Involved in the Progression of Glioma. Front. Oncol. 2021, 11, 648152. [Google Scholar] [CrossRef]
- Oldak, L.; Chludzinska-Kasperuk, S.; Milewska, P.; Grubczak, K.; Reszec, J.; Gorodkiewicz, E. Laminin-5, Fibronectin, and Type IV Collagen as Potential Biomarkers of Brain Glioma Malignancy. Biomedicines 2022, 10, 2290. [Google Scholar] [CrossRef]
- Ge, X.; Wang, Z.; Jiang, R.; Ren, S.; Wang, W.; Wu, B.; Zhang, Y.; Liu, Q. SCAMP4 is a novel prognostic marker and correlated with the tumor progression and immune infiltration in glioma. Int. J. Biochem. Cell Biol. 2021, 139, 106054. [Google Scholar] [CrossRef]
- Fujii, T.; Nishikawa, J.; Fukuda, S.; Kubota, N.; Nojima, J.; Fujisawa, K.; Ogawa, R.; Goto, A.; Hamabe, K.; Hashimoto, S.; et al. MC180295 Inhibited Epstein–Barr Virus-Associated Gastric Carcinoma Cell Growth by Suppressing DNA Repair and the Cell Cycle. Int. J. Mol. Sci. 2022, 23, 10597. [Google Scholar] [CrossRef]
- Lavin, M.F.; Yeo, A.J. Clinical potential of ATM inhibitors. Mutat. Res. 2020, 821, 111695. [Google Scholar] [CrossRef]
- Miguel, R.d.A.; Hirata, A.S.; Jimenez, P.C.; Lopes, L.B.; Costa-Lotufo, L.V. Beyond Formulation: Contributions of Nanotechnology for Translation of Anticancer Natural Products into New Drugs. Pharmaceutics 2022, 14, 1722. [Google Scholar] [CrossRef]
- Wang, Y.; Zeng, G.; Jiang, Y. The Emerging Roles of miR-125b in Cancers. Cancer Manag. Res. 2020, 12, 1079–1088. [Google Scholar] [CrossRef]
- Liu, S.; Chen, Q.; Wang, Y. MiR-125b-5p suppresses the bladder cancer progression via targeting HK2 and suppressing PI3K/AKT pathway. Hum. Cell 2020, 33, 185–194. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Y.; Huang, W.C.; Zheng, L.C. MiR-125b-1-3p Exerts Antitumor Functions in Lung Carcinoma Cells by Targeting S1PR1. Chin. Med. J. 2018, 131, 1909–1916. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Zhao, X.; Wang, J.; Lv, L.; Wang, C.; Feng, L.; Shen, L.; Ren, W. miR-125b regulates the drug-resistance of breast cancer cells to doxorubicin by targeting HAX-1. Oncol. Lett. 2018, 15, 1621–1629. [Google Scholar] [CrossRef]
- Minari, R.; Valentini, S.; Madeddu, D.; Cavazzoni, A.; La Monica, S.; Lagrasta, C.A.M.; Bertorelli, R.; De Sanctis, V.; Fassan, P.; Azzoni, C.; et al. YES1 and Amplifications as Synergistic Resistance Mechanisms to Different Generation ALK Tyrosine Kinase Inhibitors in Advanced NSCLC: Brief Report of Clinical and Preclinical Proofs. JTO Clin. Res. Rep. 2022, 3, 100278. [Google Scholar] [CrossRef] [PubMed]
- Toledano-Osorio, M.; López-García, S.; Osorio, R.; Toledano, M.; García-Bernal, D.; Sánchez-Bautista, S.; Rodríguez-Lozano, F.J. Dexamethasone and Doxycycline Doped Nanoparticles Increase the Differentiation Potential of Human Bone Marrow Stem Cells. Pharmaceutics 2022, 14, 1865. [Google Scholar] [CrossRef]
- Mohanty, A.; Uthaman, S.; Park, I.K. Utilization of Polymer-Lipid Hybrid Nanoparticles for Targeted Anti-Cancer Therapy. Molecules 2020, 25, 4377. [Google Scholar] [CrossRef]
- Liu, X.; Su, S.; Wei, F.; Rong, X.; Yang, Z.; Liu, J.; Li, M.; Wu, Y. Construction of nanoparticles based on amphiphilic copolymers of poly (γ-glutamic acid co-L-lactide)-1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine as a potential drug delivery carrier. J. Colloid Interface Sci. 2014, 413, 54–64. [Google Scholar] [CrossRef]
- Aleo, D.; Elshaer, Z.; Pfnür, A.; Schuler, P.J.; Fontanella, M.M.; Wirtz, C.R.; Pala, A.; Coburger, J. Evaluation of a Navigated 3D Ultrasound Integration for Brain Tumor Surgery: First Results of an Ongoing Prospective Study. Curr. Oncol. 2022, 29, 6594–6609. [Google Scholar] [CrossRef]
- Panda, J.; Satapathy, B.S.; Sarkar, R.; Tudu, B. A zinc ferrite nanodrug carrier for delivery of docetaxel: Synthesis, characterization and in vitro tests on C6 glioma cells. J. Microencapsul. 2022, 39, 1–19. [Google Scholar] [CrossRef]
- Medikonda, R.; Choi, J.; Pant, A.; Saleh, L.; Routkevitch, D.; Tong, L.; Belcaid, Z.; Kim, Y.H.; Jackson, C.M.; Jackson, C.; et al. Synergy between glutamate modulation and anti-programmed cell death protein 1 immutnotherapy for glioblastoma. J. Neurosurg. 2021, 136, 379–388. [Google Scholar] [CrossRef]
- Lesueur, F.; Easton, D.F.; Renault, A.-L.; Tavtigian, S.V.; Bernstein, J.L.; Kote-Jarai, Z.; Eeles, R.A.; Plaseska-Karanfia, D.; Feliubadaló, L.; Arun, B.; et al. First international workshop of the ATM and cancer risk group (4–5 December 2019). Fam. Cancer 2022, 21, 211–227. [Google Scholar] [CrossRef]
- Qu, M.; Xu, H.; Chen, J.; Xu, B.; Li, Z.; Ma, B.; Guo, L.; Ye, Q.; Xie, J. Differential comparison of genotoxic effects of aristolochic acid I and II in human cells by the mass spectroscopic quantification of γ-H2AX. Toxicol. Vitr. 2022, 81, 105349. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.H.; Lin, X.L.; Guo, D.M.; Zhang, Y.; Yuan, C.; Tan, T.P.; Chen, Y.D.; Wu, S.J.; Ye, Z.F.; He, J. Resveratrol protects cardiomyocytes from doxorubicin-induced apoptosis through the AMPK/p53 pathway. Mol. Med. Rep. 2016, 13, 1281–1286. [Google Scholar] [PubMed] [Green Version]
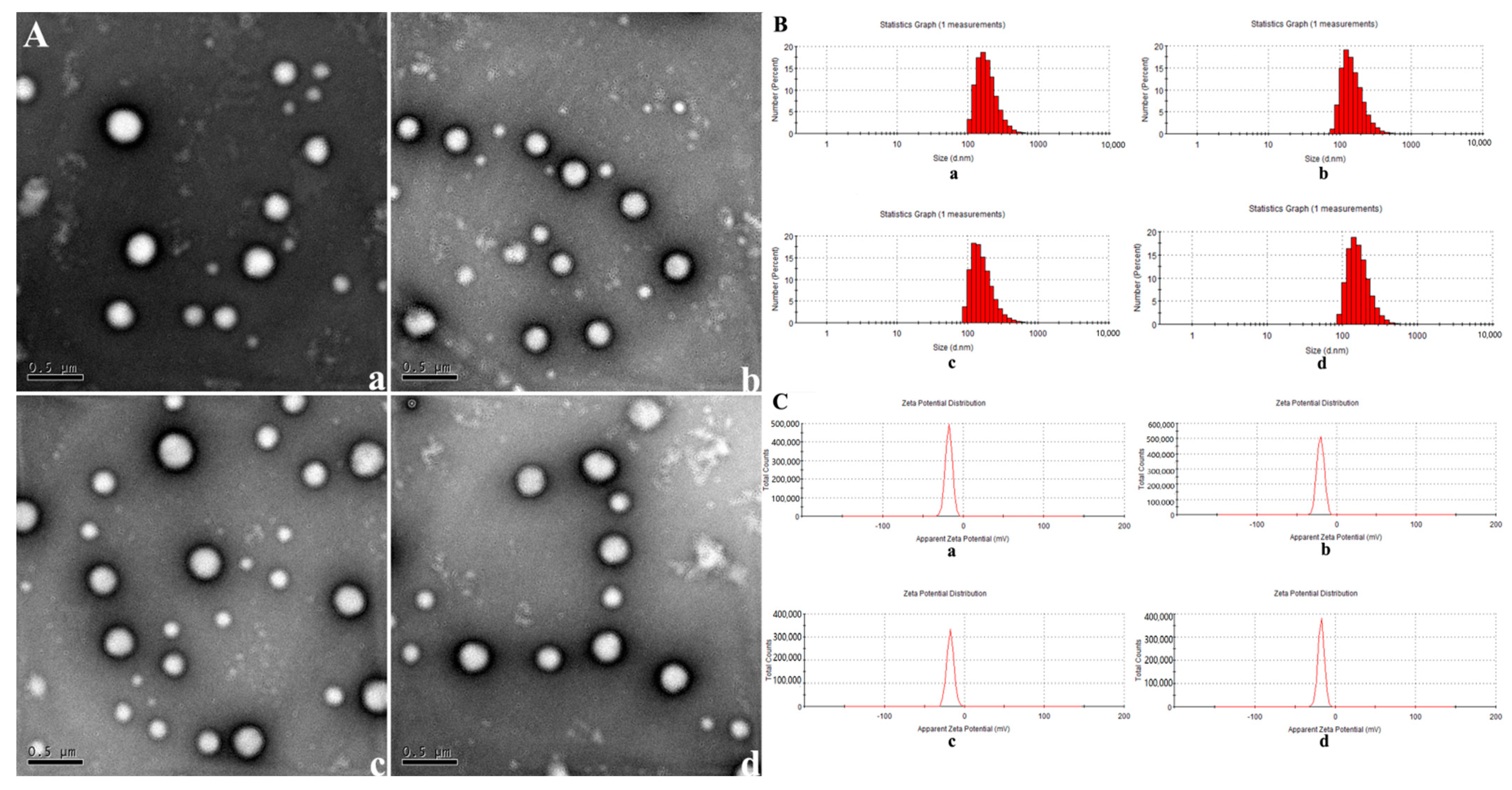




| Physicochemical Characteristics | Size (nm) | PDI | Zeta Potential (mV) |
|---|---|---|---|
| NPs | 256.08 ± 1.68 | 0.069 ± 0.03 | −17.8 ± 4.26 |
| NPs+DOX | 237.2664 ± 3.18 | 0.111 ± 0.02 | −17.5 ± 4.36 |
| NPs+miR-125b | 220.7669 ± 0.71 | 0.056 ± 0.04 | −19.9 ± 4.47 |
| DOX+NPs+miR-125b | 235.6625 ± 1.16 | 0.062 ± 0.03 | −19.1 ± 4.7 |
| DOX+NPs/miR-125b Mass Ratio | 4/1 | 6/1 | 8/1 |
|---|---|---|---|
| Encapsulation efficiency of DOX (%) | 35.2 ± 2.9 | 40.9 ± 2.5 | 48.8 ± 3.2 |
| Encapsulation efficiency of miR-125b (%) | 85 | 89 | 92 |
| Primer Name | Primer InforMation | Base Sequence (5′–3′) | Tm Value | CG% | Product Length | |
|---|---|---|---|---|---|---|
| H-GAPDH | NM_001256799.2 | Sense | CATCATCCCTGCCTCTACTGG | 59.4 | 57.1 | 259 |
| Antisense | GTGGGTGTCGCTGTTGAAGTC | 60.1 | 57.1 | |||
| H-YES1 | NM_005433.4 | Sense | TTTGTGGCCTTATATGATTATGAAG | 58.2 | 32 | 193 |
| Antisense | AATACCATTCTTCTGCCTGAATG | 58.2 | 39.1 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Pan, T.; Wang, Y.; Yu, J.; Qu, P.; Chen, Y.; Xin, H.; Wang, S.; Liu, J.; Wu, Y. Effect of Nanoparticles of DOX and miR-125b on DNA Damage Repair in Glioma U251 Cells and Underlying Mechanisms. Molecules 2022, 27, 6201. https://doi.org/10.3390/molecules27196201
Wang L, Pan T, Wang Y, Yu J, Qu P, Chen Y, Xin H, Wang S, Liu J, Wu Y. Effect of Nanoparticles of DOX and miR-125b on DNA Damage Repair in Glioma U251 Cells and Underlying Mechanisms. Molecules. 2022; 27(19):6201. https://doi.org/10.3390/molecules27196201
Chicago/Turabian StyleWang, Lin, Tingting Pan, Yan Wang, Jiewen Yu, Peiyi Qu, Yue Chen, Hua Xin, Sicen Wang, Junxing Liu, and Yan Wu. 2022. "Effect of Nanoparticles of DOX and miR-125b on DNA Damage Repair in Glioma U251 Cells and Underlying Mechanisms" Molecules 27, no. 19: 6201. https://doi.org/10.3390/molecules27196201





